Abstract
We investigated effects of short- and long-term hydrologic fluctuations on denitrification in constructed wetland basins. N2 and N2O emission rates, their ratio and a number of physicochemical variables were compared between pulsed (with annual drawdown-flooding cycles) and static (constantly flooded) wetland basins. The abundance and structure of general and denitrifying bacterial communities were compared based on quantitative PCR and terminal restriction fragment polymorphism analyses of 16S rRNA and nosZ genes, respectively. These variables significantly differed among sampling months, but not between the two long-term hydrologic treatments. The same set of variables were measured and compared among samples from different sites of one pulsed basin. The single basin experiment revealed temporal shifts in measured variables, which coincided with short-term hydrological changes. Bank sites, which had larger changes in abiotic conditions, showed greater variability in denitrifying activities than basin sites. Significant difference in N2 and N2O emission rates, 16S rRNA and nosZ gene abundance and structural composition was also identified between basin and bank sites. Our results suggest that activity and community structure of denitrifying bacteria in wetland soils are insensitive to moderate hydrology change, but can be significantly impacted by large hydrological fluctuation at both short-term and long-term scales.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Wetland soils are important sites for denitrification, the step-wise reduction of nitrate (NO3 −) to nitrous oxide (N2O) and dinitrogen (N2) gases (Tiedje et al. 1989). N2 is largely inert in the atmosphere, while N2O has a negative environmental impact and a global warming potential about 310 times that of CO2 (IPCC 2007). Therefore, the amount of N2O relative to N2 released during denitrification is environmentally important. Denitrification is carried out by diverse microbial taxa and their activity in situ is regulated by complex mechanisms, including the taxonomic structure and the presence and expression level of functional genes of the denitrifier community as well as environmental conditions (Wallenstein et al. 2006). Denitrifiers carry an assembly of genes encoding reduction of nitrate (nar), nitrite (nir), nitric oxide (nor) and N2O (nos) (Zumft 1997; Philippot et al. 2007). While some denitrifiers have a complete set of reductase genes and produce N2 as the final product, others lack nos genes and terminate denitrification with production of N2O (Zumft 1997). The distribution of denitrifying genes is not specific to certain taxa, and many denitrifying enzymes are functionally redundant (Philippot et al. 2007). Consequently, composition of the denitrifying community may (Cavigelli and Robertson 2000 and 2001; Wittebolle et al. 2009; Peralta et al. 2010) or may not be correlated with the overall denitrification rate and/or N2O:N2 ratio (rN2O) (Rich and Myrold 2004; Song et al. 2010; Salles et al. 2012).
A variety of environmental conditions influence denitrification, including soil moisture, temperature, aeration (oxygen), pH and availability of nitrate/nitrite and organic carbon (Wallenstein et al. 2006; Pinay et al. 2007; Braker et al. 2010; Bowles et al. 2012). In freshwater wetlands, these parameters vary with hydrologic fluctuations. “Wet-dry’ cycles may enhance relative emission of N2O (Priemé and Christensen 2001), likely due to shifts of redox conditions in soils (Martikainen et al. 1993; Hernandez and Mitsch 2007; Mitsch and Gosselink 2007), which may replenish substrates (nitrate and organic carbon) for denitrification (Sahrawat 2003; Venterink et al. 2002; Reddy and DeLaune 2004). At the cellular level, this is attributed to decreased abundance and/or expression of nitrous oxide reductase (nos) genes, which mediate use of N2O to produce N2 (Korner and Zumft 1989).
When the wet-dry hydrology regime is long-term (years), variation in denitrification rate and product appear to be correlated with succession in the denitrifying community (Stres et al. 2008; Kjellin et al. 2007; Bougon et al. 2009; Peralta et al. 2010). Whereas, when the hydrological change is short-term (days to weeks, such as a flood-pulse), the interconnection between functional rate and community structure is unclear (Kim et al. 2010; Song et al. 2010). No studies have simultaneously investigated both short-term and long-term effects of hydrology on denitrification and denitrifier communities. Moreover, many studies on denitrification rely on an enzyme activity assay to estimate denitrification rate, a procedure that destroys the 3-dimensional soil structure and causes a deviation from in situ rates (Tiedje et al. 1989).
In this study, we used constructed freshwater wetlands with adjustable water inflow and outflow to examine effects of both short-term flood-pulsing and long-term hydrologic regimes on denitrification, the rN2O, and bacterial and denitrifier community structure. We measured N2O and N2 production using an in situ chamber method that preserves soil microsites, and used terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rDNA and nitrous oxide reductase gene (nosZ) to track changes in bacterial and denitrifier community composition. We hypothesized that short-term flood-pulsing would increase the overall denitrification rate and relative N2O production in wetland soils with no corresponding change in functional gene or taxonomic composition of the denitrifier community, but that soils differing in long-term hydrologic regimes would harbor different denitrifier communities and differ in denitrification rate and N2O production.
Methods
Study Site and Experimental Set Up
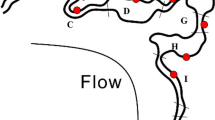
This study was conducted at Kent State University’s Herrick Aquatic Ecology Research Facility (HAERF) in Kent, OH. The facility consists of 10 constructed wetland basins that are surrounded by hardwood trees (such as Populus deltoids and Quercus palustris), shrubs and herbaceous plants (such as Viburnum recognitum and Cornus amomum). The banks of the wetland basins are covered by facultative wetland plant species such as Cyperus esculents, Elymus riparius and Eupatorium perfoliatum, while the bottoms of the basins are vegetated by obligate wetland plant species, such as Carex crinite, Eleocharis palustris and Pontederia cordata (Drinkard et al. 2011). Each wetland basin is approximately 10 m (wide) × 20 m (long) × 2 m (deep). All basins receive water inflow from the Allerton Creek, a second order perennial creek with a 1–3 cm wide and 10 cm deep water channel during base flow and approximately 1 km2 water shed. The water depth of each basin is controlled independently by inflow and outflow flashboard risers (Fig. 1; Barlett and Leff 2010). Starting in 2009, five out of the ten basins, i.e., Basins 1, 3, 6, 7 and 10, have undergone a drawdown and re-flooding treatment once or twice during each summer/fall. These wet-dry cycled basins are referred as pulsed basins; they become visibly dry over the course of the 2–3 week drawdown and then, when the flashboards are returned, they refill from the Allerton Creek. In comparison, the other five basins, i.e., Basins 2, 4, 5, 8 and 9, have been permanently flooded without drawdown events and are referred as static basins.
Schematic diagrams of constructed wetlands in HAERF facility. a An overhead view shows location of pulsed basins (light gray) that have received wet-dry cycles and static (dark gray) basins, which were permanently flooded during the study b A cross section view shows placement of gas collection chambers in relation to wetland hydrology and topography
In 2011, a multiple basin experiment was performed and samples were taken from central basin locations in all 10 basins when pulsed basins were at flooded conditions. The five static basins (Basins 2, 4, 5, 8 and 9) were flooded yearlong in 2011 (as has been the case since 2009). In contrast, five pulsed basins (Basins 1, 3, 6, 7 and 10) lacked standing water in early August 2011 for three weeks and then were re-flooded to a water depth of ~55 cm in late August 2011. Samples were collected in July, August and November 2011 under flooded conditions.
In 2012, the pulsed basin (Basin 6) that showed the largest variability in rN2O in 2011 samples was chosen to examine within basin denitrification variability due to short term hydrology change. In June–October 2012, Basin 6 was drained twice, once in July and once in September, each for 2 weeks before reflooding. In June, August and October 2012, Basin 6 was flooded to water depth ranged between 55 and 65 cm. Samples were taken along a cross basin transect on the high-bank (2 sites, 1.5 m-above the bottom of the basin, never submerged), low-bank (2 sites, 50 cm above the bottom of the basin, submerged only during flooded condition) and within the basin (7 sites, 3 m apart on the transect, submerged except during the drawdown condition) at both flooded and drawdown conditions (Fig. 1b).
Gas Sample Collection and Analysis
In situ N2O flux and denitrification rates were measured using a chamber method modified from several sources (Tiedje et al. 1989; Choudhary et al. 2002; Dhondt et al. 2004; Hernandez and Mitsch 2006). Briefly, air-tight gas collection chambers were made from PVC pipes with one end fitted with a threaded adapter at the top and the other end driven down at least 10 cm into the soil. The headspace height of each chamber was <50 cm. Variation in chamber water depth and headspace height was accounted for in the N2O concentration calculation (Knowles 1979).
Each sampling location had 6 chambers. At the time of gas sampling, three chambers were treated with 10% v/v acetylene to block the final step in denitrification (Tiedje et al. 1989; Malone et al. 1998). Afterwards, headspace samples were taken immediately, and then at 24-h intervals for a total of 4 days. The other three chambers were used to capture N2O directly from the headspace. The headspace gas was mixed before sampling by exchanging a 30 ml syringe for ten times, and then 10 ml of headspace was taken from the chamber and injected into a 5 ml serum vial. N2O in gas samples were analyzed with a Shimadzu GC-2014 gas chromatograph (Columbia, MD) with Ni63 electron capture detector.
Water Chemistry
For flooded basins, water temperature, pH, conductivity, dissolved oxygen, and redox potential were measured in situ following gas headspace sampling at 2 cm above the soil using a Hach HQD-40 portable field meter and probes (Ames, IA). Water samples were collected in acid-washed polypropylene bottles, transported to the laboratory on ice for chemical analyses.
Whole water samples were analyzed for total organic carbon (TOC) and total nitrogen (TN) using a TOC5000 analyzer (Shimadzu, Columbia, MD). The same analysis was performed for filtered water samples (0.45 μm nitrocellulose membranes; Millipore, Billerica, MA) to determine dissolved organic carbon (DOC) and dissolved nitrogen (DN). Filtered water samples were also analyzed for soluble reactive phosphorus (SRP) and nitrate plus nitrite (NOx) using a LachAT Quikchem 800 FLA+ system (LachAT Instruments, Hach Company, Loveland, CO) and for ammonium (NH4 +) using the OPA method (Holmes et al. 1999). Concentrations of total phosphorus (TP) were measured from whole water samples following the EPA method 365.3 (water.epa.gov).
For chlorophyll a analysis, whole water samples were filtered through glass microfiber, GF/F filters (GE Whatman, Pittsburgh, PA). The resulting GF/F Filters were extracted overnight with 90% acetone/1% MgCl2 and measured on a Trilogy Fluorometer (Turner Designs, Sunnyvale, CA).
Soil Chemistry
After gas sample collection, samples were taken from the top 10 cm of soil inside each chamber. Each soil sample was homogenized by stirring with a sterile spatula in a sterile beaker for 10 min. Afterwards, then half of the homogenized sample was stored in a sterile whirl-pak bag (Nasco, Fort Atkinson, WI) and stored at −20 C before molecular analysis. The other half of the sample was transferred into an acid-washed polypropylene bottle and shaken with 1 M KCl for 3 times over the course of 24 h to extract exchangeable ions (Robertson et al. 1999). The resulting soil slurry for each sample was filtered through 0.45 μm nitrocellulose membranes and analyzed for soil NOx (i.e., NOxs) as described above.
To determine soil percent moisture (MOIs%) and ash free dry mass, homogenized subsamples were weighed and dried overnight at 60 °C to determine moisture content and then combusted for 4 h at 500 °C to determine organic content (Santmire and Leff 2007).
Soil pore water was extracted by centrifugation at 4000 g for 30 min. The supernatant was filtered through 0.45 μm nitrocellulose membranes, acidified with HCl to pH < 2, and stored at 4 °C in the dark before nutrient analysis (i.e., DOCpw and NOxpw). The remaining soil sample was stored at −20 °C before subsequent DNA extractions.
Molecular Analyses
Frozen soil samples in Whirl-pak bags were thawed at 4 °C and DNAs were extracted from 0.25 g wet soil using a Powersoil DNA Extraction Kit (MoBio, Carlsbad, CA) following the manufacturer’s instructions. PCR of 16S rRNA and the nosZ genes for terminal restriction fragment length polymorphism (T-RFLP) analysis was carried out using 16S rRNA gene primers 1392-R and an equimolar mixture of Eub-338F-0-III and Eub-338F-I-II (Blackwood et al. 2005), and nosZ gene primers nosZ1F-1184/nos-R-1880 (Rich et al. 2003), respectively. Forward primers for each gene were labeled with 6-carboxyfluoroscein (FAM). PCR reactions consisted of template DNA, 0.2 μM of each primer, and PCR master mix (Promega, Madison, WI). PCR was performed with the following thermal profile: 95 °C for 3 min, followed by 30 cycles of 95 °C for 30 s, 58 °C for 1 min, and 72 °C for 1 min, then a final extension at 72 °C for 5 min. For each sample, three individual PCR reactions (25 μL) were performed; then amplicons were pooled and checked for bands of the correct size on 1% agarose gels by electrophoresis. Verified amplicons were excised from the gel and purified using an UltraClean GelSpin DNA Purification Kit (MoBio, Carlsbad, CA). Purified PCR amplicons were digested with HaeIII (New England Biolabs, Ipswich, MA) for 4 h at 37 °C. Digested PCR amplicons were cleaned by ethanol/glycogen precipitation and analyzed with a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA) in the Plant Microbe Genetics Facility at Ohio State University. T-RFLP data were standardized based on the relative peak area of each terminal restriction fragment (T-RF), which was used to estimate the relative abundance of bacterial species associated with that T-RF peak. T-RFs with <2% relative peak areas were excluded from the analysis (Mou et al. 2005). The number of T-RF was used to estimate the richness of bacterial taxa.
Quantitative PCR (qPCR) of the 16S rRNA, and nosZ genes was carried out using an Mx3005P thermocycler (Stratagene, La Jolla, CA). qPCR reactions consisted of DNA template, 0.5 μM forward and reverse primers, and Brilliant III UltraFast QPCR Master Mix with SYBR Green (Agilent Technologies, Santa Clara, CA). Primer pairs used for qPCR of 16S rDNA and nosZ genes were Eub338-F/Eub518-R, and nosZ1F/nosZ1R, respectively (Fierer et al. 2005 and Henry et al. 2006). For 16S rDNA, qPCR conditions were 95 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, 57 °C for 1 min, and 72 °C for 30 s with data acquisition. A dissociation curve followed amplification, starting at 55 °C and increasing 1 °C per cycle for 40 cycles (Fierer et al. 2005). For nosZ, the qPCR program was an initial denaturation at 95 °C for 15 min, 40 cycles of 94 °C for 30 s, 62 °C for 30 s, 72 °C for 30 s, 80 °C for 30 s (data acquisition step), followed by a dissociation curve identical to that of the 16S rDNA program (Henry et al. 2006). Standard curves for qPCR assays were generated using genomic DNA of Pseudomonas aeruginosa PAO1. Template DNA was quantified using Quant-iT PicoGreen (Invitrogen, Carlsbad, CA).
Statistical Analysis
All statistical analyses were performed using R software (version 2.15.2) with the nlme (Pinheiro et al. 2013), relaimpo (Groemping and Lehrkamp 2013) or vegan (Oksanen 2013) package, unless otherwise specified. Specifically, comparisons of physicochemical parameters and gas fluxes between pulsed and static basins were analyzed by two-way t-tests. Correlations between gas flux and environmental data were analyzed using Pearson product-moment correlation analysis. Comparisons between sampling dates and sampling locations were analyzed with repeated measures analysis of variance (ANOVA). Comparisons among samples based on environmental parameters were performed using principle components analysis (PCA). Abiotic data were natural logarithm-transformed and all T-RFLP data were square-root transformed.
Analyses of bacterial community structure data (T-RFLP) were performed using the Primer v5 (Plymouth Marine Laboratory, Plymouth, United Kingdom). Nonmetric multidimensional scaling analysis (NMDS) and analysis of similarity (ANOSIM) were done to examine similarity/differences among samples. ANOSIM index Ranosim was reported on a scale of 0 to 1. Sample groups were reported as either well separated (Ranosim > 0.75, P < 0.05), clearly different but overlapping (0.5 < Ranosim ≤ 0.75, P < 0.05), separated but strongly overlapping (0.25 < Ranosim ≤ 0.5), or not separable (Ranosim < 0.25) (Clarke and Warwick 2001). BIO-ENV analysis was done to identify the most influential abiotic variables to bacterial structure (Clarke and Warwick 2001).
Results
Multiple Basin Experiment
Physical Chemical Measurements
Principal component analysis (PCA) based on measured physical-chemical variables did not group samples based on basin type (hydrology). PCA ordinated samples along PCA1 (40.3% of variance) and PCA2 (25.3% variance) gradients into three clusters based on sampling time (Fig. 2). Cluster 1 consisted Nov-2011 samples from all 10 (both pulsed and static) basins and had lowest values of T (9.6 °C in average), water conductivity (Cond; 233.8 mg/L), Chlorophyll (Chl a; 4.7 μg/L) and of organic matter (OMs%; 1.3%) among samples (ANOVA, P < 0.05). The second cluster on the PCA plot consisted of Aug-2011 samples from the pulsed basins only; they had highest concentrations of DO (4.7 mg/L), ammonium (NH4 +; 113.1 μg/L) and total nitrogen (TN; 3.1 mg/L) in water (ANOVA, P < 0.05). Jul-2011 samples of both pulsed and static basins and Aug-2011 samples of the static basins were intermingled with each other and formed the third cluster on the PCA plot. Cluster 3 samples generally had significantly lower water DO (<0.5 mg/L) and higher amount of OMs% (> 3%) than other samples (ANOVA, P < 0.05).
ANOSIM analysis yielded consistent results with PCA ordination and showed that samples were statistically not separable based on basin hydrology differences (pulsed vs. static; Ranosim = 0.17). In terms of temporal variability, Nov-2011 samples were different from the other two months (Ranosim = 0.64); and July- and Aug-2011 samples were similar (Ranosim = 0.25). Differences between pulsed and static basin treatments were only identified within Aug-2011 samples (Ranosim = 1), but not within either July-2011 or Nov-2011 samples (Ranosim < 0.20).
Bacterial Community Structure
Abundance of the overall bacterial community and denitrifiers were estimated by quantifying 16S rRNA and nosZ genes using qPCR (Fig. 3a). The numbers of 16S rRNA and nosZ genes were significantly correlated and co-varied temporally (Pearson correlation, r = 0.90; P < 0.05). Both genes generally had higher copy numbers in Aug-2011 than the other months (repeated measures ANOVA, P < 0.05). No difference of 16S rRNA and nosZ genes was found between samples of the pulsed and static basins in any given months or overall.
Community structure of the overall bacterial community and denitrifiers were examined based on terminal restriction fragment (T-RF) patterns of 16S rRNA and nosZ genes, respectively. NMDS analysis revealed that T-RF patterns of 16S rRNA genes were significantly different among months (Fig. 4a). ANOSIM analysis confirmed the statistical significance of observed temporal variations (Ranosim = 0.88). However, no significant differences were observed based on either NMDS or ANOSIM between pulsed and static basins overall or within a given month (Ranosim < 0.03). For nosZ genes T-RF patterns (Fig. 4b), NMDS and ANOSIM identified no significant difference based on either sampling time or basin hydrology (pulsed vs. static) (Ranosim < 0.2).
BIO-ENV analysis revealed that T and OMs% together explained 71% variation of 16S rRNA gene T-RF patterns among samples. Each of these environmental variables alone could explain 55.1% and 62.5% of 16S rRNA gene T-RF variability, respectively. In contrast, environmental variables showed little correlation with nosZ gene T-RF patterns; a combination of T, Cond and DOC only explained <30% variation in nosZ gene T-RF variability.
Denitrification Rate and their Correlation with Biotic and Abiotic Variables
The emission rates of N2O (0–5 μg/m2/h) and N2 (0–5.8 μg/m2/h) were significantly correlated with each other (Pearson correlation r = 0.9, P < 0.05; Fig. 5a); and both showed no significant difference between pulsed and static basins in any given month or overall across sampling months (t-tests, P > 0.05). For both pulsed and static basins, N2O and N2 emission rates showed no significant temporal changes (repeated measures ANOVA, P > 0.05). N2O emission rates showed no significant correlation with either 16S rRNA (Pearson correlation r = 0.13, P > 0.05) or nosZ genes abundances (r = 0.40, P > 0.05). On the other hand, N2 emission rates showed no significant correlation with the abundance of 16S rRNA genes, but were significantly correlated with the abundance of nosZ genes (Pearson correlation r = 0.58, P < 0.05).
Ratios between N2O and N2 emission rates (rN2O), which reflect partitions between the two potential end points of denitrification, did not differ significantly between static and pulsed basins or among sampling months (repeated measures ANOVA, P > 0.05; Fig. 5a). In Aug-2011 pulsed basins and Nov-2011 static basins, both N2O and N2 emission rates reached the highest, rN2O were the highest (t test, P < 0.05). rN2O had no significant correlation with any measured environmental variables.
Single Basin Experiment
Physicochemical Measurements
During the single basin experiment, measurements of all 16 water and soil chemistry variables were done only at the three central basin sites where standing water was present (Fig. 1b). Significant temporal variations were only identified for NOx, NH4 + and DO concentrations, all of which had highest values in Oct-2012 and lowest values in Aug-2012 (ANOVA, P < 0.05).
Concentrations of MOIs%, OMs% and NOxs in soil samples were measured over time for all 11 sites including 7 basin, 2 low-bank and 2 high-bank sites. PCA analysis of these data also revealed groupings of samples generally based on month (Fig. 6), except Sept-2012 samples, which were spread among Jul- and Aug-2012 samples. ANOSIM analysis yielded consistent result and had Ranosim value of 0.59 for overall separation based on month. Neither PCA nor ANOSIM (Ranosim < 0.2) identified sample separation based on sampling location (basin, low-bank and high-bank) or hydrology (flooded vs. drawdown). Moreover, although high-bank and low-bank samples were indistinguishable (Ranosim = 0.05), they both were apart from the basin samples (Ranosim = 0.62 and 0.56, respectively).
Bacterial Community Structure
Abundances of 16S rRNA and nosZ gene copies were significantly correlated with each other (Pearson correlation r = 0.9, P < 0.05; Fig. 3b) and both varied significantly among sampling months (one-way ANOVA, P < 0.05). 16S rRNA and nosZ gene abundances were higher in Sep- and Oct-2012 than earlier sampling months in 2012 (one-way ANOVA, P < 0.05). At each sampling time, abundances of neither 16S rRNA or nosZ gene showed significant difference among the sampling locations (basin, low bank and high bank) or between the hydrological (drawdown and flooded) conditions (one-way ANOVA, P > 0.05).
NMDS analysis of general bacterial community16S rDNA T-RFLP data generally grouped samples in the basin together and away from bank samples (Fig. 7a). Subsequent ANOSIM analysis yielded a Ranosim of 0.51 between basin and bank sites, which confirmed the statistical significance of the NMDS ordination pattern. BIO-ENV analysis revealed that changes of MOIs% alone best explained the variance of the overall bacterial community structure (Rbio-env = 0.44). Temporal difference in 16S rDNA T-RFLP patterns was not identified based on either NMDS or ANOSIM.
NMDS analysis of denitrifying bacterial community nosZ gene T-RFLP data (Fig. 7b), also generally grouped basin samples away from bank samples, as for 16S rRNA genes. However, the extent of overlap was lower than that of the 16S rRNA gene T-RFLP patterns. This was consistent with a higher Ranosim value (0.77) for ANOSIM analysis of nosZ gene T-RFLP data. Similar to the findings of 16S rRNA genes, neither PCA nor ANOSIM analysis identified significant difference among nosZ gene T-RFLP data of different months or between drawdown and flooded conditions. BIO-ENV analysis revealed that changes of MOIs% explained 45.7% of the variance of the nosZ gene-carrying denitrifying community structure, while NOxs explained 25.9% of the variance.
Denitrification Rate and their Correlation with Biotic and Abiotic Variables
N2 and N2O emission rates were significantly correlated with each other (Pearson correlation r = 0.97; P < 0.05; Fig. 5b). The highest N2 (2552 μg N/m2/h) and N2O emission rates (1127 μg N/m2/h) co-occurred in Aug-2012 (flooded condition), which were over 200 and 50 times higher than their lowest rates that were observed in Oct-2012 (flooded condition), respectively (one-way ANOVA, P < 0.05). N2 and N2O emission rates in the two drawdown months, i.e., Jul- and Sep-2012, were similar and averaged 257 and 157 μg N/m2/h for the basin sites, respectively (t test; P > 0.05). Within month, basin sites generally had significantly higher N2 and N2O emission rates than high and low bank sites (ANOVA, P < 0.05). Both N2 and N2O emission rates were significantly correlated with a number of variables (Pearson correlation r > 0.70, P < 0.05). They were correlated positively with DOC, TP, TN, Chl, T, DO, pH, NOxs and rNosZ and negatively with NOx, NH4 +, TN, DO, and abundance of 16S rRNA genes. The variability of N2 and N2O emission rates was higher for the high and low bank sites than the basin sites.
rN2O was negatively correlated with N2O emission rates (Pearson correlation r = −0.67; P < 0.05). The correlations between rN2O and environmental variables was opposite of those of N2 and N2O emission rate for NOx, NH4 +, TN, DO, Chl a, T, DO, pH and NOxs. In contrast to N2 and N2O emission rates, rN2O were not significantly correlated with either the abundance of 16S rRNA or nosZ genes. In addition, rN2O was significantly correlated with DOCpw. Similar to the findings of the N gas emission rates, the variability of rN2O is higher for the bank sites than the basin sites.
Discussion
Effects of hydrological changes on composition and function of denitrifying communities depends on the type and duration of the hydrological regime (Bougon et al. 2009; Kim et al. 2010; Peralta et al. 2010; Song et al. 2010). We hypothesized that denitrifying community composition would be influenced primarily by differences in long-term hydrological conditions rather than by short-term changes, while denitrification activity would be influenced by short-term hydrological changes. Yet, results of our multiple basin experiment did not support the hypothesis. We found no significant difference in abundance or community composition of 16S rRNA and nosZ genes between static and pulsed basins. This suggests that long-term hydrology had little impact on overall or denitrifying bacterial communities in the experimental wetlands examined. Seasonal changes in both of abundance and composition of 16S rRNA genes largely followed changes in temperature and OMs%, in accordance with effects of temperature and organic substrate supply on bacterial community structure that has been described previously (Zogg et al. 1997; Eiler et al. 2003). Meanwhile, no significant change of nosZ gene diversity was observed among sampling time, although the nosZ gene abundance did vary temporally. The observed contrasting patterns between 16S rRNA and nosZ genes are in accordance with previous findings that the diversity of 16S rRNA genes may be much higher than that of the nosZ genes within denitrifying bacterial communities and that nosZ genes of similar sequence identities can be carried out by taxonomically different bacteria (Mounier et al. 2004; Wallenstein et al. 2006; Mao et al. 2011). The absence of structural shifts in nosZ genes could also be due to the limitation of T-RFLP, which resolution may be insufficient to detect small changes in nosZ gene sequences. High-throughput deep sequencing of nosZ genes may potentially overcome this resolution limitation. However, in a recent study that simultaneously performed T-RFLP and high-throughput pyrosequencing of nosZ genes (20,000 in total, 1600–2951 per sample of nosZ gene sequences), T-RFLP well captured community shifts of nosZ genes due to changes of soil properties, generating consistent results as revealed by high-throughput pyrosequencing (Mao et al. 2011).
While the multiple basin experiment examined differences between static and pulsed basins, the single basin experiment examined a different type of hydrology difference: between bank and basin sites in a single pulsed basin. Unlike the results of the multiple basin experiments, results of single basin experiments supported our hypothesis. The different results between the single and multiple basin experiments and between bank and basin sites are likely because differences in hydrology among bank and basin samples in the single basin experiment were greater than between pulsed and static basins in the multiple basin experiment. Despite semiannual draw downs in pulsed basins, soils in pulsed and static basins were similar with respect to moisture content, both are saturated throughout most of the year, and both displayed color and appearance characteristic of hydric soils, such as gleying, mottling and oxidized root zones. High-bank and low-bank samples, on the other hand, had lower percent moisture than basin samples, are not saturated most of the year, and displayed lighter and redder colors more characteristic of upland soils, especially the high bank samples. The single basin study also found that community composition of denitrifying bacteria was more sensitive to differences in long-term hydrology than the general bacterial community (Fig. 7).
The single basin study again showed temporal variation in abundance of nosZ genes, but not their composition. This suggests that difference of N2 and N2O emission rates and their ratio (rN2O) among sampling months is likely due to changes in abundance or growth rate of denitrifiers rather than community structure change. This finding contrasts with a previous study (Cavigelli and Robertson 2000), which found that differences in denitrifier community structure were the only explanation for differences in rN2O between two different soils. However, the above study defined differences in community composition using functional measures that would include both bacterial and fungal denitrifiers. Fungal denitrifiers make large contributions to soil denitrification, and have been shown to be the dominant denitrifiers in certain environments (Seo and DeLaune 2010; Long et al. 2013). In addition, fungal denitrifiers make large contributions to N2O emissions because they often lack the nosZ gene (Shoun et al. 1992). Since our analysis of denitrifying communities was based on the nosZ gene, the relative importance of fungi in denitrifying activities was not assessed. However, the multiple and single basin experiments did reveal a significant correlation between N2 emission and the absolute and relative abundance of nosZ gene, respectively, suggesting that bacterial community played an important role in N2 production in tested wetlands.
The single basin studies also identified significant variation in N2 and N2O emission rates and lower rN2O when the basin conditions changed between flooded and drawn-down months; similar results have been observed previously by Song et al. (2010). However, this short-term effect was not accompanied by any significant change in nosZ gene abundance or community structure. In addition, N2 and N2O emission rates and rN2O were significantly correlated with a number of water and soil variables, which had significant seasonal variations. Moreover, although MOIs% was found to track changes of denitrifying bacterial communities, it was not correlated with nitrogen gas emission rates. Therefore, observed short-term changes in gas emissions and rN2O between drawdown and flooded conditions in the single basin study were likely regulated by temporal changes rather than hydrological properties.
Denitrification rates observed in our study were within the range of published studies (Song et al. 2010); the values showed large variations within and among basins, and between sampling dates. This variation is a well-documented problem in denitrification research and is thought to be due to a number of spatially and temporally heterogeneous factors that regulate denitrification (Groffman 2012). We observed no difference in denitrification rates based on hydrology in the multiple basin experiment. If samples were also collected during the drawdown months and sample numbers were larger, there may have been clearer differences in denitrification rates based on hydrology, as seen in the single basin experiment.
In conclusion, we have found that large differences in long-term hydrologic conditions may structure denitrifying community composition and activities in nitrogen gas production and partitioning of gas production routes between N2 and N2O. In contrast, static and pulsed basins, which had moderate hydrological differences, did not differ presumably because of similarities in soil properties and other environmental conditions. Short-term hydrologic changes did correlate with shifts in denitrification rates, and not surprisingly, higher denitrification rates were associated with samples of higher temperature, soil percent moisture and nutrient supply.
References
Barlett MA, Leff LG (2010) Planktonic bacterial responses to nutrient amendments in wetland mesocosms. Wetlands 30:1161–1170
Blackwood C, Oaks A, Buyer J (2005) Phylum- and class-specific PCR primers for general microbial community analysis. Applied and Environmental Microbiology 71:6193–6198
Bougon N, Aquilina L, Auterives C, Vandenkoornhuyse P (2009) Influence of hydrological fluxes on bio-geochemical processes in a peatland. Biogeosciences Discussions 6:4829–4863
Bowles MW, Nigro LM, Teske AP, Joye SB (2012) Denitrification and environmental factors influencing nitrate removal in Guaymas Basin hydrothermally altered sediments. Frontiers in Microbiology 3:377
Braker G, Schwarz J, Ralf C (2010) Influence of temperature on the composition and activity of denitrifying soil communities. FEMS Microbiology Ecology 73(1):134–148
Cavigelli M, Robertson G (2000) The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology 81:1402–1414
Cavigelli M, Robertson P (2001) Role of denitrifier diversity in rates of nitrous oxide consumption in a terrestrial ecosystem. Soil Biology and Biochemistry 33:297–310
Choudhary MA, Akramkhanov A, Saggar S (2002) Nitrous oxide emissions from a New Zealand cropped soil: tillage effects, spatial and seasonal variability. Agriculture, Ecosystems and Environment 93:33–43
Clarke KR, Warwick RM (2001) Change in marine communities: an approach to statistical analysis and interpretation, 2nd edn. PRIMER-E Ltd, Plymouth, United Kingdom
Dhondt K, Boeckx P, Hofman G, Van Cleemput O (2004) Temporal and spatial patterns of denitrification enzyme activity and nitrous oxide fluxes in three adjacent vegetated riparian buffer zones. Biology and Fertility of Soils 40:243–251
Drinkard M, Kershner MW, Romito A, Nieset J, de Szalay FA (2011) Responses of plants and invertebrate assemblages to water-level fluctuation in headwater wetlands. Journal of the North American Benthological Scoiety 30:981–996
Eiler A, Langenheder S, Bertilsson S, Tranvik LJ (2003) Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Applied and Environmental Microbiology 69:3701–3709
Fierer N, Jackson J, Vilgalys R, Jackson R (2005) Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Applied and Environmental Microbiology 71:4117–4120
Groemping U, Lehrkamp M (2013) Relative importance of regressors in linear models. R Package, ver. 2013:2–2
Groffman PM (2012) Terrestrial denitrification: challenges and opportunities. Ecological Processes 1:1–11
Henry S, Bru D, Stres B, Hallet S, Philippot L (2006) Quatitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Applied and Environmental Microbiology 72:5181–5189
Hernandez M, Mitsch W (2006) Influence of hydrologic pulses, flooding frequency, and vegetation on nitrous oxide emissions from created riparian marshes. Wetlands 26:862–877
Hernandez M, Mitsch W (2007) Denitrification in created riverine wetlands: influence of hydrology and season. Ecological Engineering 30:78–88
Holmes R, Aminot A, Kerouel R, Hooker B, Peterson B (1999) A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Canadian Journal of Fisheries and Aquatic Sciences 56:1801–1808
IPCC (2007) Climate Change 2007: Working Group I, the physical science basis. In: Solomon S, Qin D, Manning M, Chen Z, Marquis M, Averyt K,Tignor M, Miller H(eds). Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA
Kim S, Miyahara M, Fushinobu S, Wakagi T, Shoun H (2010) Nitrous oxide emission from nitrifying activated sludge dependent on denitrification by ammonia-oxidizing bacteria. Bioresource Technology 101:3958–3963
Kjellin J, Hallin S, Worman A (2007) Spatial variations in denitrification activity in wetland sediments explained by hydrology and denitrifying community structure. Water Research 41:4710–4720
Knowles R (1979) Denitrification, acetylene reduction, and methane metabolism in lake sediment exposed to acetylene. Applied and Environmental Microbiology 38:86–493
Korner H, Zumft W (1989) Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Applied and Environmental Microbiology 55:1670–1676
Long A, Heitman J, Tobias C, Philips R, Song B (2013) Co-occurring anammonx, denitrification, and codenitrification in agricultural soils. Applied and Environmental Microbiology 79:168–176
Malone JP, Stevens RJ, Laughlin RJ (1998) Combining the 15 N and acetylene inhibition techniques to examine the effect of acetylene on denitrification. Soil Biology and Biochemistry 30:31–37
Mao Y, Yannarell A, Mackie R (2011) Changes in N-transforming Archaea and bacteria in soil during the establishment of bioenergy crops. PloS One 6:e24750
Martikainen P, Nykanen H, Crill P, Silvola J (1993) Effect of a lowered water table in nitrous oxide fluxes from northern peatlands. Nature 366:51–53
Mitsch WJ, Gosselink JG (2007) Wetlands, 4th edn. Wiley, New York
Mou X, Moran MA, Stepanauskas R, González JM, Hodson RE (2005) Flow-cytometric cell sorting and subsequent molecular analyses for culture-independent identification of bacterioplankton involved in dimethylsulfoniopropionate transformations. Applied and environmental microbiology 71:1405–1416
Mounier E, Hallet S, Cheneby D, Benizri E, Gruet Y, Nguyen C, Piutti S, Robin C, Slezack-Deschaumes S, Martin-Laurent F, Germon J, Philippot L (2004) Influence of maize mucilage on the diversity and activity of the denitrifying community. Environmental Microbiology 6:301–312
Oksanen P (2013) Vegan 2.0–8 in R version 2.15.2 2012–10-26.
Peralta A, Matthews J, Kent A (2010) Microbial community structure and denitrification in a wetland mitigation bank. Applied and Environmental Microbiology 76:4207–4215
Philippot L, Hallin S, Schloter M (2007) Ecology of denitrifying prokaryotes in agricultural soil. Advances in Agronomy 96:249–305
Pinay G, Gumiero B, Tabacchi E, Gimenez O, Tabacchi-Planty A, Hefting M, Burt T, Black V, Nilsson C, Iordache V, Bureau F (2007) Patterns of denitrification rates in European alluvial soils under various hydrological regimes. Freshwater Biology 52:252–266
Pinheiro J, Bates D, DebRoy S, Sarkar D (2013) Nlme version 3.1.111 in R version 2.15.2 2012–10-26.
Priemé A, Christensen S (2001) Natural perturbations, drying–wetting and freezing–thawing cycles, and the emission of nitrous oxide, carbon dioxide and methane from farmed organic soils. Soil Biology and Biochemistry 33:2083–2091
Reddy K, Delaune R (2004) Biogeochemistry of wetlands: science and applications. CRC Press, Boca Raton, FL
Rich J, Myrold D (2004) Community composition and activities of denitrifying bacteria from adjacent agricultural soil, riparian soil, and creek sediment in Oregon, USA. Soil Biology and Biochemistry 36:1431–1441
Rich J, Heichen R, Bottomley P, Cromack K, Myrold D (2003) Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils. Applied and Environmental Microbiology 69:5974–5982
Robertson GP, Greenberg J, Gosz JR (1999) Soil CO2, N2O, and CH4 exchange. Standard soil methods for long-term ecological research. 185
Sahrawat K (2003) Organic matter accumulation in submerged soils. Advances in Agronomy 81:169–201
Salles JF, Le Roux X, Poly F (2012) Relating phylogenetic and functional diversity among denitrifiers and quantifying their capacity to predict community functioning. Frontiers in Microbiology 3:1–15
Santmire J, Leff L (2007) The effect of sediment grain size on bacterial communities in streams. Journal of the North American Benthological Society 26:601–610
Seo DC, DeLaune RD (2010) Fungal and bacterial mediated denitrification in wetlands: influence of sediment redox condition. Water Research 44:2441–2450
Shoun H, Kim DH, Uchiyama H, Sugiyama J (1992) Denitrification by fungi. FEMS Microbiology Letters 94:277–282
Song K, Lee S, Mitsch W, Kang H (2010) Different responses of denitrification rates and denitrifying bacterial communities to hydrologic pulsing in created wetlands. Soil Biology and Biochemistry 42:1721–1727
Stres B, Danevčič T, Pal L, Fuka MM, Resman L, Leskovec S, Hacin J, Stopar D, Mahne I, Mandic-Mulec I (2008) Influence of temperature and soil water content on bacterial, archaeal and denitrifying microbial communities in drained fen grassland soil microcosms. FEMS microbiology ecology 66:110–122
Tiedje JM, Simkins S, Groffman PM (1989) Perspectives on measurement of denitrification in the field including recommended protocols for acetylene based methods. Plant and Soil 115:261–284
Venterink H, Davidsson T, Kiel K, Leonardson L (2002) Impact of drying and re-wetting on N, P and K dynamics in a wetland soil. Plant and Soil 243:119–130
Wallenstein M, Myrold D, Firestone M, Voytek M (2006) Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecological Applications 16:2143–2152
Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, De Vos P, Verstraete W, Boon N (2009) Initial community evenness favors functionality under selective stress. Nature 458:623–626
Zogg GP, Zak DR, Ringelberg DB, White DC, MacDonald NW, Pregitzer KS (1997) Compositional and functional shifts in microbial communities due to soil warming. Soil Science Society of America Journal 61S:475–481
Zumft W (1997) Cell biology and molecular basis of denitrification. Microbiology and Molecular Biology Reviews 61:533–616
Acknowledgements
We thank X. Lu, D. Jones, P. Zuccaro and R. Maggiore for assistant with lab work, O. Valverde for assistant with statistical analysis, and A. Hopson and D. Johnson for assistant with sampling. This work is funded by Kent State Farris Family award to XM.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Brower, S., Leff, L. & Mou, X. Effects of Short and Long-Term Hydrological Regimes on the Composition and Function of Denitrifying Bacteria in Experimental Wetlands. Wetlands 37, 573–583 (2017). https://doi.org/10.1007/s13157-017-0891-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13157-017-0891-7