Abstract
The correct identification of species diversity of small single-celled green coccoid microalgae still causes difficulties, since their relatively simple morphology hides a high physiological, ecological and genetic diversity. The use of molecular genetic methods has revolutionized the study of the true biodiversity of so-called “small green balls,” allowing the discovery of numerous new taxa. This article presents the results of a study of strains recently isolated from small freshwater urbanized lakes (Vasilievsky Lakes system, Samara region, Russian Federation). Morphologically, these strains were close to the genus Meyerella: spherical cells, cup-shaped, or wide girdle-shaped parietal chloroplast without pyrenoid. Analysis of the 18S–ITS1–5.8S–ITS2 sequences also showed that the studied strains belong to this genus. Comparison of morphological characteristics, habitat and lifestyle, analysis of tree topology, genetic distances and secondary structures of the ITS1 and ITS2 spacers of the Meyerella members, as well as the delimitation results using the Automatic Barcode Gap Discovery (ABGD) method, the Poisson Tree Processes (PTP) model, the Generalized Mixed Yule The Coalescent (GMYC) method allowed us to establish that the studied strains ACSSI 346, ACSSI 362 and ACSSI 363 are representatives of a new species – M. similis sp. nov.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The first representatives of the Chlorella-clade were described more than 130 years ago (Beijerinck, 1890). However, so far, this group is characterized by a high critical diversity and there are still quite a few “blank spots” in its taxonomy (Pröschold et al., 2021). So the genus Meyerella with the type species M. planktonica was described by Fawley et al. (2005) using the example of strains selected in Lake Itasca (Minnesota, USA). A distinctive feature of this genus from other Chlorella-clade members is the absence of a pyrenoid. The representatives of this genus are mainly free-living freshwater planktonic organisms. The vast majority of them are inhabitants of freshwater reservoirs: Lake Itasca (Minnesota, USA), Arrowwood Lake (North Dakota, USA), Kuibyshev reservoir (Russian Federation), and some urban lakes (St. Petersburg, Kazan, Russian Federation) (Fawley et al., 2005; Lanzoni et al., 2016; Frolova & Sverdrup, 2019; Sverdrup & Frolova, 2020). At the same time, Fučikova et al. (2014) described a strain of Meyerella sp. from desert soil crusts (Virginia Park, Utah, USA). Also Lanzoni et al. (2016) found Meyerella strains which were facultative symbionts of ciliates. The study of this genus representatives is of practical interest, since they can be raw materials for biofuels due to the high growth rate combined with a high lipid content (Karpagam et al., 2015). Despite some findings indicating that the genus Meyerella is not monotypic (Fučikova et al., 2014; Lanzoni et al., 2016), no new species have been described so far. This is probably partly due to the fact that the 18S–ITS1–5.8S–ITS2 fragment is read only for an extremely small number of M. planktonica strains. As a rule, identification of algae was performed either by V4 region of the 18S rRNA gene or by metabarcoding results, which is still not enough for a full description of a new species. Currently, the best approach for identification and creating a new, more natural classification of “little green balls” is considered to be a polyphasic approach combining morphological, ecophysiological, and molecular phylogenetic methods. In this regard, within this study, we propose a description of the new Meyerella species isolated from freshwater lakes (Russia) using several methods, including morphological and ultrastructural observations, analysis of phylogeny (18S rRNA, ITS1, ITS2), DNA-based species delimitation (the Automatic Barcode Gap Discovery (ABGD) method, the Poisson Tree Processes (PTP) model, the Generalized Mixed Yule The Coalescent (GMYC) method), and ecology.
Materials and methods
Isolation and cultivation of algal strains
The studied strains were isolated from small urbanized reservoirs of Tolyatti (Samara region, Russian Federation) in 2019. Strain ACSSI 346 was isolated from ciliate Pseudoblepharisma sp., the strain ACSSI 362 — from ciliate Holophrya sp. from Lake Prudovikov (53°31′46.4″N, 49°30′58.4″E), whereas strain ACSSI 363 was isolated from the pelagic zone (depth 3 m) of Lake Vos’merka (53°30′4.1″N, 49°30′8.1″E). There are small urban eutrophic lakes. Area of Lake Prudovikov is 22400 m2, average depth – 1.7 m, maximum depth – 6.5 m. Lake Vos’merka area is 128800 m2, average depth – 3.1 m, and maximum depth – 6.8 m (Nomokonova et al., 2001). For isolation of their green algae endosymbionts, single ciliate cells were washed 6 times and transferred into fresh liquid BG-11 medium (pH = 7.2). After starvation and digestion of food for 72 h, ciliate were wasted again through 0.5% streptomycin to reduce the risk of bacterial contamination and then were transferred on to BG-11 medium (2% agar, pH = 7.2). After being placed on an agarized plate, the ciliate cell was pierced with a sterile needle. Further cultivation of endosymbionts was carried out under standard conditions. To isolate free-living organisms, a drop of unfiltered lake water was spread onto BG-11 (2% agar, pH = 7.2), and then individual colonies were repeatedly subcultured. All new strains were deposited at the Algal Collection of the Soil Science Institute (ACSSI, Pushchino, Russia). The studied strains were grown on BG-11 medium (Stanier et al., 1971) in a growth chamber at 23–25 °C under a 12-h:12-h light–dark regime with a photon fluence rate of 60–75 μmol quanta m–2 s–1.
Microscopy
The morphology and life cycles of the strains were studied using light microscopy with Leica DM750 and Carl Zeiss Axio Scope A1 microscopes (Germany) in the Collective Use Center, Institute of Physicochemical and Biological Problems in Soil Science, Russian Academy of Sciences. The results were documented using drawings and photographs obtained with Carl Zeiss MRc 5 (Germany) color digital cameras. The observations of the strains were performed for 2 weeks to 6 months. Also algal cells imaging was performed on a Leica THUNDER 3D Cell Culture Imaging system (Leica Microsystems, Germany) using the following THUNDER Large Volume Computational Clearing. For electron microscopy, the same sample preparation protocol as before was used (Egorova et al., 2018). Cells were viewed and photographed with a transmission electron microscope Libra 120 + (Carl Zeiss, Germany) of the Core Facility Center “Cell and Molecular Technologies in Plant Science” at the Komarov Botanical Institute RAS (St. Petersburg, Russia).
DNA isolation, amplification, purification, and sequencing
Total DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, USA) according to the manufacturer’s protocol. The primers for amplification of the 18S rRNA gene, internal transcribed spacer 1 (ITS1), 5.8S rRNA gene, and internal transcribed spacer 2 (ITS2) were described by Katana et al. (2001), White et al. (1990), and Johnson et al. (2007). Amplification conditions were optimized when needed (Table 1). The target PCR products were detected using electrophoresis in a 1% agarose gel. Further amplicon purification was performed using the Cleanup Mini kit (Evrogen, Russia). Samples for sequencing were processed by the commercial company Evrogen (Russia).
Molecular phylogenetic analysis
To analyze the phylogeny and clarify the taxonomic position of the studied strains, the homology of the18S–ITS1–5.8S–ITS2 nucleotide sequences was searched using the BLASTn algorithm in GenBank (https://blast.ncbi.nlm.nih.gov). The selection of sequences was carried out based on the criteria of maximum identity (similarity ≥ 95%), reading quality, reading length (at least 2300 bp), and belonging to type species and authentic strains. The sample for phylogenetic analysis included 123 strains (ESM_1). The representatives of Parachlorella-clade (Trebouxiophyceae, Chlorophyta), was chosen as an outgroup. To compare the topology of trees, we used data from articles by Krienitz et al. (2004), Luo et al. (2006), Hoshina et al. (2010, 2017, 2021), Pröschold et al. (2010, 2011, 2020, 2021), Bock et al. (2010, 2011), Hoshina and Fujiwara (2013), Hoshina and Nakada (2018), and Pröschold and Darienko (2020). The names of taxa are given according to the International electronic Database AlgaeBase (Guiry & Guiry, 2021). In the BioEdit program, multiple alignment was performed using the ClustalW algorithm. The V4 and V9 region was found using the primer sequences reported by Bradley et al. (2016). An ultrametric Bayesian phylogenetic tree was used to visualize the analysis results. A sample of ultra-fast bootstrap analysis trees obtained in the IQ-TREE program was combined with the topology of a Bayesian ultrametric tree to calculate bootstrap support by the maximum likelihood method (ultra-fast bootstrap analysis). Thus, the support of the ultrametric Bayesian tree topology was evaluated by the Bayesian posterior probabilities (PP) and bootstrap proportion (BP). To calculate bootstrap support, we used an algorithm previously developed by us (Temraleeva et al., 2018), implemented using the functions of the “APE” package (Paradis et al., 2004) for the R statistical software environment v. 3.4.4. The distribution of genetic distances was visualized as a histogram in the R statistical software environment v.3.4.4. Genetic differences between nucleotide sequences were characterized using genetic distances (K2P distances), which were calculated in the MEGA 6.0 program.
The phylogenetic tree reconstructed by the maximum likelihood (ML) method in the IQ-TREE program (with an assessment of the reliability of the topology by ultra-fast bootstrap analysis and testing of the evolutionary model using the BIC criterion) was used to distinguish species using the Poisson tree processes (PTP) algorithm on an online server https://species.h-its.org/. To distinguish species in the data array, the method of automatic search for interspecific gap in genetic distances (automatic barcode gap discovery – ABGD) (Puillandre et al., 2012) was used on an online server https://bioinfo.mnhn.fr/abi/public/abgd/. For ABGD, a matrix of genetic distances was used, calculated using the maximum likelihood method in the IQ-TREE program. When using the ABGD method, the results were analyzed both in the initial partition mode and in the recursive partition mode. Another one method was a generalized mixed Yule model taking into account the integrity of species (general mixed Yule coalescent model, GMYC) (Fujisawa & Barraclough, 2013), implemented in the “splits” package for the R programming language v. 3. 4. 4 (https://www.R-project.org/). For GMYC analysis, an ultrametric tree was used, reconstructed in the BEAST v. 2.6.2 program (Bouckaert et al., 2019). The reconstruction of the tree in BEAST was carried out using two speciation models: the Birth–death speciation model (Lambert & Steel, 2013) with a strict molecular clock; a model of speciation of the birth–death of species with a relaxed molecular clock with the rates of evolution distributed according to the lognormal distribution. The birth – death speciation model was chosen on the assumption that both speciation and species extinction events took place in the studied group of organisms. For the reconstruction of the tree in the BEAST program 50,000,000 generations for Markov chains with saving the results of every 2500 generations were used. With these parameters of the number of generations, all the values of the ESS statistics (the convergence indicator of the BEAST analysis) were more than 200. The selection between strict and relaxed molecular clock was made based on the value of the coefficient of substitution rate variation (CV) in BEAST relaxed molecular clock calculation. If 95% highest posterior density (HPD) of CV includes the zero value, the strict molecular clock is most suitable for the data set (Drummond & Rambaut, 2007).
Folding of ITS1 and ITS2 was performed using the RNAfold web server (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi) in accordance with the principle of minimum energy. When assessing the correctness of the prediction of the secondary structure, ITS1 and ITS2 were guided by A. Coleman (2015) and Caisova et al. (2013), respectively. The comparison of the secondary structure of spacers between strains, the search for conservative motives and compensatory base changes (CBCs) was carried out in the 4SALE program (Seibel et al., 2006, 2008). In the analysis of ITS2 for the species distinguishing, special attention is paid to the approach of sensu A. Coleman (2000, 2007, 2009, 2015), according to which the presence of even one CBC in conservative regions of ITS2 (5 bp of helix I, 10 bp of helix II, all helix III) in two microalgae correlates with their sexual incompatibility. The secondary structures of spacers are visualized in the PseudoViewer3 program. The analysis of the genetic differences of ITS2 is carried out by 2 methods. According to the first method, the nucleotide sequences of ITS2 were aligned taking into account the secondary structure in the 4SALE program. Then the genetic distances were calculated in the in the MEGA 6.0 program (using the Kimura 2-parameter model). As the second method, the method proposed by Hoshina and Fujiwara (2013), Hoshina et al. (2021) was used, when gaps counted as fifth character.
Results
Morphological observations by light microscopy
The strains ACSSI 346, ACSSI 362, and ACSSI 363 had similar morphological characteristics typical for members of the genus Meyerella (Fig. 1a–c, g). None of them produced bristles or colonies. The vegetative cells were spherical or broad oval and 1.2–2.7 × 1.3–2.8 μm without mucilaginous covering. Chloroplast was single, parietal, cup-shaped or wide girdle-shaped, sometimes was divided into two lobes in adult or old cells (Fig. 1d). Pyrenoid was absent. Reproduction by 2 − 4 autospores. Sporangium size 3–5 μm in diameter. Spherical or oval autospores of all strains were equal in size (0.6–1.0 × 0.6–1.1 μm in diameter) and exhibited liberation by rupture of the sporangium cell wall. Zoospores and sexual reproduction not observed.
Morphology of strains ACSSI 346 a, ACSSI 362 b, and ACSSI 363 c cells. The inset shows a more detailed image of an adult cell. The autospores are shown with arrows. THUNDER image of strain ACSSI 346 d. The ciliates with Meyerella cells: Pseudoblepharisma sp. e, Holophrya sp. f. Drawings of light microscopical characters of studied strains (g). 1 – young cell; 2– adult cell; 3,4 – autosporangium. Scale bar a–f: 10 μm, g: 2 μm
Ultrastructural observations by transmission electron microscopy
The cells of strain ACSSI 346 were examined by transmission electron microscopy (TEM). They are covered by a very thin two-contoured trilaminar cell wall usually ranging 20–50 nm, and sometimes reaching 100 nm (Fig. 2). The inner lamella is usually darker (Fig. 2a, e). It is closely adjacent to the plasma membrane. In dividing cells (Fig. 2b) and sporangia (Fig. 2c) the mother cell wall peels off. A single chloroplast is parietal, cup-, or wide girdle-shaped, it occupies 3/4 of the cell volume in the opposite position from the cytoplasm containing the nucleus (Fig. 2a, c, d). It is clearly delineated by a chloroplast envelope and mostly filled with long bundles of thylakoids, numbering from 3(4) to 15 units in each (Fig. 2c). The thylakoid-free regions of the chloroplast stroma contain numerous ribosomes and a small number of large starch grains. Pyrenoid is not detected. A single nucleus is located in a rather small portion of cytoplasm, close to the plasma membrane. It is surrounded by a double nuclear membrane with pores. Nucleoplasm contains weakly aggregated chromatin located along the nuclear periphery as a thick rim and in the form of loose lumps throughout the nucleus (Fig. 2a, d). One Golgi body with a large portion of cisternae is seen in just divided cells (Fig. 2b). Mitochondria are very small, and are barely visible in the concave side of the chloroplast, close to the nucleus (Fig. 2a, c), or at the periphery of the cell. Vacuoles are rather large, with transparent or fibrillar contents (Fig. 2b, c).
Cellular ultrastructure of strain ACSSI 346. a Young vegetative cell with trilaminar cell wall (long black arrows), large nucleus (N), parietal chloroplast (C), and small mitochondrion (transparent arrowhead). b Just divided two cells with developed cell walls and the surrounding mother wall (small black arrows); note the Golgi tanks (G). c Two autospores with vacuoles, starch (S) in the chloroplast; note thylakoid bundles (T), lying just parallel. d The autospore with the well-developed cell wall, cup-shaped chloroplast; it still remains inseparable from another autospore of the common sporangium. e Enlarged cell fragment with a cell wall. Scale bar: 0.5 μm
Phylogenetic analysis
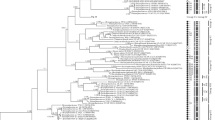
The best model of DNA evolution for the studied dataset of nucleotide sequences (18S–ITS1–5.8S–ITS2) is GTR + I + G4 (BIC = 47,097), which was used for all further calculations. The results of Bayesian calculations with relaxed molecular clock showed 95% highest posterior density (HPD) of coefficient of substitution rate variation (CV) from 0.4 to 0.71 (HPD does not include 0 value) which is indicative of the relaxed molecular clock hypothesis (different rates of substitution accumulation in different evolutionary lines). For all further calculations and visualizations, the results of Bayesian calculations with relaxed molecular clock were used. The phylogenetic tree of the Chlorella-clade based on the fragments 18S–ITS1–5.8S–ITS2 (ESM_2) was shown in Fig. 3. The results of the phylogenetic analysis were consistent with the modern taxonomy of the Chlorella-clade (Chae et al., 2019; Heeg & Wolf, 2015; Hoshina et al., 2021; Krivina & Temraleeva, 2020). Thus, the genera Hindakia, Heynigia, Didymogenes, Carolibrandtia, Micractinium, Actinastrum, Hegewaldia, and Meyerella had high statistical support (PP–1.00, BP 99–100%). Also, the polyphyly of the genus Chlorella was once again confirmed. The studied strains ACSSI 346 (OL619997), ACSSI 362 (OL619998), ACSSI 363 (OL619999) were included in the genus Meyerella, the distinctive feature of which within the Chlorella-clade is the absence of pyrenoid (PP – 1.00, BP – 100%). In contrast to sister species M. planktonica all studied strains ACSSI 346, ACSSI 362, and ACSSI 363 had one group I intron (length – 416 bp) at S1367 (the homologous position in the Escherichia coli SSU rRNA). The genetic distances of the 18S–ITS1–5.8S–ITS2 between the ACSSI strains were low and did not exceed 0.1%. At the same time, the level of genetic differences between the studied strains and representatives of M. planktonica was 2.4%.
Comparative analyses of the V4 and V9 regions of the 18S rRNA gene
The V4 region length of Meyerella representatives was 376 nucleotides. Within the Chlorella-clade, the molecular signature of the genus Meyerella was discovered — A (152 bp), A (153 bp), A (190 bp), and T (242 bp) (Fig. 4A, ESM_3). The ACSSI strains, unlike other Meyerella strains, also had a specific signature — G (56 bp), G (60 bp), T (88 bp), and C (89 bp). The V9 region length of the for all studied Meyerella members was 106 bp. Within this region, a unique nucleotide combination was also found for ACSSI strains, distinguishing them from other Meyerella strains — A (41 bp), C (46 bp), T (66 bp), and G (79 bp) (Fig. 4B, ESM_4).
a A rooted ultrametric phylogenetic tree of Chlorella-clade green microalgae, constructed by the Bayes inference (BI), based on the 18S–ITS1–5.8S–ITS2 sequences (2650 bp). As statistical support for the nodes of the tree, a posterior probabilities (PP) and bootstrap values (BP), respectively, are indicated; the values of PP < 0.7 and BP < 70% are not shown. The model of nucleotide substitutions: GTR + I + G4. Note: studied ACSSI strains are highlighted in bold; * – authentic strains; (T) – type species. The rectangles indicate clustering by various methods of species delimitation: ABGD, GMYC, mlPTP, bPTP. b The fragment of rooted ultrametric phylogenetic tree of Chlorella-clade green microalgae, corresponding to the genus Micractinium. The symbols see in Fig. 3b
The ITS1 secondary structures of the studied strains corresponded to the generally accepted model of eukaryotic organisms, which was developed by A. Coleman (2000, 2007, 2015). All of them included 4 unbranched helices. Helices I − III were located next to each other. Helix IV was separated from the rest by unpaired nucleotides. Following helix IV, there was a single-stranded A-rich region adjacent to the 5.8S rRNA gene. The ITS1 length of the strains ACSSI 346, ACSSI 362, and ACSSI 363 was 238 n. (Fig. 5). The ITS1 length of M. planktonica strains was greater (259 n.). All the studied ACSSI strains had a short helix I, the length of which was only 12 n. and was the smallest in the Chlorella-clade whereas strains of M. planktonica had a length of 30 n. Also strains ACSSI 346, ACSSI 362, and ACSSI 363 had 1 CBC (4 bp, helix I) compared with sister species. Should be noted this combination (A–U in 4 bp, helix I) was found only for these ACSSI strains within the Chlorella-clade. So it could be considered as their “molecular signature.”
The ITS2 secondary structures of these strains under consideration had common features specific for green microalgae (Chlorophyta): four unbranched helices, a pyrimidine-pyrimidine mismatch of the helix II and conservative motif GGUAGG on the 5′-side of helix III (Coleman, 2000, 2009, 2015). The ITS2 length of studied ACSSI strains was 252 n. (Fig. 6) whereas ITS2 length of M. planktonica was 242 n. Comparative analysis of ITS2 secondary structures showed that 1 CBC was found between the strains ACSSI 346, ACSSI 362, and ACSSI 363 and M. planktonica strains in the conservative region (third nucleotide pair) of the helix II. Classic analysis of ITS2 aligned sequences genetic distances (with secondary structure) showed that genetic differences between strains ACSSI 346, ACSSI 362, and ACSSI 363 were 0.3%, between studied strains and M. planktonica were 6.7% (ESM_5). Analysis by the Hoshina et al. method showed that the level of differences between the studied strains was 0%, between ACSSI strains and strains of M. planktonica – 15%.
Species delimitation
The method ABGD identified 40 MOTUs (molecular operational taxonomic units) of the species level in the Chlorella-clade, not counting the outgroup. The ABGD distance of species differentiation in the pairwise comparison of sequences was 0.038. The ABGD results in the range of species distinction distances according to the variants of the algorithm of initial delimitation (initial partition) and recursive delimitation (recursive partition) coincided with each other. Using the method GMYC, 58 MOTUs were identified (the delimitation distance of species is 0.0053). Statistical support for the results of differentiation P = 3.885781e- − 15 < 0.05; therefore, there is enough data in the array to obtain reliable results. Using the mlPTP, 63 MOTUs were identified, which is close to the results of the method GMYC. However, only 39 from them have high statistical support (> 0.95). According to the bPTP results the largest number of MOTUs was identified – 79. Of these, the same 39 MOTUs had high statistical support as in the previous analysis. The results of species delimitation by ABGD, GMYC, mlPTP, and bPTP methods are shown on the phylogenetic tree (Fig. 3). All delimitation methods confirmed that the studied strains do not relate to species M. planktonica and belong to a new species. According to the PTP results, ACSSI strains can be divided into 2 species clusters. However, these clusters have low statistical support.
Ecology
The studied strains ACSSI 346, ACSSI 362, and ACSSI 363 are facultative endosymbionts. All of them were isolated from small highly eutrophic urban lakes. They can independently exist in the plankton of freshwater reservoirs, but they can also enter into relations of optional endosymbiosis with various ciliates – Pseudoblepharisma sp. (Fig. 1e) and Holophrya sp. (Fig. 1f). Genus Pseudoblepharisma includes free-living elongate heterotrich ciliates with short anterior adoral zone of membranelles. Members of Pseudoblepharisma were found in thickets of higher aquatic vegetation, where the greatest amount of organic matter was noted. These ciliates are mixotrophic in that it filter-feeds on bacteria from the water while hidden in its lorica, and it also likely receives nutrients from its algal symbionts, a relationship well documented for representatives of this genus (Esteban et al., 2010; Hines et al., 2022). Members of Holophrya are free-living holotrich ciliates, ellipsoidal, sometimes even spherical, slightly asymmetrical, with some asymmetrically positioned mouth and a transparent alveolar layer clearly visible around the perimeter of the body. As in the previous case, they are mixotrophs, in the cytoplasm of which symbiotic algae can often be found. These ciliates, as a rule, prefer waters with moderate or high degree of organic pollution (Foissner, 2019). Returning to the ACSSI strains, we should be note, that all of them can grow on BG11-N + medium without adding vitamins. This can be seen as another proof that symbiosis is exactly facultative.
Discussion
Currently, the most effective approach for the identification and classification of algae from Chlorella-clade is considered to be the polyphasic approach, which combines morphological, ecophysiological, and molecular phylogenetic methods (Hoshina & Nakada, 2018; Hoshina et al., 2021; Krivina & Temraleeva, 2020; Pröschold et al., 2020, 2021).
Morphologically, studied strains ACSSI 346, ACSSI 362, and ACSSI 363 resemble representatives of genera such as Mychonastes (Krienitz et al., 2011; Simpson & Valkenburg, 1978), Picochlorum (Henley et al., 2004; Krasovec et al., 2018), Pseudochloris (Somogyi et al., 2014), Edaphochloris (Temraleeva et al., 2022), and other similar “small green balls,” unicellular, with predominantly spherical cells with one parietal chloroplast that lack a pyrenoid. Within Chlorella-clade, a similar morphotype is typical for only genus Meyerella (Fawley et al., 2005). However, it is impossible to establish belonging to this clade and this genus only on the basis of the results of light microscopy without using methods of molecular genetic analysis. In general, the morphological and ultrastructural characteristics of the studied strains were close to those of M. planktonica (Table 2). However, there were still a number of differences. The cell sizes of the ACSSI strains were slightly smaller than those of the M. planktonica strains. There were also differences in the cell shape and the type chloroplast. According to the original description of Fawley et al. (2005) M. planktonica had mostly cylindrical with rounded cell shape ends and only sometimes spherical, so cell shape of ACSSI strains usually were spherical or broad oval. At the same time, the chloroplast of M. planktonica was plate-shaped in adult cells and trough-shaped in older cells, whereas chloroplast of ACSSI strains was mainly cup-shaped or wide girdle-shaped and sometimes is divided into two lobes in adult or old cells.
Phylogenetic analysis of the 18S–ITS1–5.8S–ITS2 fragment confirmed that the studied strains belong to the genus Meyerella, but separated from the species M. planktonica. The genetic distances between strains ACSSI 346, ACSSI 362, and ACSSI 363 did not exceed 0.1% and corresponded to intraspecific level within Chlorella-clade (Krivina & Temraleeva, 2020). Genetic differences between these strains and the sister species M. planktonica clearly indicate the interspecific level (2.4%). Within the Chlorella-clade, the issue of establishing clear and unambiguous boundaries is difficult, largely due to the polyphyly of the genus Chlorella proper. At the same time, in the genus Micractinium, which is characterized by high species richness, but is not polyphyletic, the interspecific level of genetic differences ranged from 0.3 to 4.2%, also in the genus Hindakia – 0.7–0.9%, in the genus Didymogenes – 1.5–1.9%.
All studied ACSSI strains have included the 416 nucleotide insertion in the 18S rRNA gene as mentioned above but this intron was not presented in none of the M. planktonica strains. The use of intron composition and its position in the 18S rRNA gene as a tool for distinguishing of the algae species is quite common (Gaonkar et al., 2018; Hoshina et al., 2010; Krivina et al., 2021b; Pröschold et al., 2021; Spanner et al., 2020; Vorobyev et al., 2009). Thus, for example, Vorobyev et al. (2009) in the process of studying symbiotic and free-living algae of Chlorella-clade came to the conclusion that introns in the 18S rRNA gene in members of this clade are good candidates for a taxonomic marker, as well as promising objects for evolutionary research. Furthermore, Gaonkar et al. (2018) using the example of the planktonic diatoms Chaetocerotaceae concluded that introns could be used as markers to different cryptic species within a morphospecies. However, sometimes the presence/absence of an intron, as well as differences in its structure or length, can be observed between strains of the same species. So Hoshina et al. (2021) found significant differences in the length of the intron between different populations of Chlorella variabilis. In this regard, within this study, the presence/absence of an intron in the 18S rRNA gene is considered as one of the additional characteristics confirming the difference between ACSSI strains and sister species M. planktonica.
Recently, high-throughput sequencing of phylogenetic markers (metabarcoding) has become increasingly popular in the study of microbial diversity in the environment. Metabarcoding of microbial eukaryotes usually targets short variable regions V4 and V9 of the 18S rRNA gene (Decelle et al., 2014; Sogin et al., 2006; Sverdrup & Frolova, 2020). Comparative analysis has shown that these regions are very conservative for the Chlorella-clade members. The V4 region of the 18S rRNA gene contained 90% conservative sites, 10% variable sites, and 8% parsimony-informative sites. The V9 region contained 83% conservative sites, 17% variable sites, and 13% parsimony-informative sites. Nevertheless, these regions do not allow to distinguish many species within the Chlorella-clade. However, a molecular signature characteristic of all currently described representatives of the genus Meyerella was found in the V4 region. Moreover, both in the V4 region and in the V9 region, we found stable motifs characteristic only of representatives of ACSSI strains. These molecular signatures allow us to use these barcodes to identify ACSSI strains in environmental samples and, in our opinion, are evidence of their independent species status.
The internal transcribed spacers ITS1 and ITS2 as more variable markers were also analyzed. As a result of this, 1 CBC was found between the studied strains and M. planktonica in the helix I of ITS1, which can be considered as the “molecular signature” of new species. Also 1 CBC was found in the conservative region in the helix II of ITS2. According to Coleman (2000), it is clear evidence that taxa belong to different species. The genetic differences of ITS2 (with secondary structure) between ACSSI strains and M. planktonica was 6.7%. This value within the Chlorella-clade is interspecific. For comparison, interspecific genetic distances for representatives of the genus Micractinium were 1.9–23.4%, Didymogenes – 3.1–5.3%, Hindakia – 3.6%. Additionally, Hoshina et al. (2010, 2021) found that differences in the ITS2 nucleotide sequences (with gaps counted as fifth character) usually less than 2% for representatives of the same species and more than 10% for different species. When analyzed by this method, the genetic differences between ACSSI strains were 15%, which also clearly corresponded to the interspecific level.
More recently, the methods for recognizing new species or testing species hypotheses became popular such as ABGD, GMYC, and PTP. Nevertheless among green algae, only few genera, for example Coccomyxa (Malavasi et al., 2016), Chlorella (Zou et al., 2016a), Micractinium (Krivina et al., 2021b), Parachlorella (Krivina et al., 2021a), Prasiola (Garrido-Benavent et al., 2017), and Scenedesmus (Zou et al., 2016b), were subjected to these delimitation algorithms. As a rule, for the analysis of groups with a low degree of genetic divergence (for example, the Parachlorella-clade, genera Chlorella, Coccomyxa, Micractinium), topological algorithms GMYC and PTP are more suitable (Krivina et al., 2021a; Malavasi et al., 2016; Zou et al., 2016a, 2016b). At the same time, it should be remembered that the isolated clusters (MOTUs), i.e. represent only preliminary hypotheses about the validity of the species, which need to be confirmed by other characteristics. The use of various methods to delimit species in the Chlorella-clade has shown that the ABGD method was less “sensitive.” In our opinion, GMYC more realistically reflected the modern taxonomy of Chlorella-clade while PTP method led to excessive differentiation. Notably all the delimitation methods used have confirmed the independent species status of the studied ACSSI strains.
The Chlorella-clade includes organisms with different lifestyles. Most of them are free-living planktonic inhabitants of various reservoirs. Some species are obligate endosymbionts, for example Chlorella variabilis (Hoshina et al., 2010, 2021; Pröschold et al., 2011), C. heliozoae (Pröschold et al., 2011), Micractinium conductrix (Hoshina et al., 2010; Pröschold et al., 2011), and Carolibrandtia ciliaticola (Hoshina et al., 2017), Hoshina and Nakada (2018). During cultivation, such organisms have a need for vitamins in vitamins B1 and B12 for the normal implementation of vital processes, since in natural conditions they receive them from the host (Hoshina & Nakada, 2018; Hoshina et al., 2010, 2017; Pröschold et al., 2011; Vorobyev et al., 2009). The such species as Chlorella vulgaris and Micractinium tetrahymenae are facultative endosymbionts (Pröschold et al., 2020). They are able both to form symbiotic associations with various protozoa, most often with ciliates, and to survive independently in the environment. All representatives of M. planktonica, whose species identity is beyond doubt, are free-living freshwater organisms. All of them are isolated from the small glacial Lake Itasca (Itasca State Park). The studied ACSSI strains are facultative endosymbionts and were found both in symbiotic associations with Pseudoblepharisma and Holophrya ciliates and in the composition of the free-living freshwater plankton. The fact that these strains are facultative endosymbionts is further evidenced by their successful cultivation on BG-11 medium without the addition of vitamins and/or casamino acids. In addition, strains not identified as any species have also been described within the genus Meyerella. One of them Meyerella sp. OL-2016 is an endosymbiont of Paramecium chlorelligerum (Lanzoni et al., 2016). The other strain BCP-CNP1VF19 was isolated from the desert soil crust (Fučikova et al., 2014). However, for these strains, only not full 18S rRNA gene were read, the length of which was 1223 bp and 1519 bp, respectively. The short length of the fragments and the presence of degenerate nucleotides make it difficult to identify these strains. Nevertheless, the analysis of the available material showed that the genetic distances of each of these strains with M. planktonica, ACSSI strains and among themselves are not less than 0.4%. This indicates that these strains can be considered as potentially new species, but for a final decision, their detailed study is necessary, including a qualitative reading of the full 18S rRNA gene and internal transcribed spacers.
Thus, the studied strains ACSSI 346, ACSSI 362, and ACSSI 363 are members of a new species of the genus Meyerella within the Chlorella-clade.
Diagnosis
Meyerella similis E. Krivina, A. Temraleeva, O. Boldina & Yu. Bukin sp. nov.
Etymology
The name refers to the similarity between the species and its sister species, M. planktonica.
Specific diagnosis
Cells solitary, planktonic, spherical, or broad oval, without bristles, 1.2–2.7 × 1.3–2.8 μm. Mucilage absent. Chloroplast single, parietal, cup-shaped, wide girdle-shaped, sometimes has two lobes in adult or old cells, without pyrenoid. Reproduction by 2 − 4 autospores. Sporangium size 3–5 μm in diameter. Autospores of all strains were oval, broad oval or spherical, equal in size (0.6–1 × 0.6–1.1 μm) and exhibited liberation by rupture of the sporangium cell wall. Zoospores and sexual reproduction not observed.
Differs from morphologically similar M. planktonica by genetic signatures in the 18S–ITS1–5.8S–ITS2 sequences.
Holotype
The authentic strain ACSSI 346 was deposited at the IPPAS collection of microalgae and cyanobacteria under the designation IPPAS C-2054 (metabolically inactive cryopreserved subculture).
Habitat
Facultative endosymbiont of ciliates or free-living in freshwater reservoirs.
Type locality
Freshwater urban lake, Lake Prudovikov, Tolyatti, Samara region, Russian Federation.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- ABGD:
-
Automatic Barcode Gap Discovery
- ACOI:
-
Coimbra Collection of Algae, Portugal
- ACSSI:
-
Algal Collection of the Soil Science Institute, Russia
- BI:
-
Bayesian inference
- BP:
-
Bootstrap proportion
- CBC:
-
Compensatory base change
- CCALA:
-
The Culture Collection of Autotrophic Organisms, The Czech Republic
- CCAP:
-
The Culture Centre Algae and Protozoa, UK
- CCMP:
-
The Culture Collection of Marine Phytoplankton, USA
- CV:
-
Coefficient of substitution rate variation
- GMYC:
-
Generalized mixed yule coalescent
- HPD:
-
Posterior density
- IPPAS:
-
Collection of microalgae and cyanobacteria IPPAS, Russia
- ITS1:
-
First internal transcribed spacer
- ITS2:
-
Second internal transcribed spacer
- LM:
-
Light microscopy
- ML:
-
Maximum likelihood
- MOTU:
-
Molecular operational taxonomic units
- NIES:
-
Microbial Culture Collection at the National Institute for Environmental Studies, Japan
- PP:
-
Posterior probability
- PCR:
-
Polymerase chain reaction
- PTP:
-
Poisson tree processes
- SAG:
-
Sammlung von Algenkulturen at the University of Göttingen, Germany
- TEM:
-
Transmission electron microscopy
- UTEX:
-
Culture Collection at the University of Texas at Austin, USA
References
Beijerinck, M. W. (1890). Culturversuche mit Zoochlorellen, Lichenengonidien und anderen niederen Algen. Botanische Zeitung, 47, 725–739, 741–754, 757–768, 781–785.
Bock, C., Krienitz, L., & Pröschold, T. (2011). Taxonomic reassessment of the genus Chlorella (Trebouxiophyceae) using molecular signatures (barcodes), including description of seven new species. Fottea, 11(2), 293–312. https://doi.org/10.5507/FOT.2011.028
Bock, C., Proschold, T., & Krienitz, L. (2010). Two new Dictyosphaerium-morphotype lineages of the Chlorellaceae (Trebouxiophyceae): Heynigia gen. nov. and Hindakia gen. nov. European Journal of Phycology, 45(3), 267–277. https://doi.org/10.1080/09670262.2010.487920
Bouckaert, R., Vaughan, T. G., Barido-Sottani, J., et al. (2019). BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLOS Computational Biology, 15(4), e1006650. https://doi.org/10.1371/journal.pcbi.1006650
Bradley, I. M., Pinto, A. J., & Guest, J. S. (2016). Design and evaluation of Illumina MiSeq compatible primers for the 18S rRNA gene for improved characterization of mixed microalgal communities. AEM, 82(19), 5878–5891. https://doi.org/10.1128/AEM.01630-16
Caisová, L., Marin, B., & Melkonian, M. (2013). A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist, 164, 482–496. https://doi.org/10.1016/j.protis.2013.04.005
Chae, H., Lim, S., Kim, H., Choi, H.-G., & Kim, J. H. (2019). Morphology and phylogenetic relationships of Micractinium (Chlorellaceae, Trebouxiophyceae) taxa, including three new species from Antarctica. Algae, 34(4), 267–275. https://doi.org/10.4490/algae.2019.34.10.15
Coleman, A. W. (2000). The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist, 151(1), 1–9. https://doi.org/10.1078/1434-4610-00002
Coleman, A. W. (2007). Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. NAR, 35, 3322–3329. https://doi.org/10.1093/nar/gkm233
Coleman, A. W. (2009). Is there a molecular key to the level of ‘biological species’ in eukaryotes? A DNA guide. Molecular Phylogenetics and Evolution, 50, 197–203. https://doi.org/10.1016/j.ympev.2008.10.008
Coleman, A. W. (2015). Nuclear rRNA transcript processing versus internal transcribed spacer secondary structure. Trends in Genetics, 31(3), 157–163. https://doi.org/10.1016/j.tig.2015.01.002
Decelle, J., Romac, S., Sasaki, E., Not, F., Mahé, F., & Lovejoy, C. (2014). Intracellular diversity of the V4 and V9 regions of the 18S rRNA in marine protists (Radiolarians) assessed by high-throughput sequencing. PLoS ONE, 9(8), e104297. https://doi.org/10.1371/journal.pone.0104297
Drummond, A. J., & Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7(1), 214. https://doi.org/10.1186/1471-2148-7-214
Egorova, I. N., Mincheva, E. V., & Boldina, O. N. (2018). Ataktogamous green microalgae of the genus Chlorosarcinopsis Herndon (Chlorophyceae, Chlorophyta) from Zabaikalskiy region (Russia). Phytotaxa, 343(1), 1–19. https://doi.org/10.11646/phytotaxa.343.1.1
Esteban, G. F., Fenchel, T., & Finlay, B. J. (2010). Mixotrophy in ciliates. Protist, 161, 621–641. https://doi.org/10.1016/j.protis.2010.08.002
Fawley, M. W., Fawley, K. P., & Owen, H. A. (2005). Diversity and ecology of small coccoid green algae from Lake Itasca, Minnesota, USA, including Meyerella planktonica, gen. et sp. nov. Phycologia, 44, 35–48. https://doi.org/10.2216/0031-8884(2005)44[35:DAEOSC]2.0.CO;2
Foissner, W. (2019). A detailed description of a Brazilian Holophrya teres (Ehrenberg, 1834) and nomenclatural revision of the genus Holophrya (Ciliophora, Prostomatida). European Journal of Protistology. https://doi.org/10.1016/j.ejop.2019.125662
Frolova, L. L., & Sverdrup, A. E. (2019). Diversity of Verhniy Kaban lake by 18S rRNA of hydrobionts on next-generation sequecing method. Bioscience Biotechnology Research Communications, 12(5), 329–335.
Fučíková, K., Lewis, P. O., & Lewis, L. A. (2014). Widespread desert affiliation of trebouxiophycean algae (Trebouxiophyceae, Chlorophyta) including discovery of three new desert genera. Phycological Research, 62(4), 294–305. https://doi.org/10.1111/pre.12062
Fujisawa, T., & Barraclough, T. G. (2013). Delimiting species using single-locus data and the generalized mixed Yule coalescent approach: A revised method and evaluation on simulated data sets. Systematic Biology, 62(5), 707–724. https://doi.org/10.1093/sysbio/syt033
Gaonkar, C. C., Piredda, R., Minucci, C., et al. (2018). Annotated 18S and 28S rDNA reference sequences of taxa in the planktonic diatom family Chaetocerotaceae. PLoS ONE, 13(12), e0208929. https://doi.org/10.1371/journal.pone.0208929
Garrido-Benavent, I., Pérez-Ortega, S., & de Los Ríos, A. (2017). From Alaska to Antarctica: Species boundaries and genetic diversity of Prasiola (Trebouxiophyceae), a foliose chlorophyte associated with the bipolar lichen-forming fungus Mastodia tessellate. Molecular Phylogenetics and Evolution, 107, 117–131. https://doi.org/10.1371/10.1016/u.ympev.2016.10.013
Guiry, M. D., & Guiry, G. M. (2021). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. Retrieved December 8, 2021, from http://www.algaebase.org
Heeg, J. S., & Wolf, M. (2015). ITS2 and 18S rDNA sequence-structure phylogeny of Chlorella and allies (Chlorophyta, Trebouxiophyceae, Chlorellaceae). Plant Gene, 4, 20–28. https://doi.org/10.1016/j.plgene.2015.08.001
Henley, W. J., Hironaka, J. L., Guillou, L., Buchheim, M. A., Buchheim, J. A., Fawley, M. W., & Fawley, K. P. (2004). Picochlorum oklahomensis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta). Phycologia, 43, 641–652. https://doi.org/10.2216/i0031-8884-43-6-641.1
Hines, H. N., McCarthy, P. J., & Esteban, G. F. (2022). A Case Building Ciliate in the Genus Pseudoblepharisma Found in Subtropical Fresh Water. Diversity, 14, 174. https://doi.org/10.3390/d14030174
Hoshina R., & Fujiwara,Y. (2013). Molecular characterization of Chlorella cultures of the National Institute for Environment Studies culture collection with description of Micractinium inermum sp. nov., Didymogenes sphaerica sp. nov. and Didymogenes soliella sp. nov. (Chlorellaceae, Trebouxiophyceae). Phycological Research, 61(2), 124–132. https://doi.org/10.1111/pre.12010
Hoshina, R., Iwataki, M., & Imamura, N. (2010). Chlorella variabilis and Micractinium reisseri sp. nov. (Chlorellaceae, Trebouxiophyceae): Redescription of the endosymbiotic green algae of Paramecium bursaria (Peniculia, Oligohymenophorea) in the 120th year. Phycological Research, 58(3), 188–210. https://doi.org/10.1111/j.1440-1835.2010.00579.x
Hoshina, R., Kobayashi, M., Suzaki, T., & Kusuoka, Y. (2017). Brandtia ciliaticola gen. et sp. nov. (Chorellaceae, Trebouxiophyceae) a commom symbiotic green coccoid of various ciliate species. Phycological Research, 66(1), 76–81. https://doi.org/10.1111/pre.12194
Hoshina, R., & Nakada, T. (2018). Carolibrandtia nom. nov. as a replacement name for Brandtia Hoshina (Chlorellaceae, Trebouxiophyceae). Phycological Research, 66(1), 82–83. https://doi.org/10.1111/pre.12208
Hoshina, R., Tsukii, Y., Harumoto, T., & Suzaki, T. (2021). Characterization of a green Stentor with symbiotic algae growing in an extremely oligotrophic environment and storing large amounts of starch granules in its cytoplasm. Scientific Reports, 11, 2865. https://doi.org/10.1038/s41598-021-82416-9
Johnson, J. L., Fawley, M. W., & Fawley, K. P. (2007). The diversity of Scenedesmus and Desmodesmus (Chlorophyceae) in Itasa State Park, Minnesota, USA. Phycologia, 46, 214–229. https://doi.org/10.2216/05-69.1
Karpagam, R., Preeti, R., Jawahar, R.K., Saranya, S., Ashokkumar, B., & Varalakshmi, P. (2015). Fatty acid biosynthesis from a new isolate Meyerella sp. N4: molecular characterization, nutrient starvation, and fatty acid profiling for lipid enhancement. Energy & Fuels, 29(1), 143–149. https://doi.org/10.1021/ef501969a
Katana, A., Kwiatowski, J., Spalik, K., Zakryś, B., Szalacha, E., & Szymańska, H. (2001). Phylogenetic position of Koliella (Chlorophyta) as inferred from nuclear and chloroplast small subunit rDNA. Journal of Phycology, 37(3), 443–451. https://doi.org/10.1046/j.1529-8817.2001.037003443.x
Krasovec, M., Vancaester, E., Rombauts, S., et al. (2018). Genome analyses of the microalga Pichochlorum provide insights into the evolution of thermotolerance in the green lieage. Genome Biology and Evolution, 10(9), 2347–2365. https://doi.org/10.1093/gbe/evy167
Krienitz, L., Bock, C., Dadheech, P. K., & Proeschold, T. (2011). Taxonomic reassessment of the genus Mychonastes (Chlorophyceae, Chlorophyta) including the description of eight new species. Phycologia, 50(1), 89–106. https://doi.org/10.2216/10-15.1
Krienitz, L., Hegewald, E. H., Hepperle, D., Huss, V. A. R., Rohr, T., & Wolf, M. (2004). Phylogenetic relationship of Chlorella and Parachlorella gen. nov. (Chlorophyta, Trebouxiophyceae). Phycologia, 43, 529–542. https://doi.org/10.2216/i0031-8884-43-5-529.1
Krivina, E., & Temraleeva, A. (2020). The difficulty identifying and the cryptic diversity of Chlorella-clada microalgae (Chlorophyta). Microbiology, 89(6), 714–727. https://doi.org/10.1134/S0026261720060107
Krivina, E., Temraleeva, A., & Bukin, Y. S. (2021a). Species delimitation and cryptic diversity analysis of Parachlorella-clade microalgae (Chlorophyta). Microbiology, 90, 455–469. https://doi.org/10.1134/S0026261721040081
Krivina, E., Temraleeva, A., & Sinetova, A. (2021b). New species Micractinium kostikovii (Chlorellaceae, Trebouxiophyceae) from Russia. Phycological Research. https://doi.org/10.1111/pre.12469
Lambert, A., & Steel, M. (2013). Predicting the loss of phylogenetic diversity under non-stationary diversification models. Journal of Theoretical Biology, 337, 111–124. https://doi.org/10.1016/j.jtbi.2013.08.009
Lanzoni, O., Fokin, S. I., Lebedeva, N., Migunova, A., Petroni, G., & Potekhin, A. (2016). Rare freshwater ciliate Paramecium chlorelligerum Kahl, 1935 and its macronuclear symbiotic bacterium candidatus Holospora parva. PLoS ONE, 11(12), e0167928. https://doi.org/10.1371/journal.pone.0167928
Luo, W., Pflugmacher, S., Pröschold, T., Walz, N., & Krienitz, L. (2006). Genotype versus phenotype variability in Chlorella and Micractinium (Chlorophyta, Trebouxiophyceae). Protist, 157, 315–333. https://doi.org/10.1016/j.protis.2006.05.006
Malavasi, V., Škaloud, P., Rindi, F., Tempesta, S., Paoletti, M., Pasqualetti, M., & Fontaneto, D. (2016), DNA-based taxonomy in ecologically versatile microalgae: A re-evaluation of the species concept within the coccoid green algal genus Coccomyxa (Trebouxiophyceae, Chlorophyta). PLoS ONE, 11(3), e0151137. https://doi.org/10.1371/journal.pone.0151137
Nomokonova, V. I., Vykhristyuk, L. A., & Tarasova, N. G. (2001). Trophic state of Vasilievskiy lakes of Togliatti suburb. Izvestiya of the Samara Russian Academy of Sciences Scientific Center, 3(2), 274–283.
Paradis, E., Claude, J., & Strimmer, K. (2004). APE: Analyses of phylogenetics and evolution in R language. Bioinformatics, 20(2), 289–290. https://doi.org/10.1093/bioinformatics/btg412
Pröschold, T., Bock, C., Luo, W., & Krienitz, L. (2010). Polyphyletic distribution of bristle formation in Chlorellaceae: Micractinium, Diacanthos, Didymogenes and Hegewaldia gen. nov. (Trebouxiophyceae, Chlorophyta). Phycological Research, 58, 1–8. https://doi.org/10.1111/j.1440-1835.2009.00552.x
Pröschold, T., & Darienko, T. (2020). Choricystis and Lewiniosphaera gen. nov. (Trebouxiophyceae, Chlorophyta), two different green algal endosymbionts in freshwater sponges. Symbiosis, 82(3),1–14. https://doi.org/10.1007/s13199-020-00711-x
Pröschold, T., Darienko, T., Silva, P. C., Reisser, W., & Krienitz, L. (2011). The systematics of Zoochlorella revisited employing an integrative approach. Environmental Microbiology, 13, 350–364. https://doi.org/10.1111/j.1462-2920.2010.02333.x
Pröschold, T., Pitsch, G., & Darienko, T. (2020). Micractinium tetrahymenae (Trebouxiophyceae, Chlorophyta), a new endosymbiont isolated from Ciliates. Diversity, 12, 200. https://doi.org/10.3390/d12050200
Pröschold, T., Rieser, D., Darienko, T., et al. (2021). An integrative approach sheds new light onto the systematics and ecology of the widespread ciliate genus Coleps (Ciliophora, Prostomatea). Scientific Reports, 11, 5916. https://doi.org/10.1038/s41598-021-84265-y
Puillandre, N., Lambert, A., Brouillet, S., & Achaz, G. (2012). ABGD, automatic barcode gap discovery for primary species delimitation. Molecular Ecology, 21, 1864–1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x
Seibel, P.N., Müller, T., Dandekar, T., Schultz, J., & Wolf, M. (2006). 4SALE: a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformatics, 7, 1−498. https://doi.org/10.1186/1471-2105-7-498
Seibel, P.N., Müller, T., Dandekar, T., & Wolf, M. (2008). Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Research Notes, 1, 1−91. https://doi.org/10.1186/1756-0500-1-91
Simpson, P. D., & Van Valkenburg, S. D. (1978). The ultrastructure of Mychonastes ruminatus gen. et sp. nov., a new member of the Chlorophyceae isolated from brackish water. British Phycological Journal, 13(2), 117–130. https://doi.org/10.2216/10-15.1
Sogin, M. L., Morrison, H. G., Huber, J. A., et al. (2006). Microbial diversity in the deep sea and the underexplored rare biosphere. Proceedings of the National Academy of Sciences, 103, 12115–12120. https://doi.org/10.1073/pnas.0605127103
Somogyi, B., Felföldi, T., Solymosi, K., Flieger, K., Márialigeti, K., Böddi, B., & Vörös, L. (2014). One step closer to eliminating the nomenclatural problems of minute coccoid green algae: Pseudochloris wilhelmii, gen. et sp. nov. (Trebouxiophyceae, Chlorophyta). European Journal of Phycology, 48(4), 427–436. https://doi.org/10.1080/09670262.2013.854411
Spanner, C., Darienko, T., Biehler, T., Sonntag, B., & Pröschold, T. (2020). Endosymbiotic green algae in Paramecium bursaria: A new isolation method and a simple diagnostic PCR approach for the identification. Diversity, 12(6), 240. https://doi.org/10.3390/d12060240
Stanier, R. Y., Kunisawa, R., Mandel, M., & Cohen-Bazire, G. (1971). Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriology Reviews, 35, 171–205. https://doi.org/10.1128/br.35.2.171-205.1971
Sverdrup, A. E., & Frolova, L. L. (2020). Saprobity identification of hydrobiont species of Verhniy Kaban lake of Kazan by 18S rRNA marker gene. Scientific Notes of V.I. Vernadsky Crimean Federal University. Biology Chemistry, 4, 127–142. https://doi.org/10.37279/2413-1725-2020-6-4-127-142
Temraleeva, A., Krivina, E., & Boldina, O. (2022) Edaphochloris gen. nov.: A new genus of soil green algae (Trebouxiophyceae, Chlorophyta) with simple morphology. Plant Systematics and Evolution. https://doi.org/10.1007/s00606-021-01795-8
Temraleeva, A., Moskalenko, S., Mincheva, E., Bukin, Y., & Sinetova, M. (2018). Spongiosarcinopsis terrestris gen. et sp. nov. (Chlorophyta, Chlorophyceae): A new genus of green algae from gray forest soil, Russia. Phytotaxa, 376(6), 291–300. https://doi.org/10.11646/phytotaxa.376.6.4
Vorobyev, K., Andronov, E., Rautian, M., Skoblo, I., Migunova, A., & Kvitko, K. (2009). An atypical Chlorella symbiont from Paramecium bursaria. Protistology, 6(1), 39–44.
White, T. J., Bruns, T., Lee, S., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR Protocols: A Guide to Methods and Applications (pp. 315–322). Academic Press.
Zou, S., Fei, C., Song, J., Bao, Y., He, M., & Wang, C. (2016a). Combining and comparing coalescent, distance and character-based approaches for barcoding microalgaes: A test with Chlorella-like species (Chlorophyta). PloS O, 11(4), e0153833. https://doi.org/10.1371/journal.pone.0153833
Zou, S., Fei, C., Wang, C., Gao, Z., Bao, Y., He, M., & Wang, C. (2016b). How DNA barcoding can be more effective in microalgae identification: A case of cryptic diversity revelation in Scenedesmus (Chlorophyceae). Science and Reports, 9(6), 36822. https://doi.org/10.1038/srep36822
Acknowledgements
We thank the Bioline company for the opportunity to use of the Leica THUNDER 3D Cell Culture Imaging system (Leica Microsystems, Germany).
Funding
The work was funded by the Russian Foundation for Basic Research (RFBR), project no.19–34-60002 (sampling, isolation, cultivation and morphological observation, DNA extraction, PCR, phylogenetic analysis) and the Ministry of Science and Higher Education of the Russian Federation project no.0279–2021-0010 (species delimitation), project no.N121021600184-6 (ultrastructural observations by transmission electron microscopy), project no.121040800126–5 (maintaining algae strains in the ACSSI collection).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Human and animals participation
Our research did not involve human or animals participants.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Krivina, E.S., Boldina, O.N., Bukin, Y.S. et al. Species delimitation polyphasic approach reveals Meyerella similis sp. nov.: a new species of “small green balls” within the Chlorella-clade (Trebouxiophyceae, Chlorophyta). Org Divers Evol 23, 25–40 (2023). https://doi.org/10.1007/s13127-022-00590-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-022-00590-8