Abstract
Cycliophorans are characterized by a complex life cycle that involves an asexual generation and a sexual generation. The most prominent life cycle stage in both generations is the so-called feeding stage. Using RNA-Seq, we profiled differential gene expression between feeding stages from asexual and sexual generations to study this intergenerational shift. For this, we also generated a reference transcriptome for the cycliophoran Symbion pandora. We found that a total of 2660 contigs (more than 10% of the total transcriptome) correspond to genes that are expressed differentially in the feeding stages from the asexual generation as compared to the sexual feeding stages. Among these, 1236 genes are upregulated in the asexual stages as compared to the sexual stages. Conversely, 1424 genes are upregulated in the sexual stages as compared to the asexual stages. The asexual stages express genes predominantly related to RNA processing and splicing as well as protein folding, which suggests a high degree of regulation at the transcriptional and post-transcriptional levels. In marked contrast, the sexual stages highly express genes related to signal transduction and neurotransmission. This is the first time that a large whole-transcriptome RNA-Seq expression dataset has been generated for any cycliophoran. Moreover, this study provides important information for further studies on the molecular mechanisms that are involved in the shift from asexual to sexual generations in this still enigmatic group.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cycliophorans are microscopic invertebrates that have been found as epizoic on the segmented mouthparts of only three nephropid decapod species (Funch and Kristensen 1995, 1997). Although regarded as highly host specific, recent observations have shown that cycliophorans are also capable of establishing a symbiotic relationship with harpacticoid copepods (Neves et al. 2014). Currently, only two species of Cycliophora have been described; Symbion pandora, Funch and Kristensen, 1995 is found on the Norway lobster, Nephrops norvegicus, while Symbion americanus Obst, Funch and Kristensen, 2006 lives on the American lobster, Homarus americanus. Moreover, studies based on molecular data suggest the existence of cryptic species in S. americanus (Obst et al. 2005; Baker and Giribet 2007; Baker et al. 2007), and an undescribed cycliophoran species lives commensally on the European lobster, Homarus gammarus (Funch and Kristensen 1997).
The cycliophoran life cycle involves an asexual generation and a sexual generation, both of which are characterized by complex sequences of various life cycle stages (see Supp. Fig. 1; Funch and Kristensen 1999; Obst and Funch 2003, 2006). The so-called feeding stage (Fig. 1a) is the largest and most prominent of all of these life cycle stages. As its name implies, it is the only cycliophoran stage with a digestive tract; during feeding activity, this stage uses a ciliated buccal funnel to filter small food particles from the water (Funch et al. 2008). The buccal funnel and gut are recurrently regenerated and replaced by new structures generated inside the trunk (Funch and Kristensen 1995; Wanninger and Neves 2015). Importantly, the feeding stage is the only life cycle stage type occurring in both the asexual and sexual generations.
Symbion pandora, light micrographs. a Feeding stage in asexual generation (i.e. without Prometheus larvae attached), sitting on a seta (se) of the host’s mouthpart. The closed buccal funnel (bf) is facing upwards. b Feeding stage in sexual generation with attached Prometheus larva (apl). st stalk, tr trunk
The feeding stage in the asexual generation produces only a daughter stage type that participates in the asexual phase of the life cycle (Funch and Kristensen 1997). This so-called Pandora larva is produced by a budding process inside the asexual feeding stage. Subsequently, the Pandora larva leaves the maternal feeding stage, settles down on the host’s mouthparts and develops into a new feeding stage closing the asexual loop of the life cycle.
The feeding stage in the sexual generation produces two different daughter stage types that participate in the sexual phase of the life cycle. These are the female and the so-called Prometheus larva. The latter leaves the maternal feeding stage, subsequently settles down on the trunk of a feeding stage and generates one to three dwarf males inside its body (Obst and Funch 2003; Neves et al. 2010a; Neves and Reichert 2015) (Fig. 1b). The female is impregnated by the dwarf male, settles down on the host’s mouthparts and encysts (Neves et al. 2010b; Neves et al. 2012). A larval dispersal stage, the so-called chordoid larva, hatches from the cyst, settles on a new host individual and develops into a new feeding stage, consequently closing the sexual loop of the life cycle (Funch 1996).
The shift from asexual to sexual generations is of great importance in the life history of cycliophorans. This is because the decapod host is a moulting animal that recurrently sheds its cuticle and, in consequence, discards its resident cyclophoran population (Castro 1992). Thus, the asexual expansion of the cycliophoran population is limited by the length of the intermolt period of the host, and a shift to sexual reproduction and the resulting generation of dispersal types are essential during the moulting period. Currently, however, nothing is known about the mechanisms that mediate the shift from asexual to sexual generations in cycliophorans. Indeed, at the molecular level, virtually nothing is known about any aspect of the life cycle stages in the asexual generation or in the sexual generation. This complete lack of information on the molecular features that characterize and differentiate the two principle life cycle types in Cycliophora is a major stumbling block for understanding the biology of this remarkable yet enigmatic animal phylum.
As a first step towards a more comprehensive molecular analysis of Cycliophora, we carried out a comparative whole-transcriptome analysis of life cycle stages in asexual versus sexual generations. For this, we first generated a reference transcriptome for S. pandora based on RNA-Seq analysis of transcripts pooled from various known life cycle stages of this cycliophoran (see Fig. 2). This reference transcriptome comprises over 24,000 different consensus transcripts, for which we find 14,541 functionally annotated sequences. Subsequently, we compared transcript profiles of sexually immature feeding stages, i.e. in the asexual generation and without attached Prometheus larvae, with the transcript profiles of feeding stages including attached Prometheus larvae, i.e. in the sexual generation. This transcriptome profiling analysis reveals surprising differences in differential gene expression of asexual versus sexual life cycle stages comprising over 2500 annotated genes and, hence, more than 10% of the total expressed transcriptome.
Scheme of the methodology employed in this study. In a first approach, the reference transcriptome (workflow in grey) was assembled de novo from three distinct life cycle stages: the feeding stage alone (asexual generation; note that in young feeding stages, the buccal funnel is located inside in the trunk), the feeding stage with Prometheus larva(e) attached to its trunk (sexual generation) and the free-swimming chordoid larva. Secondly, in the differential gene expression analysis (workflow in black), only two different conditions were investigated: feeding stages with Prometheus larva(e) attached to its trunk and feeding stages alone. Finally, sequenced reads were mapped to the reference transcriptome
Material and methods
Collection and preservation of specimens
Specimens of the Norway lobster, N. norvegicus, were caught in the Gullmarsfjorden (Sweden) in July 2014 and kept for several days in running seawater without food supply. Lobster mouthparts were dissected from the host, and setae with feeding stages of S. pandora were removed from the mouthparts and kept overnight in filtered seawater to avoid contamination. The next day, feeding stages were detached from the host’s setae and transferred with an Irwin loop into a tube containing RNAlater (Ambion, New York, USA). In addition, free-swimming chordoid larvae were also collected from the aliquots of filtered seawater containing the mouthparts and stored in RNAlater. Asexual feeding stages (some of them were recently settled Pandora larvae in which the buccal funnel was still inside the trunk) and sexual feeding stages along with their attached Prometheus larvae as well as chordoid larva were separated into different subsamples, and these were processed further independently. In addition, a subsample comprising a mix of all of these life cycle stages was produced and also processed separately (a total of 11 subsamples in 11 tubes containing specimens immersed in RNAlater were prepared; for more details, see Table 1). All four types of subsamples were kept in tubes at −20 °C and transported to Switzerland where isolation and RNA-Seq library preparation were carried out (Microsynth AG, Switzerland). A simplified schematic description of the methodology followed during this study is provided in Fig. 2.
RNA extraction and cDNA library construction
The four different subsample types (asexual feeding stages, sexual feeding stages with attached Prometheus larvae, chordoid larvae and mix of these three) were processed separately for RNA extraction and sequencing. Subsamples were thawed and centrifuged at 16.000×g for 2 min, and the RNAlater (Ambion) solution was carefully removed. The specimens were then resuspended with 500 μl RLT Plus lysis buffer (Qiagen) containing 10 μl of β-mercaptoethanol (14.3 M) per millilitre. For disruption of their body cuticle, the specimens were further processed in a TissueLyser II (Qiagen) in vials containing 2.8-mm ceramic beads, 0.5-mm ceramic beads and 0.1-mm glass beads two times during 1 min, at 30 Hz. Subsequently, total RNA was isolated using the RNeasy Plus 96 Kit (Qiagen) according to the manufacturer’s recommendations. Concentration and purity of the total RNA extracted were assessed using RiboGreen measurement (Quant-iT™ RiboGreen® RNA Reagent and Kit, Invitrogen) and the Agilent 2100 Bioanalyzer system (Agilent Technologies), respectively.
De novo transcriptome assembly and functional annotation
In the absence of a sequenced genome for S. pandora, we assembled a de novo reference transcriptome for mapping and quantifying the expression of transcripts in this cycliophoran species. To maximize transcript coverage for this reference transcriptome, we pooled the RNA from three different life cycle stages, namely the asexual feeding stages, including recently settled Pandora larvae in which the buccal funnel was still inside the trunk, the sexual feeding stages with attached Prometheus larvae and the chordoid larvae. Note that RNA from free-swimming Pandora larvae, females and Prometheus larvae was not pooled (see Table 1).
A library for messenger RNA (mRNA) sequencing was prepared according to the “TruSeq RNA sample preparation v2” protocol from Illumina (Illumina Inc.) with minor modifications. Briefly, poly-A mRNA was isolated from total RNA using oligo-dT attached magnetic beads, fragmented with ultrasound (two pulses of 30 s at 4 °C) and then used to construct a complementary DNA (cDNA) library. The cDNA was synthesized using Superscript II Reverse Transcriptase as indicated in the Illumina protocol. First-strand cDNA synthesis was primed with a N6 randomized primer, and Illumina TruSeq sequencing adapters were ligated to the 5′ and 3′ ends of the cDNA. The cDNA was then amplified with PCR (25 cycles) using a proofreading enzyme, and subsequently, the cDNA library was sequenced on a MiSeq using a v3 2 × 300-cycle kit. The produced paired-end reads which passed Illumina’s chastity filter were subject to demultiplexing and trimming of Illumina adaptor residuals (no further refinement or selection). The quality of the reads was checked with the software FastQC (version 0.10.1; see http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
The sequenced reads were de novo assembled into contigs using Trinity version 2.0.6 (Grabherr et al. 2011). The parameters used were as follows: paired-end library input, in silico read trimming (with default parameters) and in silico read normalization (with default parameters), and the minimum length of a contig to be reported was set to 300 bp. Subsequently, the raw assembly was subject to two different methodological approaches in order to reduce redundancy. In the first method—a clustering approach—we used Cluster_fast from the USEARCH software suite (version v8.1.1861_i86 Linux64) with sequence identity set to 90% and word length set to 8. In an alternative method—an assembly approach—we used Cap3 (version date: 2 October 2015) with standard parameters to further assemble the contigs produced by Trinity. USEARCH software suite produced the fewest resulting transcripts and was selected as the representation of the transcriptome for further use.
The software TransDecoder (version 2.0.1) was used to predict coding regions in the transcript sequences. Long open reading frames (TransDecoder.LongOrfs) were extracted, and their homology to known proteins was determined. Two different methods were used. The first was a protein search in a protein database (blastp with E value <1 × 10−5; BLAST 2.2.31+ against Swiss-Prot database, (http://www.ncbi.nlm.nih.gov/books/NBK6234)), while the second was a search of protein domains in a protein profile database (hmmscan with default parameters (HMMER 3.1b2) against Pfam-A database). Both databases were downloaded March 2016 (see Supp. Table 1). The long open reading frames were then submitted to TransDecoder.Predict, which predicts the final coding regions of the assembled transcripts by also considering the supplement information gathered from Swiss-Prot and Pfam-A. The transcripts were then annotated using the information gained from Swiss-Prot and Pfam-A. The ID-mapping files provided by UniProt were used to link gene ontologies (GO) (see in the following) and KEGG IDs to the matched Swiss-Prot protein IDs as well as to the transcripts themselves. In addition, a general feature format (gff) file was created to be used as an interface between the de novo transcriptome and its predicted coding regions and the differential gene expression analysis.
Differential expression analysis
A differential expression analysis was carried out to determine the differentially expressed genes in asexual feeding stages as compared to sexual feeding stages with attached larvae. For this, the corresponding RNA samples—three replicates for each of the two conditions (see Table 1)—were poly-A selected and then used to construct TruSeq cDNA libraries. The libraries were sequenced on a NextSeq 500 (Illumina Inc., San Diego, CA, USA) using single-ended sequencing and 75 cycles. The past filter single-end reads produced (i.e. the reads that pass the internal Illumina system filter) were subject to demultiplexing and trimming of Illumina adaptor residuals (no further refinement or selection; quality of the reads was checked with the software FastQC as described previously) and subsequently mapped (with an average mapping rate of 92.96% across all six samples) to the final version of the transcriptome (described previously) using the software Bowtie 2 (version 2.2.6) with very sensitive “local” mapping presettings (Langmead and Salzberg 2012). Then, htseq-count from the HTSeq framework (version 0.6.1p1) was used in conjunction with the gff file and the following parameters: reverse (for TruSeq stranded libraries), minimum mapping quality of 10 and “union” as the overlap resolution mode (Anders et al. 2015). Htseq-count produces a table in which each annotated feature (e.g. gene) as well as the corresponding number of mapped reads is listed. Finally, the produced raw count files and the software DESeq2 (version 1.8.1) were used to estimate differential expression of transcripts between the two different reproductive stages by using three biological replicates for each of the populations of interest. DESeq2 was used with standard parameters (Love et al. 2014).
In order to robustly classify genes as upregulated or downregulated based on the fold changes of mapped reads to that gene between the two conditions, we used a conservative threshold of false discovery rate (FDR) <0.01. This method provides a way to filter out genes that may seem upregulated or downregulated as a result of performing multiple tests (type I errors). By enforcing a conservative FDR <0.01, we only have a 1% probability that the selected upregulated or downregulated genes were incorrectly chosen due to multiple comparisons being performed. Moreover, we use the terms upregulated and downregulated depending if the abundance of gene expression is relatively greater or lower in one condition than in the other, respectively.
Enrichment analysis
The association between a given protein-coding transcript/gene and the respective GO terms was derived from the BLAST hits against the Swiss-Prot database (see previously mentioned). To compute significant GO enrichments, we perform a Fisher’s exact test using as background the list of all confidently annotated genes from the reference transcriptome (i.e., 14,541 genes). Overrepresented ontologies were selected using a false discovery rate of 1% and are shown in Figs. 4, 5 and 6 and Supp. Figs. 2, 3 and 4.
Results
De novo transcriptome assembly and functional annotation
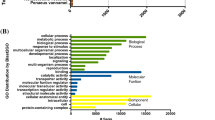
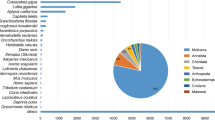
The final assembly of the transcriptome was performed with 18,165,497 trimmed paired-end reads after removal of ribosomal RNAs. We identified 24,583 consensus sequences comprising a total of 18,729 different genes and multiple isoforms. Overall, 14,541 contigs had highly confident BLAST matches on databases of known protein sequences (E values ≤1 × 10−5) (Supp. Fig. 2; Supp. Table 1; see “Material and methods” section). These sequences correspond to proteins in 394 different genera and 546 different species, of which Homo sapiens and Mus musculus are the two most represented (Supp. Fig. 3). Based on sequence homology, these contigs were categorized into groups belonging to three main classes of GO: biological function, molecular function and cellular components (see “Material and methods” section). In addition, sequence reads have been deposited to the NCBI sequence read archive (SRA) under the study accession number SRP068479.
The analysis of GO terms related to biological functions showed that the majority of the annotated genes are involved in processes such as transcription of DNA and its regulation, protein transport and metabolic process of small molecules (Supp. Fig. 4a). Further biological functions are those involved in cell division, translation, signal transduction and DNA repair. The top 10 enriched GO terms in molecular functions correspond to ATP binding, metal ion binding, zinc ion, DNA binding, binding, poly-A RNA binding, Ca2+ binding, RNA binding, GTP binding, nucleotide binding and protein kinase activity (Supp. Fig. 4b). Finally, the results for GO terms in cellular components show a high proportion of genes related to cytoplasm, nucleus and membrane, although the extracellular vesicular exosome, mitochondrion and the nuclear subdomain nucleolus are also highly represented (Supp. Fig. 4c).
Differential transcript expression in asexual versus sexual stages
The establishment of this first reference transcriptome for a cycliophoran is an essential initial step towards a comprehensive understanding of genome-wide gene expression in this still enigmatic phylum. As a next step, we used this reference transcriptome for mapping and quantifying differential gene expression in the asexual versus sexual generations through comparative transcript profiling. For simplicity, in the following, we refer to the asexual feeding stages including recently settled Pandora larvae in which the buccal funnel was still inside the trunk as “asexual stages” and to the sexual feeding stages including their attached Prometheus larvae as “sexual stages”.
Gene expression profiles for the sexual and asexual stages revealed that ca. 76.5% of the transcripts on the reference transcriptome (18,800 out of 24,583) are expressed during these two stages. We found 479 (∼1.9% of all genes) and 394 (∼1.6% of all genes) genes that are mutually exclusively expressed in the sexual and asexual stages, respectively. This suggests a remarkable level of gene regulation during these two reproductive stages.
Using a stringent cut-off (FDR <0.01), we found a total of 2660 contigs corresponding to genes that are expressed differentially in the asexual stages as compared to the sexual stages (see Supp. Table 2). Among these, the expression of 1236 genes (1030 of which were successfully annotated) is upregulated in the asexual stages as compared to the sexual stages. Conversely, the expression of 1424 genes (793 of which were successfully annotated) is upregulated in the sexual stages as compared to the asexual stages. An overview of the mRNA fold changes, between sexual and asexual stages, as a function of the average expression level of a contig is shown in Fig. 3.
Differential transcript expression analysis. Log ratio versus abundance plot for the feeding stage with Prometheus larva(e) attached (sexual generation) versus the feeding stage alone (asexual generation). For a false discovery rate <0.01, 1424 genes were found to be upregulated in the feeding stages with Prometheus larva(e) attached (red dots), while in the feeding stages alone 1236 genes are upregulated (blue dots) (Colour figure online)
In the sexual stages, the biological GO categories corresponding to the differentially expressed transcripts that are enriched compared to asexual stages are predominantly related to signal transduction and the G protein-coupled receptor signalling pathways (Fig. 4a). Other enriched biological categories include innervation and nervous system communication, as well as cation transmembrane transport, including the release of Ca++ into the cytosol. In terms of molecular functions, the enriched GO categories correspond to Ca++ binding, metalloendopeptidase activity, voltage-gated potassium channel and calcium channel activity (Fig. 5a). The predominant enriched categories for cellular components comprise transcript products related to the plasma membrane and cell junction (Fig. 6a). Other enriched terms for cellular components are the post-synaptic and presynaptic membrane, Z disc (i.e. the protein band marking the boundaries between adjacent sarcomeres in striated muscle fibres) and the voltage-gated calcium channel complex.
In the asexual stages, the biological GO categories corresponding to the differentially expressed transcripts that are enriched compared to sexual stages are predominantly related to protein folding; formation of translation preinitiation complex; and RNA processing, translation, splicing and maturation, which suggests a high degree of regulation at the transcriptional and post-transcriptional levels (Fig. 4b). An additional enriched biological process identified is cilium assembly. The enriched molecular GO categories are mainly poly-A mRNA binding and small nucleolar RNA binding as well as translation initiation factor activity, which is in line with the high degree of regulation at the transcriptional and post-transcriptional levels observed in the asexual stage (Fig. 5b). The enriched GO categories for cellular components comprise transcript products related to the nucleolus and the mitochondrial small ribosomal subunit as well as to the eukaryotic translation initiation factor 3 complex (Fig. 6b). Other enriched terms for cellular components are eukaryotic 48S and 43S preinitiation complexes, proteasome complex, small-subunit processome, chaperonin-containing T-complex and catalytic step 2 spliceosome.
Taken together, the results of this transcriptome profiling analysis reveal surprising differences in differential gene expression of sexual versus asexual stages of the cycliophoran. A total of over 2500 genes and, hence, more than 10% of the total transcriptome are differentially expressed in the two types of life cycle stages.
Discussion
In this report, we present a whole-transcriptome analysis of the cycliophoran S. pandora. This analysis comprises the generation of a reference transcriptome as well as the comparative transcriptome profiling of sexual versus asexual stages. In the following, we discuss the significance and implications of our analysis for further work aimed at achieving a more in-depth understanding of the biology of this enigmatic group of bilaterians.
The reference transcriptome indicates that 24,583 consensus sequences comprising 18,729 different genes and multiple isoforms are expressed in the principle life cycle stages of Symbion (excluding free-swimming Pandora larvae, females and Prometheus larvae). Among these, 14,541 could be functionally annotated based on known protein sequences in other species including humans. Given the fact that virtually no information was previously available on any molecular aspect of Cycliophora, this extensive information on expressed genes in Symbion, and the corresponding homologues in other more well-studied animals, is likely to be a useful starting point for subsequent investigations of the molecular and cellular features of this remarkable yet still largely uncharted phylum.
As a first step in this direction, we have utilized this reference transcriptome for Symbion to characterize differential gene expression in asexual versus sexual feeding stages by means of comparative transcript profiling. Interestingly, over two thirds of the whole transcriptome is expressed in the two stages taken together. Among these, 2660 are differentially expressed in either one stage or the other; this corresponds to more than 10% of the reference transcriptome. Thus, while the majority of the expressed transcripts are shared in the asexual (i.e. without Prometheus larvae attached) and the sexual stages (i.e. with Prometheus larvae attached), a significant fraction of the transcripts is differentially expressed in the two different life cycle stages.
Among the biological GO categories corresponding to the differentially expressed transcripts that are enriched in the sexual feeding stage (i.e. with Prometheus larvae attached) compared to asexual stage are signalling, innervation and nervous system communication. In the sexual stages, both the female and the dwarf males generated inside the attached Prometheus larvae have relatively (in relation to body size) conspicuous nervous systems thought to be important for free-swimming behaviour and the mating process (Neves et al. 2012; Neves and Reichert 2015). However, currently nothing is known about the molecular mechanisms involved in the establishment and function of these nervous systems. Hence, it will now be interesting to determine if the corresponding differentially expressed transcripts identified in our study encode proteins which are expressed in the female and dwarf male nervous systems and, if so, which nervous system-specific functions they fulfil.
Among the biological GO categories corresponding to the differentially expressed transcripts that are enriched in the asexual stage versus the sexual stage are RNA processing, translation and protein folding. A key feature in the biology of cycliophorans is the rapid asexual reproduction and subsequent organismal growth that is essential for the fast growth of the population that inhabits a decapod host (Funch and Kristensen 1997). This fast rapid asexual reproduction is required since overall reproduction in the population is limited by the length of the transient intermolt period of the host (Castro 1992). Given that intense RNA processing, translation and protein folding are likely to be required to generate the protein levels needed for rapid reproduction and growth, future studies on the role of the specific differentially expressed transcripts identified in this investigation may lead to important insight into the molecular mechanisms involved in asexual reproduction/growth of Symbion.
The striking differences between asexual and sexual stages are also evident in the categories molecular function and cellular components of GO terms, which corroborate the results for the category biological processes. For example, the enriched terms for molecular functions found in the sexual stages reflect enzymatic activity as well as ion transmembrane transport. By contrast, in the asexual stages all enriched terms composing the list of molecular functions are related to the processing of mRNA as well as regulation of the translation process.
Conclusion
This study presents the first large transcriptomic RNA-Seq expression dataset for Cycliophora. Our results provide important insights into the expression of transcripts in the feeding stage with Prometheus larvae attached, which participates in the sexual part of the cycliophoran life cycle. Our findings inform us of the variety of biological processes and molecular functions occurring in this sexual stage, such as the establishment and function of the nervous system, formation of the body musculature and cell-cell communication. In addition, we performed a differential gene expression analysis between this stage and the asexual feeding stage, which does not have Prometheus larvae attached to its trunk. Our results show that, in contrast to sexual stages, asexual stages undergo a process of intense transcriptional regulation. In the future, a deeper understanding of the gene expression profiles in the sexual and asexual parts of the life cycle will require more experiments in order to include all the other life cycle stages not specifically represented in the analysis presented here, i.e. the female, the dwarf male, the chordoid larva, the Prometheus larva and the Pandora larva.
References
Anders, S., Pyl, P. T., & Huber, W. (2015). HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics, 31(2), 166–169.
Baker, J. M., & Giribet, G. (2007). A molecular phylogenetic approach to the phylum Cycliophora provides further evidence for cryptic speciation in Symbion americanus. Zoologica Scripta, 36, 353–359.
Baker, J. M., Funch, P., & Giribet, G. (2007). Cryptic speciation in the recently discovered American cycliophoran Symbion americanus; genetic structure and population expansion. Marine Biology, 151, 2183–2193.
Castro, M. (1992). A methodology for obtaining information on the age structure and growth rates of the Norway lobster, Nephrops norvegicus (L.) (Decapoda, Nephropoidea). Crustaceana, 63, 29–43.
Funch, P. (1996). The chordoid larva of Symbion pandora (Cycliophora) is a modified trochophore. Journal of Morphology, 230, 231–263.
Funch, P., & Kristensen, R. M. (1995). Cycliophora is a new phylum with affinities to Entoprocta and Ectoprocta. Nature, 378, 711–714.
Funch, P., & Kristensen, R. M. (1997). Cycliophora. In F. W. Harrison & R. M. Woollacott (Eds.), Microscopic anatomy of invertebrates, lophophorates, entoprocta and cycliophora (Vol. 13, pp. 409–474). New York: Wiley-Liss.
Funch, P., & Kristensen, R. M. (1999). Cycliophora. In E. Knobil & J. D. Neill (Eds.), Encyclopaedia of reproduction (Vol. 1, pp. 800–808). New York: Academic.
Funch, P., Thor, P., & Obst, M. (2008). Symbiotic relations and feeding biology of Symbion pandora (Cycliophora) and Triticella flava (Bryozoa). Vie et Milieu, 58, 185–188.
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nature Biotechnology, 29, 644–652.
Langmead, B., & Salzberg, S. (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods, 9, 357–359.
Love, M. I., Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12), 550.
Neves, R. C., & Reichert, H. (2015). Microanatomy and development of the dwarf male of Symbion pandora (phylum Cycliophora): new insights from ultrastructural investigation based on serial section electron microscopy. PloS One, 10, e0122364.
Neves, R. C., Cunha, M. R., Kristensen, R. M., & Wanninger, A. (2010a). Comparative myoanatomy of cycliophoran life cycle stages. Journal of Morphology, 271, 596–611.
Neves, R. C., Cunha, M. R., Funch, P., Wanninger, A., & Kristensen, R. M. (2010b). External morphology of the cycliophoran dwarf male: a comparative study of Symbion pandora and S. americanus. Helgoland Marine Research, 64, 257–262.
Neves, R. C., Kristensen, R. M., & Funch, P. (2012). Ultrastructure and morphology of the cycliophoran female. Journal of Morphology, 273, 850–869.
Neves, R. C., Bailly, X., & Reichert, H. (2014). Are copepods secondary hosts of Cycliophora? Organisms Diversity & Evolution, 14, 363–367.
Obst, M., & Funch, P. (2003). Dwarf male of Symbion pandora (Cycliophora). Journal of Morphology, 255, 261–278.
Obst, M., & Funch, P. (2006). The microhabitat of Symbion pandora (Cycliophora) on the mouthparts of its host Nephrops norvegicus (Decapoda: Nephropidae). Marine Biology, 148, 945–951.
Obst, M., Funch P., & Giribet G. (2005). Hidden diversity and host specificity in cycliophorans: a phylogeographic analysis along the North Atlantic and Mediterranean Sea. Molecular Ecology, 14, 4427–4440.
Obst, M., Funch, P., & Kristensen, R. M. (2006). A new species of Cycliophora from the mouthparts of the American lobster, Homarus americanus (Nephropidae, Decapoda). Organisms Diversity & Evolution, 6, 83–97.
Wanninger, A., & Neves, R. C. (2015) Cycliophora. In A. Wanninger (Ed.), Evolutionary developmental biology of invertebrates (Vol. 2: Lophotrochozoa (Spiralia), pp. 79–87). Wien: Springer-Verlag.
Acknowledgements
We are grateful to the generous support by the staff from The Sven Lovén Centre for Marine Sciences at the University of Gothenburg. The authors also thank Birgitte Rubak and Stine Elle (both Copenhagen) for providing the line drawings. We thank Kevin Kocot (University of Alabama, USA) for his helpful comments on a preliminary version of this article.
Author’s contributions
RCN and HR conceived of the study and participated in its design and coordination and drafted the manuscript. RCN is the main coordinator of the project, prepared the material for sequencing, coordinated the molecular genetic studies and analysed the data. SS analysed the data. JCG performed statistical analysis and helped to draft the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Supplementary Figure 2.
(A) Number of annotated contigs as a function of the stringency of the E-value threshold applied. Note that the lower the E-value, the more similar is the match between the contig and the annotated gene. (B) BLAST annotation of transcriptome assembly for E-values ≤1 × 10−5. Note that 14,541 contigs have high similarity with known proteins. (TIFF 166 kb)
Supplementary Figure 3.
Annotation of transcriptome assembly. Bar charts represent the level of similarity to proteins that belong to other (A) genera and (B) species. (TIFF 367 kb)
Supplementary Figure 4.
Annotation of transcriptome assembly. Bar charts represent the categorization of the contigs (E-values ≤1 × 10−5) into functional groups belonging to three main GO ontologies: (A) biological processes, (B) molecular functions and (C) cellular components. (TIFF 588 kb)
Supplementary Table 1.
List of all BLAST hits for the long open reading frames (ORF) predicted for the contigs present in the reference transcriptome. The table shows the following information for each of the long ORFs: Swiss-Prot accession and description of the protein hit, BLAST hit statistics such as percent identity, alignment length, mismatches, gap opens, E-value and bit score. The next columns for each ORF list associated IDs from different publicly available databases (EMBL, Ensembl and RefSeq). The last two columns of the table show the KEGG and GO IDs which could be associated with the Swissprot protein hit by the queried ORF. All IDs are formatted as hyperlinks into the respective databases for easy reference. (XLSX 4084 kb)
Supplementary Table 2.
List of all differentially expressed contigs/transcripts during the life cycle of Symbion pandora. We find 2660 genes that are significantly differentially expressed (P-adjusted <0.01) in the feeding stages from the asexual generation as compared to the sexual feeding stages. Among these, 1424 genes are upregulated in the sexual stages as compared to the asexual stages (upper part of the table). Conversely, 1236 genes are upregulated in the asexual stages as compared to the sexual stages (lower part of the table). Gene ontology (GO) class, description, and official GO identifier are listed for all the genes for which we were able to identify homologues in other species. Note that each contig appears as many times as it is assigned to a GO term. (XLSX 5031 kb)
Rights and permissions
About this article
Cite this article
Neves, R.C., Guimaraes, J.C., Strempel, S. et al. Transcriptome profiling of Symbion pandora (phylum Cycliophora): insights from a differential gene expression analysis. Org Divers Evol 17, 111–119 (2017). https://doi.org/10.1007/s13127-016-0315-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-016-0315-1