Abstract
Most extant New World marsupials belong in the Didelphidae, which comprises ca. 110 currently recognized species of opossums. Didelphids are small mammals with their mean body mass, at species level, ranging from ca. 7 g to 2.2 kg. The largest species belong in a single clade, while substantial variation remains scattered across the remaining groups. We seek out to explore the details of this mass variation in an evolutionary framework. To this end, we first reconstructed the phylogeny of didelphids based on an extensive, although fragmentary sample of sequences from ten genes. We recovered a fully resolved, highly robust phylogeny that tested and confirmed most previously reported groupings, providing a simultaneous depiction of phylogenetic relationships for 81 % of currently recognized species and all relevant supra-specific clades. As much as 69 % of total body mass variation in didelphids was explained by this phylogenetic hypothesis. Mapped on it, mass variation evolved as much as 6.8 kg of total changes, starting from a reconstructed ancestral body mass range of 22–33 g. No single, family-wide pattern was evident; in fact, the dominant pattern for mass variation was that of increases in body mass along a few successive branches, or phyletic giantism, followed by apomorphic nanism, i.e., decreases localized in single terminal branches. Phyletic trends indicated the persistence of gradual, directional changes along considerable spans of geological time and show that substantial variation of interest resides in this and perhaps most groups of small mammals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Marsupialia (Metatheria) conforms a particular group of mammals, not only with regard to anatomical and physiological traits (e.g., osteology, dentition, reproductive system; Dickman 2005), but also given their distinct biogeographic and evolutionary history. With seven orders and some 330 living species, marsupials comprise ca. 7 % of the extant mammalian diversity (Dickman 2005). These mammals have lived in all continents, but since the Middle Miocene, they became restricted to the Australasian region east to the Wallace line (Groves 2005) and the New World (Gardner 2005). In the Americas, marsupials are represented by three extant orders (Microbiotheria, Paucituberculata, and Didelphimorphia), each one including a single extant family (Microbiotheriidae, Caenolestidae, and Didelphidae, respectively). Microbiotheriidae groups several extinct taxa and the single extant species Dromiciops gliroides; likewise, Caenolestidae includes many extinct but just six extant species classified in three genera. Presently, Didelphidae comprises the majority of the extant diversity of New World marsupials (Gardner 2008), with ca. 110 currently recognized species in 18 genera (after synonymy of Micoureus with Marmosa; Voss et al. 2004; Gardner 2005; Voss et al. 2005; Solari 2007; Teta et al. 2009; Voss and Jansa 2009; Gutiérrez et al. 2010; Voss et al. 2012; Paglia et al. 2012; Solari et al. 2012; Caramaschi et al. 2011; Pavan et al. 2012; Voss et al. 2013; Giarla and Jansa 2014; Martínez-Lanfranco et al. 2014; Palma et al. 2014; Pavan et al. 2014; Voss et al. 2014; Vilela et al. 2015; Astúa 2015).
The evolutionary history of New World marsupials is richly documented in the fossil record (see McKenna and Bell 1997). This history has been shaped by extinction events (e.g., Dickman and Vieira 2005) and major changes in key characters, remarkably body mass and conspicuous functional features associated to diet and locomotion (e.g., Argot 2003, 2004a, b). However, didelphids in particular exhibit a comparatively poor fossil record (see McKenna and Bell 1997) and have experienced relatively modest changes along their own evolutionary history. As a consequence, didelphids are often perceived as remarkably conservative in body structure; e.g., the didelphid molar model is only a slight modification of an ancient one already found in Cretaceous metatherians (Fox 1987; Chemisquy et al. 2015). For this reason, didelphids, and particularly the type genus Didelphis, have been traditionally considered a useful functional model of generalized metatherians and hence of primitive mammals (e.g., Crompton and Hiiemae 1970). However, this conservatism is only apparent (e.g., Astúa 2009), as didelphids exhibit interesting systematic variation in relevant morphological characters that contribute synapomorphies to both the group and most inner clades (e.g., Voss and Jansa 2009; Flores 2009). Here, we focus on macroevolutionary variation in body mass in this natural group. Size is the single most important factor affecting the biology of mammals (McNab 2007 and Citations therein), and the particular way in which body mass varies in different groups, or in mammals as a group, has been a constant source of research in the Class (e.g., Meiri et al. 2008; Cooper and Purvis 2010; Venditti et al. 2011; Giannini et al. 2012; Slater 2013). Gould and MacFadden (2004) describe evolutionary body mass variation in terms of apomorphic versus phyletic change (i.e., changes traceable to a single branch versus changes accumulated along several successive branches of a phylogeny), as well as increases versus decreases. These compose, in combination, the possible outcomes of apomorphic or phyletic giantism or nanism, in addition to stasis. Gould and MacFadden (2004) studied macroevolutionary size variation in large animals, specifically equids among mammals. While the largest specimens of Didelphis virginiana can exceptionally reach 7 kg (although only in urban environments; McManus 1974; Cerqueira and Tribe 2007) and some extinct forms were seemingly larger (e.g., Thylophorops; Simpson 1972, Goin et al. 2009; Hyperdidelphis, Goin and Pardiñas 1996), the great majority of extant didelphids are rather small mammals (Birney and Monjeau 2003). To our knowledge, small mammals seldom have been examined in the framework proposed by Gould and McFadden (2004), perhaps only bats (Giannini et al. 2012).
In preliminary grounds, several aspects of body mass variation in didelphids are noteworthy and invite further investigation. First, the observed interspecific variation in the average of body mass in extant didelphid species ranges from 7.6 g in Gracilinanus emiliae to 2195.5 g in Didelphis virginiana (data from Smith et al. 2003) or an impressive 289-fold difference between the average of the smallest and the largest species. Second, cranial morphometric studies have revealed that didelphids are among the most morphologically highly integrated, least modular groups of mammals (Shirai and Marroig 2010); here, body mass is important because that magnitude of integration arises when most morphological variation is highly correlated with size variation (Porto et al. 2009; see also Astúa 2009). Third, a single group of didelphids, Didelphini sensu Steiner et al. (2005) contains Metachirus and all the largest species in four genera (the so-called 2n = 22 clade, composed of Chironectes, Lutreolina, Philander, and Didelphis; Flores 2009; Voss and Jansa 2009). This strongly suggests a key role of phylogenetic legacy in the evolution of body mass in the group. Naturally, discovering the specific way in which body mass evolved in didelphids demands investigation within an explicit phylogenetic framework.
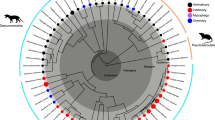
Recent studies made significant advances in the phylogeny of the group. First, although the monophyly of this group was never seriously challenged within a cladistic context, all modern phylogenies successively accrued strong support for a didelphid clade from both molecular (e.g., Voss and Jansa 2003; Jansa and Voss 2005; Gruber et al. 2007; Voss and Jansa 2009) and morphological data (e.g., Horovitz and Sánchez‐Villagra 2003; Voss and Jansa 2003; Jansa and Voss 2005; Flores 2009). Second, and contrary to earlier studies, the new phylogenies revealed that internal relationships among didelphids are remarkably congruent across different sources of evidence (morphology, karyotypes, and gene sequences; Flores 2009; Voss and Jansa 2009). These results, defining the branching pattern of extant didelphids, have been consistently reflected in the current systematics of the family (Fig. 1; Voss and Jansa 2009; de la Sancha et al. 2012; Palma et al. 2014; Voss et al. 2014).
Here, we seek to investigate in detail the evolution of body mass in this group with relatively modest, albeit interesting variation. To this end, we contribute a comprehensive phylogeny that includes 81 % of currently recognized species. This phylogeny was built upon molecular data including sequences from ten genes from recent studies, so constructed in order to increase the taxonomic density and, thus, the reliability of both the phylogenetic framework used and the estimation of evolutionary change in body mass. We show (1) that the current phylogenetic hypothesis of didelphid relationships (Fig. 1) is extraordinarily robust to the test of significant (near 2-fold) increase in both character and taxonomic sampling and (2) that while research has been centered around trends that comprise relatively large changes in wide body mass ranges (e.g., Cope’s rule or the island rule; Lomolino 2005), macroevolutionary variation in small mammals such as didelphids reveals highly interesting patterns with significant implications in the perceived evolution of body mass in mammals.
Materials and methods
Taxonomic sampling
We constructed a data matrix that includes 89 currently recognized species following Gardner (2005) and other authorities (Voss et al. 2004; Voss et al. 2005; Solari 2007; Teta et al. 2009; Voss and Jansa 2009; Gutiérrez et al. 2010; Voss et al. 2012; Paglia et al. 2012; Solari et al. 2012; Caramaschi et al. 2011; Pavan et al. 2012; Voss et al. 2013; Giarla and Jansa 2014; Martínez-Lanfranco et al. 2014; Palma et al. 2014; Pavan et al. 2014; Voss et al. 2014; Vilela et al. 2015). This matrix includes all the species represented in the GenBank database as of November 2014 (see taxonomic list and accession numbers in Table SI.1, Supplementary Information) for the gene sequences selected for this study (see below). All didelphid genera and species groups are represented in this sample (Table 1), except for the monotypic Chacodelphys for which no sequences are available. Five taxa were added as outgroups: the caenolestids Rhyncholestes raphanurus, Lestoros inca, and Caenolestes fuliginosus (Paucituberculata: Caenolestidae) and Dromiciops gliroides (Microbiotheria: Microbiotheriidae), which together with didelphids represent the extant higher-level diversity of New World marsupials, plus one basal representative of the large native Australian marsupial clade, Dasyurus geoffroii (Dasyuromorphia: Dasyuridae). Thereby, the analysis comprised a total of 94 terminals.
Characters and matrix compilation
We selected sequences from four mitochondrial and six nuclear genes. The mitochondrial genes included the protein-coding cytochrome b (Cyt-b) and the cytochrome c oxidase subunit 1 (COI) and the ribosomal subunits 12S and 16S. The nuclear genes included the interphotoreceptor retinoid binding protein (IRBP), the breast cancer susceptibility protein 1 (BRCA1, exons 10 and 11), the dentin matrix protein 1 (DMP1), the recombination activating protein 1 (RAG1), the von Willebrand factor (vWF, exon 28), and the transthyretin intron 1 (TTR). These sequences are of proven systematic utility in didelphids, and they have been generated and used, in different combinations, by several previous authors (e.g., Palma and Spotorno 1999; Steiner et al. 2005; Jansa et al. 2006; Gruber et al. 2007; Voss and Jansa 2009). Sequences of these genes were available for at least 23 of the study species (Tables 2 and SI.1 for accession numbers).
Sequences were compiled with the program GenBank to TNT (GB2TNT; Goloboff and Catalano 2012), a pipeline for creating large molecular matrices that selects sequences included in a GenBank file (by using filters defined by the user), creates a Fasta file, produces the alignment (by calling an external program defined by the user), and generates the data matrix in TNT (Goloboff et al. 2008b) format. From this format, we migrated manually to PHYLIP format used by RAxML (Stamatakis et al. 2008). The completeness and accuracy of the dataset so generated were checked against the original GenBank depository to avoid missing sequences, duplicates, and other mistakes, which include, for instance, syntactic ambiguities in the species entries. Sequences were aligned using the program MAFFT (version 7, Katoh and Standley 2013) with default settings. In all analyses (see below), indels were treated as missing values, and the australidelphian Dasyurus geoffroii was designated to root the trees.
Phylogenetic analyses
We conducted two series of phylogenetic analyses with different optimality criteria. First, a maximum likelihood (ML) analysis was performed on the total dataset using the server version of RAxML 7.2.8 (Stamatakis et al. 2008). This program approximates the finding of ML trees using a sample of 100 bootstrap replicates. The substitution model applied was GTR + GAMMA + I.
Second, we performed parsimony (maximum parsimony (MP)) analyses for tree search and branch support using the program TNT 1.1 (Goloboff et al. 2008b). We executed heuristic searches based on 200 random addition sequences (RAS) each followed by Tree Bisection Reconnection (TBR) branch swapping, saving up to 15 trees per replicate. The search was refined with an additional TBR round applied to all trees kept in memory. Strict consensus tree was calculated from the set of most parsimonious trees obtained. All characters were treated as unordered and using both equal weighting (default option) and implied weighting (Goloboff 1993, 2014), as implemented in the current version of TNT. Implied weighting is a traditional technique that parsimoniously weights against homoplasy during tree search (Goloboff 1993), and it has been shown to outperform equally weighted parsimony (Goloboff et al. 2008a). This technique was applied with default settings (e.g., concavity constant k = 3; Goloboff et al. 2008a). No constraints were enforced during searches. Branch stability and support were estimated with a symmetric resampling (jackknife) analysis based on 1000 replicates and sequential Bremer absolute values. We followed Giannini and Bertelli (2004) to calculate Bremer values from ten successive samplings of up to 2000, one-step-longer suboptimal trees. This search strategy was applied to each gene and to concatenated datasets, specifically nuclear, mitochondrial, total, and a nine-gene dataset excluding RAG1 sequences ran to test the impact of the homoplasy problem caused by convergence in CG content on the third position of this gene (Gruber et al. 2007).
Evolution of body mass
We obtained body mass data, in grams, from specific sources (Mondolfi and Pérez-Hernández 1984; Catzeflis et al. 1997; Smith et al. 2003; Voss et al. 2004; Rossi 2005; Voss et al. 2005; Cáceres et al. 2007; Flores et al. 2008; Forero-Medina and Vieira 2009; Leiner and Silva 2009; Perez-Carusi et al. 2009; Voss and Jansa 2009; Lim et al. 2010; Rossi et al. 2010; Gutiérrez et al. 2011; Paglia et al. 2012; Pavan et al. 2012; Voss et al. 2012; Voss et al. 2013; Barrera-Niño and Sánchez 2014; Martínez-Lanfranco et al. 2014; and Flores, Teta, Solari and Voss, personal communication; Table SI.2). In addition, we gathered data from 11 didelphid species housed in the Colección Mamíferos Lillo (CML; Tucumán, Argentina; Table SI.2 and see Appendix 1 for specimen vouchers, Supplementary Information).
We approached the macroevolutionary analysis of body mass in didelphids using two contrasting methodologies. First, we aimed at assessing the global phylogenetic effect on body mass in the group, by means of estimating the fraction of total mass variance explained by significant tree partitions. This method, Canonical Phylogenetic Ordination (hereafter CPO; Giannini 2003), considers the comparative data as dependent variable(s), here the vector of body mass data for each didelphid species, in a linear model; tree partitions represent the independent part of the model and are equivalent to clades of a rooted tree. Tree partitions use a presence-absence (binary) code to represent membership of species, and each constitutes an explanatory variable that is tested using unrestricted Monte Carlo permutations (here, 4999 permutations were used). These tests of individual tree partitions are followed by a forward stepwise selection procedure that seeks to produce a maximally explanatory, but non-redundant model that is a linear combination of selected partitions (clades). These analyses were executed using the program CANOCO version 4.0 (ter Braak and Šmilauer 1998).
Second, we chose to apply a local, node-by-node descriptive approach that allowed an appropriate evaluation of evolutionary changes in the framework proposed by Gould and McFadden (2004). The body mass information was arranged in TNT data matrix format and mapped as a continuous character (see Goloboff et al. 2006) with a single value (the species average) per terminal, as implemented in TNT. Following Giannini et al. (2012), we chose to interpret nodal location, sign (increase or decrease), and magnitude of change, with the latter being the net change or the amount of body mass increase or decrease common to all reconstructions for a given branch. We looked for macroevolutionary patterns in body mass variation, in terms of phyletic or apomorphic changes, giantism or nanism, and their corresponding combinations (see Gould and McFadden 2004). To assess the persistence of the phyletic pattern, we defined a simple metric, the order of phyletic change, as the number of consecutive branches composing the phyletic pattern, so the higher the order, the more persistent the pattern; for instance, increases of body mass along three consecutive branches represented a case of third-order phyletic giantism.
Results
Phylogeny
Our molecular phylogeny includes the most comprehensive taxonomic sampling to date, comprising 81 % of extant didelphid diversity at the species level (i.e., 89 currently recognized species; Table 1). The alignment produced 14,489 nucleotide characters distributed in 4451 and 10,038 bp of mitochondrial and nuclear sequences, respectively (Table 2 and see Table SI.1 for accession numbers). Clades recovered from the various analyses in this study (see below) are summarized in Table 3, and parenthetical trees are shown in Appendix 2 (Supplementary Information).
Individual genes
The nuclear genes BRCA1, IRBP, DMP1, and vWF and the mitochondrial gen Cyt-b recovered all (or almost all) the phylogenetic structure of the total dataset analysis (see below “Total molecular evidence” section). RAG1 and TTR recovered clades to approximately genus level, while 12S and 16S so did at a higher taxonomic level (i.e., family, subfamilies). Finally, COI exhibited a good performance chiefly within Marmosini (sensu Steiner et al. 2005).
Mitochondrial and nuclear datasets
Both the mitochondrial and nuclear analyses yielded highly resolved, supported trees largely in agreement with the total molecular evidence analysis (see below), but the latter recovered more monophyletic groups. Except for the position of Lestodelphys halli, which was nested within Thylamys, the global structure of the nuclear dataset result was almost identical to that of total molecular evidence phylogeny (see below), strongly suggesting that this data partition dominated the combined analysis.
Total molecular evidence
The maximum likelihood (ML) best tree topology with bootstrap values is shown in Fig. 2. Subfamilies Caluromyinae and Didelphinae, as well as Marmosini, Didelphini, and Thylamyini clades (sensu Steiner et al. 2005), were recovered as strongly supported monophyletic groups. Besides, Didelphini and Thylamyini were reconstructed as sister groups, and Hyladelphys kalinowskii was the basalmost taxon of Didelphinae.
Best tree resulting from total dataset (ten genes) maximum likelihood analysis. Bootstrap support values are indicated for each node and are also reflected on degree of branch thickness. Asterisks: Pavan et al. (2014) included Monodelphis theresa in Monodelphis scalops, Monodelphis sorex in Monodelphis dimidiata, and Monodelphis umbristriata in Monodelphis americana
Didelphini was strongly supported, with Metachirus nudicaudatus recovered as sister to the other genera subsequently branching in the order of Chironectes, Lutreolina, and Philander + Didelphis. Within Thylamyini, Marmosops was sister of a clade that included Gracilinanus + Cryptonanus and Lestodelphys + Thylamys. In turn, Thylamys consisted of five inner clades: (1) (Thylamys velutinus, Thylamys karimii), (2) (Thylamys venustus (Thylamys cinderella, Thylamys sponsorius)), (3) Thylamys macrura, (4) (Thylamys tatei (Thylamys elegans, Thylamys pallidior)), and (5) (Thylamys citellus (Thylamys pulchellus, Thylamys pusillus)). Bootstrap values in Thylamyini were very high (>80), with the exception of two small terminal clades. Finally, Marmosini consisted of two main clades, the specious genus Monodelphis and the clade formed by Tlacuatzin + Marmosa. The latter included five clades, all matching the subgenera recognized by Voss et al. (2014): (1) subgenus Eomarmosa: Marmosa rubra, (2) subgenus Exulomarmosa: (Marmosa simonsi (Marmosa robinsoni, Marmosa xerophila) (Marmosa mexicana (Marmosa zeledoni, Marmosa isthmica))), (3) subgenus Stegomarmosa: Marmosa lepida, (4) subgenus Marmosa: ((Marmosa macrotarsus, Marmosa waterhousei) (Marmosa tyleriana, Marmosa murina)), and (5) subgenus Micoureus: ((Marmosa constantiae, Marmosa regina) (Marmosa paraguayanus (Marmosa alstoni, Marmosa demerarae))). The majority of bootstrap values were high (>70) in these groups. Monodelphis consisted of five inner clades: (1) (Monodelphis scalops, Monodelphis theresa), (2) (Monodelphis sorex, Monodelphis dimidiata), (3) (Monodelphis gardneri (Monodelphis iheringi (Monodelphis umbristriata, Monodelphis americana))), (4) ((Monodelphis domestica (Monodelphis glirina, Monodelphis sanctaerosae)) (Monodelphis arlindoi (Monodelphis touan (Monodelphis brevicaudata, Monodelphis palliolata)))), and (5) (Monodelphis kunsi ((Monodelphis adusta, Monodelphis reigi) (Monodelphis peruviana (Monodelphis osgoodi, Monodelphis handleyi)))). The Monodelphis groups themselves (and the relationships within them) were strongly supported (bootstrap values >75), but this was not the case with their interrelationships (bootstrap values <50).
Parsimony analysis of the total dataset using implied weighting (IW) is shown in Fig. SI.1 (Supplementary Information). The resulting topology is the strict consensus of the three most parsimonious trees obtained, which shows almost identical relationships as compared with the ML tree, with just a few differences concentrated in small terminal clades. The single politomy in the consensus involved four species of Marmosops (Marmosops dorothea, Marmosops invictus, Marmosops impavidus, and Marmosops noctivagus). The parsimony total dataset analysis using equal weighting (EW) generated 216 most parsimonious trees. The strict consensus was quite similar to that of IW analysis, except for the greater number of politomies. This was replicated in the remainder of EW analysis. For that reason, we used only IW topologies for the comparison of phylogenetical results (Table 3).
Concatenated partition excluding RAG1
The global topology recovered with this analysis was very similar to that of the total analysis. The main differences consisted of the following: (1) the position of Tlacuatzin canescens as sister to Marmosa + Monodelphis, instead of the Marmosa clade; (2) the relative position among monophyletic groups within Monodelphis (although the groups remained the same) and the position of Monodelphis emiliae (in a more nested clade); and (3) the relative positions of monophyletic groups within Thylamys (although the groups remained the same) and position of Thylamys macrura (sister to the “venustus” group).
Evolution of body mass
We obtained average values of body mass from the literature and museum specimens that covered all terminal taxa (Table SI.2). The log-transformed distribution of mass in didelphids is shown in Fig. 3, with calculated mean = 179 g, median = 51 g, and mode = 17 g. This distribution was discontinuous due to species of Didelphini and Caluromyinae excluding Glironia, which appeared as a separate group in Fig. 3.
Phylogeny explained much of the total body mass variation, but the global, non-redundant effect of phylogeny on body mass was quite restricted. Specifically, ten clades were individually significant at alpha = 0.01 (defined to be intermediate between the true Bonferroni-corrected value and the conventional 0.05 significance level). However, when submitted to forward stepwise selection procedure, a single clade was retained in the model (F = 206.17, P = 0.0002). This clade was Didelphis, and the variation explained by this single-variable model was as high as 69 %. This result represents the gross-scale effect of phylogeny on the body mass variation in didelphids.
The fine-scale, detailed reconstruction of body mass evolution is shown in Fig. 4. Total amount of change, i.e., the reported length for the body mass character on the retrieved phylogeny (steps), was 6760 g, but total net change, i.e., the sum of changes common to all reconstructions, was 4921 g. The ancestral node of Didelphidae was assigned states 22–33 g. There was no net change along the backbone, so the first changes to occur were reconstructed within each of the main clades (i.e., Caluromyinae, Marmosini, Didelphini, Thylamyini). The largest absolute increase in any didelphid branch was located at the node of Didelphis virginiana (+971 g), whereas the largest increase in relative terms was reconstructed on the branch leading to Didelphini (967 % increase in body mass with respect to the reconstructed hypothetical ancestor). The largest absolute decrease was located at the Didelphis imperfecta branch (−430 g), while in relative terms, the largest decrease was reconstructed at the Marmosa lepida branch (−264 % with respect to the descendant).
A summary with frequency percentages of stasis, increases, and decreases is shown in Table 4. A prevalence of stasis was observed both at global level (62.7 %) as well as within particular groups. With regard to the frequency of net changes, although increases predominated over decreases globally, both types of change were relatively balanced within groups, with the exception of Caluromyinae where only increases were reconstructed. However, the magnitude of increases was twice as much (or more) as the magnitude of decreases, both at global level as in particular groups (Table 5).
With respect to the observed patterns of body mass evolution, no clear global pattern (e.g., Cope’s rule or the like) was detected. However, multiple cases of either phyletic and apomorphic changes were reconstructed in particular clades or branches, being the first type nearly twice as common as the second type (Table 6). Considering these specific patterns, the more frequent combinations were phyletic giantism and apomorphic nanism, both at global level and within principal clades (Table 6). The most notable cases of phyletic giantism were, as expected, concentrated in the Didelphis clade (Fig. 4), specifically Didelphis virginiana, Didelphis marsupialis, and Didelphis pernigra, representing a fourth-order case of phyletic giantism. In the other hand, the most notable cases of apomorphic nanism were located in Marmosini (Fig. 4), with Marmosa lepida (−37 g or 264 % decrease with respect to its descendant) and Monodelphis sanctaerosae (−43 g or 187 % decrease).
Discussion
In this research, we aimed at understanding body mass evolution in a group of small mammals of our interest, the didelphid marsupials. This is particularly important because didelphid marsupials are morphologically highly integrated taxa (see Porto et al. 2009), meaning that they are evolutionarily less flexible than most other mammals (Marroig et al. 2009) so that their evolutionary responses to selection are basically aligned with size variation (Marroig et al. 2009; Shirai and Marroig 2010). Macroevolutionary patterns, such as this one of body mass evolution, can only be discerned in an explicit phylogenetic framework (Gould and MacFadden 2004; Hone et al. 2005; Butler and Goswami 2008). Thus, we contributed a comprehensive, strong phylogeny based on unconstrained tree searches that included 89 currently recognized didelphid species. Previously, the most comprehensive phylogenetic hypothesis of the group was the marsupial supertree provided by Cardillo et al. (2004), containing 62 didelphid species; however, this supertree lacks resolution particularly for New World marsupials. Here, we carried out our phylogeny of didelphids from a supermatrix approach, with no constraints on recovered clades. This analysis contains at least twice as many species included in recent supermatrix phylogenies (e.g., Jansa et al. 2006; Gruber et al. 2007; Voss-Jansa 2009; Flores et al. 2009; Astúa 2009; May-Collado et al. 2015). We were able to provide a fully resolved, highly supported phylogenetic hypothesis for the Didelphidae. This result is somewhat unexpected given the level of character conflict apparent in earlier studies of didelphid systematics (e.g., Kirsch 1977; Kirsch and Archer 1982; Kirsch and Palma 1995; Reig et al. 1987); by contrast, our phylogeny and its systematic implications were generally consistent with all recent analyses, both at a suprageneric and infrageneric level. Moreover, the two distinct analyses performed (ML and parsimony) produced almost identical results. The tree reflected, for each major group, the resolution found in specific previous studies (see below). Although this may seem trivial given that sequences came from those studies, results need not to be congruent globally when data are gathered together from different partial studies attacking specific subgroups, but we obtained a remarkable result of high congruence when all taxa and data were analyzed together, as we discussed next. We recovered Hyladelphys as sister to all didelphids as in Jansa and Voss (2005), and the internal structure of the remaining clades, particularly Marmosa, and the recently proposed subdivision in five subgenera (see Gutiérrez et al. 2010; de la Sancha et al. 2012; Voss et al. 2014), Monodelphis (Caramaschi et al. 2011; Carvalho et al. 2011; Pavan et al. 2012, 2014), Philander (Chemisquy and Flores 2012), and Thylamys (Giarla et al. 2010; Giarla and Jansa 2014; Palma et al. 2014). Our tree differs slightly from others only in poorly supported clades (e.g., inside Monodelphis; see Vilela et al. 2015). This means that the data accrued to date by many independent researchers proved to be of high quality and congruence. It should be noted that the global topology was not significantly modified when RAG1 sequences were excluded from the supermatrix, thus contrasting with Gruber et al. (2007) whose results substantially improved with the exclusion of the RAG1 third position. This might be due to the fact that homoplasy by convergence in CG content on this position, which Gruber et al. (2007) claimed to cause the inconsistent topology problem in the group, would be compensated by the congruent signal of many other independent characters. In summary, we simultaneously tested, with more taxa and characters, the monophyly of all didelphid groups proposed to date, found strong support for most previously recognized groups in a single, robust phylogenetic hypothesis, and thus offered a strong confirmation to the current classification of the Didelphidae. This provides a firm basis for future taxonomic, evolutionary, and biogeographic studies in the group.
A first application of this remarkable phylogenetic result is precisely our analysis of body mass evolution. In spite of being small and therefore limited to the lower range of the whole mammalian variation, didelphids revealed considerable and highly interesting variation. This is both in terms of comparisons between observed extremes, with a ca. 289-fold difference between the average mass of smallest and largest species, and in evolutionary terms (see below). We detected one major tree partition with large, significant differences between the set of its respective members, specifically Didelphis, which explained as much as 69 % of total interspecific average mass variation in the group. We also reconstructed, by means of character mapping, many changes that allowed a detailed analysis of body mass evolution in the group. The ancestral interval, estimated at 22–33 g, is quite narrow, so the confidence on its reconstructed range increases. From this small root value, dated at a point estimate of 31.4 mya in the Oligocene (or range estimated in 23–38 mya, between late Eocene and Oligocene; Meredith et al. 2011), didelphids evolved as much as 6760 g of total mass changes. This scenario differs from a larger previous estimate of ancestral body mass based on morphometric data, corresponding to the mass of Marmosa (subgenus Micoureus) or Glironia (Astúa 2009), that is between 75 and 130 g (Table SI.2). In all likelihood, this difference with our ancestral estimate (at 22–33 g, see above) may be due to the fact that the taxonomic sample in Astúa (2009) lacked small outgroups (caenolestids not included) and small relevant basal taxa (particularly Hyladelphis; 16 g, Table SI.2). The amount of evolutionary change that we reconstructed did not unfold in a specific pattern for didelphids as a group. We see, as discussed next, that evolution of body mass was group-specific, and this reflects, within a particular clade of mammals, a pattern found at the level of the Class (e.g., Venditti et al. 2011).
The total magnitude of changes in absolute terms was greater for increases than for decreases, as expected. This bias toward a greater magnitude of increases is not an artifact of the reconstruction technique but a property of the character scale (restricted to the set of positive rational numbers). Large increases are more likely to occur than large decreases; e.g., from any small value, like the root at 27.5 g (middle between 22 and 33 g), it is possible to have a large evolutionary increase of, say 200 g, thus producing a descendant at 227.5 g, but it is not possible to have the same absolute amount of decrease. By contrast, the frequencies of increases and decreases were relatively balanced within groups (except in Caluromyinae). A major difference was though that increases (giantism in Gould and McFadden’s (2004) terminology) were arranged more commonly in a phyletic pattern, while decreases (nanism) were more often single, or apomorphic, changes. In this way, the largest values of body mass observed in extant Didelphidae were reconstructed as the cumulative evolution along successive branches through time, while the smallest values of body mass were often the product of single evolutionary events. Given this pattern, the most persistent case of phyletic giantism was that of Didelphis. Particularly in Didelphis virginiana, four changes along six branches added a total of 2032 g, that is, a ca. 62-fold increase from the ancestral Didelphini and a ca. 80-fold increase between the reconstructed ancestral body mass at 22–33 g (mean 27.5 g) and the largest terminal (Didelphis virginiana at average 2195 g). Still, 82 % of that change occurred at the ancestral Didelphis and its descendant species. If we assume a basal position to the oldest known Didelphis specimen, which is also the smallest member of the genus, of late Miocene (Huayquerian) age from the Solimões formation in the Brazilian Amazon (Cozzuol et al. 2006), it shows that this trend of increasing body mass has persisted for at least 9–6.8 my. The rate of change, estimated between 2032 g/9 my = 225.7 g my−1 and 2032 g/6.8 my = 298.8 g my−1, was very high (4-to-6-fold the median value for the family at 51 g). This trend is consistent with fast but gradual (phyletic) change and suggests directional selection for increased body mass sustained over substantial geological time. This also holds if extinct forms (e.g., the largest known didelphid, the late Pliocene Thylophorops lorenzinii, Goin et al. 2009) are considered.
This evolution must have affected the biology of Didelphis at a critical geologic age. Modern Didelphis appeared shortly before the mammalian peak of geodispersal during the Great American Biotic Interchange (GABI; Cozzuol et al. 2006; also see Bacon et al. 2015) and must have speciated during the latest GABI phases. At this time, placental carnivores gradually entered South America, and no other marsupials as large or larger than Didelphis escaped extinction; Didelphis not only survived, but also speciated and even expanded its range to North America (Giannini et al. 2011, and Citations therein). This marsupial clade accumulated defensive morphologies and skills (e.g., presence of hypertrophied spinous processes in cervical vertebrae that function as neck armor, catatonia or death feigning, secretion of anal glands, etc.; see Giannini et al. 2011), shifted to more generalized feeding and locomotor habits, and achieved a relatively large body mass as compared with most of their ancestors and extinct close relatives (cf. Goin et al. 2009). Possibly, this body mass permitted to be large enough for accessing vertebrate prey and for promoting the development of efficient defensive traitsFootnote 1 but, at the same time, to be still small enough for avoiding potential placental omnivore competitors (e.g., Procyonidae), and for maintaining those generalized habits (Astúa 2009). Thus, we speculate that phyletic giantism in didelphids is associated with long-term, persistent changes in diet (e.g., from insectivory to increased frugivory or omnivory, as in caluromines and didelphines) and/or locomotion (see Astúa 2009). By contrast, evolution of body mass showed that the most prominent decreases were more recent (i.e., near the terminals), specifically within Marmosini (see above). Apomorphic nanism is possibly related to specific limitations (e.g., physiological or ecological) for reducing mass in a mammal whose ancestor was already small.
Previous studies on marsupials relate body mass with risk of extinction (e.g., Cardillo and Bromhan 2001; Fisher et al. 2003; Johnson and Isaac 2009). These investigations attempted to test the controversial hypothesis of “critical weight range” (CWR) proposed by Burbidge and McKenzie (1989), who stated that species of non-volant terrestrial mammals of intermediate body mass (between 35 and 5500 g) have declined most severely. However, these studies were restricted to Australasian species, and the picture seems to be different for American marsupial species. On preliminary grounds, the surviving species have a small to intermediate body mass (being the last successfully widespread), while the large clades (order Sparassodonta) became all extinct (see above). The traditional explanation for this pattern has been the competition with placental carnivorans, which entered from North America during the GABI and caused the decline and extinction of large carnivorous marsupials. However, this idea has been questioned, criticized, or rejected in recent years, as there is no evidence of temporal overlap between carnivorans and their sparassodont ecological counterparts (Prevosti et al. 2013), so apparently, carnivorans occupied a niche left vacant by extinct sparassodonts (a passive replacement pattern), as already advanced by Marshall (1978) and commented by Vieira and Astúa de Moraes (2003). Whatever the cause, the association between risk of extinction and body mass in American marsupial species appears to follow a different pattern from that of Australian species and deserves further investigation.
In conclusion, here, we provided a robust phylogenetic basis for evolutionary comparisons of relevant life history traits in a group of particular biological interest, the largest New World radiation of marsupials, the didelphids. On this basis, we revealed interesting patterns of body mass evolution in a group of typical small mammals, including no clade-wide dominating trend, predominance of phyletic increases and apomorphic decreases, and persistent clade-specific trends consistent with directional selection over extended periods of geological time. This is particularly relevant given the constraint imposed by mass on morphological evolutionary responses to directional selection in highly integrated mammals, such as didelphids (Marroig et al. 2009). We conclude that evolutionary body mass variation of substantial interest may reside in groups of small mammals; this should be explored further in more groups in the light of robust phylogenetic reconstructions, as demonstrated here with didelphids.
Notes
An anonymous reviewer suggested that large body mass may also be physiologically advantageous for colonization of cold areas, as is the case of Didelphis virginiana in its northern distributional limit.
References
Argot, C. (2003). Functional adaptations of the postcranial skeleton of two Miocene borhyaenoids (Mammalia, Metatheria), Borhyaena and Prothylacinus, from South America. Palaeontology, 46, 1213–1267.
Argot, C. (2004a). Functional-adaptive features and paleobiologic implications of the postcranial skeleton of the late Miocene sabretooth borhyaenoid Thylacosmilus atrox (Metatheria). Alcheringa, 28, 229–266.
Argot, C. (2004b). Functional-adaptive analysis of the postcranial skeleton of a Laventan Borhyaenoid, Lycopsis longirostris (Marsupialia, Mammalia). Journal of Vertebrate Paleontology, 24, 689–708.
Astúa, D. (2009). Evolution of scapula size and shape in didelphid marsupials (Didelphimorphia: Didelphidae). Evolution, 63(9), 2438–2456.
Astúa, D. (2015). Family Didelphidae (Opossums). In: D.E. Wilson and R.A. Mittermeier (Orgs.), Handbook of the mammals of the world, 1st ed., Vol. 5 - Monotremes and Marsupials, pp: 70–18. Barcelona, Lynx Editions.
Bacon, C. D., Silvestro, D., Jaramillo, C., Smith, B. T., Chakrabarty, P., & Antonelli, A. (2015). Biological evidence supports an early and complex emergence of the Isthmus of Panama. Proceedings of the National Academy of Sciences, 112(19), 6110–6115.
Barrera-Niño, V., & Sánchez, F. (2014). Forrajeo de Didelphis pernigra (Mammalia: Didelphidae) en un área suburbana de la Sabana de Bogotá, Colombia. Therya, 5(1), 289–302.
Birney, E.C., & Monjeau, J.A. (2003). Latitudinal patterns in South American marsupial biology. Carnivorous with pouches: biology of carnivorous marsupials, 293–313.
Burbidge, A. A., & McKenzie, N. L. (1989). Patterns in the modern decline of Western Australia’s vertebrate fauna: causes and conservation implications. Biological Conservation, 50(1), 143–198.
Butler, R. J., & Goswami, A. (2008). Body size evolution in Mesozoic birds: little evidence for Cope’s rule. Journal of Evolutionary Biology, 21(6), 1673–1682.
Cáceres, N. C., Napoli, R. P., Lopes, W. H., Casella, J., & Gazeta, G. S. (2007). Natural history of the marsupial Thylamys macrurus (Mammalia, Didelphidae) in fragments of savannah in southwestern Brazil. Journal of Natural History, 41(29–32), 1979–1988.
Caramaschi, F. P., Nascimento, F. F., Cerqueira, R., & Bonvicino, C. R. (2011). Genetic diversity of wild populations of the grey short-tailed opossum, Monodelphis domestica (Didelphimorphia: Didelphidae), in Brazilian landscapes. Biological Journal of the Linnean Society, 104(2), 251–263.
Cardillo, M., & Bromham, L. (2001). Body size and risk of extinction in Australian mammals. Conservation Biology, 15(5), 1435–1440.
Cardillo, M., Bininda‐Emonds, R. P., Boakes, E., & Purvis, A. (2004). A species-level phylogenetic supertree of marsupials. Journal of Zoology, 264(1), 11–31.
Carvalho, B. D. A., Oliveira, L. F., Langguth, A., Freygang, C. C., Ferraz, R. S., & Mattevi, M. S. (2011). Phylogenetic relationships and phylogeographic patterns in Monodelphis (Didelphimorphia: Didelphidae). Journal of Mammalogy, 92(1), 121–133.
Catzeflis, F., Richard-Hansen, C., Fournier-Chambrillon, C., Lavergne, A., & Vié, J. (1997). Biométrie, reproduction et sympatrie chez Didelphis marsupialis et D. albiventris en Guyane française (Didelphidae: Marsupialia). Mammalia, 61(2), 231–244.
Cerqueira, R., & Tribe, C. J. (2007). Genus Didelphis Linnaeus, 1758. Mammals of South America, 1, 17–25.
Chemisquy, M. A., & Flores, D. A. (2012). Taxonomy of the southernmost populations of Philander (Didelphimorphia, Didelphidae), with implications for the systematics of the genus. Zootaxa, 3481, 60–72.
Chemisquy, M. A., Prevosti, F. J., Martin, G., & Flores, D. A. (2015). Evolution of molar shape in didelphid marsupials (Marsupialia: Didelphidae): analysis of the influence of ecological factors and phylogenetic legacy. Zoological Journal of the Linnean Society, 173(1), 217–235.
Cooper, N., & Purvis, A. (2010). Body size evolution in mammals: complexity in tempo and mode. The American Naturalist, 175(6), 727–738.
Cozzuol, M. A., Goin, F., Reyes, M. D. L., & Ranzi, A. (2006). The oldest species of Didelphis (Mammalia, Marsupialia, Didelphidae), from the late Miocene of Amazonia. Journal of Mammalogy, 87(4), 663–667.
Crompton, A. W., & Hiiemae, K. (1970). Molar occlusion and mandibular movements during occlusion in the American opossum, Didelphis marsupialis. Zoological Journal of the Linnean Society, 49, 21–47.
de la Sancha, N. U., D’Elía, G., & Teta, P. (2012). Systematics of the subgenus of mouse opossums Marmosa (Micoureus)(Didelphimorphia, Didelphidae) with noteworthy records from Paraguay. Mammalian Biology-Zeitschrift für Säugetierkunde, 77(4), 229–236.
Dickman, C.R. (2005). Introduction. In R.M. Nowack (ed.), Walker’s marsupials of the world. Johns Hopkins University Press, 226 pp.
Dickman, C. R., & Vieira, E. M. (2005). Ecology and life histories. In P. J. Armati, C. R. Dickman, & I. D. Hume (Eds.), Marsupials. Cambridge: Cambridge University Press.
Fisher, D. O., Blomberg, S. P., & Owens, I. P. (2003). Extrinsic versus intrinsic factors in the decline and extinction of Australian marsupials. Proceedings of the Royal Society of London B: Biological Sciences, 270(1526), 1801–1808.
Flores, D. A. (2009). Phylogenetic analyses of postcranial skeletal morphology in didelphid marsupials. Bulletin of the American Museum of Natural History, 320, 1–81.
Flores, D. A., Barquez, R. M., & Díaz, M. M. (2008). A new species of Philander Brisson, 1762 (Didelphimorphia, Didelphidae). Mammalian Biology-Zeitschrift für Säugetierkunde, 73(1), 14–24.
Forero-Medina, G., & Vieira, M. V. (2009). Perception of a fragmented landscape by neotropical marsupials: effects of body mass and environmental variables. Journal of Tropical Ecology, 25(01), 53–62.
Fox, R. C. (1987). Paleontology and the early evolution of marsupials. Possums and Opossums: Studies in Evolution, 1, 161–169.
Gardner, A.L. (2005). Order Didelphimorphia, Paucituberculata and Microbiotheria. In D.E. Wilson and D.M. Reeder (eds.), Mammals species of the world: a taxonomic and geographic reference, 3rd ed., Vol. 1, 743 pp. Johns Hopkins University Press.
Gardner, A.L. (2008). Cohort Marsupialia. In A.L. Gardner (ed.), Mammals of South America, Volume 1, marsupials, xenarthrans, shrews, and bats, 669 pp. The University of Chicago Press.
Giannini, N. P. (2003). Canonical phylogenetic ordination. Systematic Biology, 52(5), 684–695.
Giannini, N. P., & Bertelli, S. (2004). Phylogeny of extant penguins based on integumentary and breeding characters. The Auk, 121(2), 422–434.
Giannini, N. P., Gaudioso, P., Flores, D. A., & Gaudin, T. J. (2011). A possible function for an enigmatic synapomorphy of Didelphis. Mammalian Biology-Zeitschrift für Säugetierkunde, 76(4), 512–514.
Giannini, N.G., Gunnell, G.F., Habersetzer, J. & Simmons, N.B. (2012). Early evolution of body size in bats. In G.F. Gunnell and N.B. Simmons (eds.), Evolutionary history of bats: fossils, molecules, and morphology (530–555 pp). Cambridge University Press.
Giarla, T. C., & Jansa, S. A. (2014). The role of physical geography and habitat type in shaping the biogeographical history of a recent radiation of Neotropical marsupials (Thylamys: Didelphidae). Journal of Biogeography, 41(8), 1547–1558.
Giarla, T.C., Voss, R.S., & Jansa, S.A. (2010). Species limits and phylogenetic relationships in the didelphid marsupial genus Thylamys based on mitochondrial DNA sequences and morphology. Bulletin of the American Museum of Natural History, 1–67.
Goin, F. J., & Pardiñas, U. F. J. (1996). Revisión de las especies del género Hyperdidelphis Ameghino 1904 (Mammalia, Marsupialia, Didelphidae). Su significación filogenética, estratográfica, y adaptativa en el Neógeno del Cono Sur sudamericano. Estudios Geológicos, 52, 327–359.
Goin, F. J., Zimicz, N., de los Reyes, M., & Soibelzon, L. (2009). A new large didelphid of the genus Thylophorops (Mammalia: Didelphimorphia: Didelphidae) from the Late Tertiary of the Pampean Region (Argentina). Zootaxa, 2005, 35–46.
Goloboff, P. A. (1993). Estimating character weights during tree search. Cladistics, 9(1), 83–91.
Goloboff, P. A. (2014). Extended implied weighting. Cladistics, 30(3), 260–272.
Goloboff, P. A., & Catalano, S. A. (2012). GB‐to‐TNT: facilitating creation of matrices from GenBank and diagnosis of results in TNT. Cladistics, 28(5), 503–513.
Goloboff, P. A., Mattoni, C. I., & Quinteros, A. S. (2006). Continuous characters analyzed as such. Cladistics, 22(6), 589–601.
Goloboff, P. A., Carpenter, J. M., Arias, J. S., & Esquivel, D. R. M. (2008a). Weighting against homoplasy improves phylogenetic analysis of morphological data sets. Cladistics, 24(5), 758–773.
Goloboff, P. A., Farris, J. S., & Nixon, K. C. (2008b). TNT, a free program for phylogenetic analysis. Cladistics, 24(5), 774–786.
Gould, G. C., & MacFadden, B. J. (2004). Gigantism, dwarfism, and Cope’s rule: “nothing in evolution makes sense without a phylogeny”. Bulletin of the American Museum of Natural History, 285, 219–237.
Groves, C.P. (2005). Order Notoryctemorphia, Dasyuromorphia, Peramelemorphia and Diprotodontia. In D.E. Wilson and D.M. Reeder (eds.), Mammals species of the world: a taxonomic and geographic reference, 3rd ed., Vol. 1, 743 pp. Wilson Johns Hopkins University Press.
Gruber, K. F., Voss, R. S., & Jansa, S. A. (2007). Base-compositional heterogeneity in the RAG1 locus among didelphid marsupials: implications for phylogenetic inference and the evolution of GC content. Systematic Biology, 56(1), 83–96.
Gutiérrez, E.E., Jansa, S.A., & Voss, R.S. (2010). Molecular systematics of mouse opossums (Didelphidae: Marmosa): assessing species limits using mitochondrial DNA sequences, with comments on phylogenetic relationships and biogeography. American Museum Novitates, 1–22.
Gutiérrez, E. E., Soriano, P. J., Rossi, R. V., Murillo, J. J., Ochoa-G, J., & Aguilera, M. (2011). Occurrence of Marmosa waterhousei in the Venezuelan Andes, with comments on its biogeographic significance. Mammalia, 75(4), 381–386.
Hone, D. W. E., Keesey, T. M., Pisani, D., & Purvis, A. (2005). Macroevolutionary trends in the Dinosauria: Cope’s rule. Journal of Evolutionary Biology, 18(3), 587–595.
Horovitz, I., & Sánchez‐Villagra, M. R. (2003). A morphological analysis of marsupial mammal higher-level phylogenetic relationships. Cladistics, 19(3), 181–212.
Jansa, S. A., & Voss, R. S. (2005). Phylogenetic relationships of the marsupial genus Hyladelphys based on nuclear gene sequences and morphology. Journal of Mammalogy, 86(5), 853–865.
Jansa, S. A., Forsman, J. F., & Voss, R. S. (2006). Different patterns of selection on the nuclear genes IRBP and DMP-1 affect the efficiency but not the outcome of phylogeny estimation for didelphid marsupials. Molecular Phylogenetics and Evolution, 38(2), 363–380.
Johnson, C. N., & Isaac, J. L. (2009). Body mass and extinction risk in Australian marsupials: the ‘critical weight range’ revisited. Austral Ecology, 34(1), 35–40.
Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780.
Kirsch, J. A. W. (1977). The comparative serology of Marsupialia, and a classification of marsupials. Australian Journal of Zoology, 52, 1–152.
Kirsch, J.A.W., & Archer, M. (1982). Polythetic cladistics, or, when parsimony’s not enough: the relationships of carnivorous marsupials. In M. Archer (ed.), Carnivorous marsupials, Vol. 2, 595–620 pp. Mosman, NSW: Royal Society of New South Wales.
Kirsch, J. A. W., & Palma, R. E. (1995). DNA/DNA hybridization studies of carnivorous marsupials. V. A further estimate of relationships among opossums (Marsupialia: Didelphidae). Mammalia, 59, 403–425.
Leiner, N. O., & Silva, W. R. (2009). Short communication: territoriality in females of the slender opossum (Marmosops paulensis) in the Atlantic forest of Brazil. Journal of Tropical Ecology, 25, 671–675.
Lim, B. K., Engstrom, M. D., Patton, J. C., & Bickham, J. W. (2010). Molecular phylogenetics of Reig’s short-tailed opossum (Monodelphis reigi) and its distributional range extension into Guyana. Mammalian Biology-Zeitschrift für Säugetierkunde, 75(4), 287–293.
Lomolino, M. V. (2005). Body size evolution in insular vertebrates: generality of the island rule. Journal of Biogeography, 32(10), 1683–1699.
Marroig, G., Shirai, L. T., Porto, A., de Oliveira, F. B., & De Conto, V. (2009). The evolution of modularity in the mammalian skull II: evolutionary consequences. Evolutionary Biology, 36(1), 136–148.
Marshall, L. G. (1978). Evolution of the Borhyaenidae, extinct South American predaceous marsupials. University of California Publications in Geological Sciences, 117, 1–87.
Martínez-Lanfranco, J. A., Flores, D., Jayat, J. P., & D’Elía, G. (2014). A new species of lutrine opossum, genus Lutreolina Thomas (Didelphidae), from the South American Yungas. Journal of Mammalogy, 95(2), 225–240.
May-Collado, L., Kilpatrick, C. W., & Agnarsson, I. (2015). Mammals from “down under”: a multi-gene species-level phylogeny of marsupial mammals. PeerJ, 3, e805. doi:10.7717/peerj.805.
McKenna, M.C., & Bell, S.K. (1997). Classification of mammals above the species level. M.C. McKenna and S.K. Bell (eds.), 631 pp. Columbia University Press.
McManus, J. J. (1974). Didelphis virginiana. Mammalian Species, 40, 1–6.
McNab, B.K. (2007). The evolution of energetics in birds and mammals. In D.A. Kelt, E.P. Lessa, J. Salazar-Bravo, and J.L. Patton (eds.), The quintessential naturalist: honoring the life and legacy of Oliver P. Pearson, 67–110 pp. University of California Publications.
Meiri, S., Meijaard, E., Wich, S. A., Groves, C. P., & Helgen, K. M. (2008). Mammals of Borneo—small size on a large island. Journal of Biogeography, 35(6), 1087–1094.
Meredith, R. W., Janečka, J. E., Gatesy, J., Ryder, O. A., Fisher, C. A., Teeling, E. C., Goodbla, A., Eizirik, E., Simão, T. L. L., Stadler, T., Rabosky, D. L., Honeycutt, R. L., Flynn, J. J., Ingram, C. M., Steiner, C., Williams, T. L., Robinson, T. J., Burk-Herrick, A., Westerman, M., Ayoub, N. A., Springer, M. S., & Murphy, W. J. (2011). Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science, 334(6055), 521–524.
Mondolfi, E., & Pérez-Hernández, R. (1984). Una nueva subespecie de zarigüeya del grupo Didelphis albiventris (Mammalia: Marsupialia). Acta Científica Venezolana, 35, 407–413.
Paglia, A.P., Fonseca, G.A.B., Rylands, A.B., Herrmann, G., Aguiar, L.M.S., Chiarello, A.G., Leite, Y.L.R., Costa L.P., Siciliano, S., Kierulff, M.C.M., Mendes, S.L., Tavares, V. da C., Mittermeier, R.A. y Patton, J.L. (2012). Lista Anotada dos Mamíferos do Brasil / Annotated Checklist of Brazilian Mammals. 2ª Edição / 2nd Edition. Occasional Papers in Conservation Biology, 6, 76 pp.
Palma, R. E., & Spotorno, A. E. (1999). Molecular systematics of marsupials based on the rRNA 12S mitochondrial gene: the phylogeny of Didelphimorphia and of the living fossil microbiotheriid Dromiciops gliroides Thomas. Molecular Phylogenetics and Evolution, 13(3), 525–535.
Palma, R. E., Boric-Bargetto, D., Jayat, J. P., Flores, D. A., Zeballos, H., Pacheco, V., Cancino, R. A., Alfaro, F. D., Rodríguez-Serrano, E. & Pardiñas, U. F. J. (2014). Molecular phylogenetics of mouse opossums: new findings on the phylogeny of Thylamys (Didelphimorphia, Didelphidae). Zoologica Scripta, 43, 217–234.
Pavan, S. E., Rossi, R. V., & Schneider, H. (2012). Species diversity in the Monodelphis brevicaudata complex (Didelphimorphia: Didelphidae) inferred from molecular and morphological data, with the description of a new species. Zoological Journal of the Linnean Society, 165(1), 190–223.
Pavan, S. E., Jansa, S. A., & Voss, R. S. (2014). Molecular phylogeny of short-tailed opossums (Didelphidae: Monodelphis): taxonomic implications and tests of evolutionary hypotheses. Molecular Phylogenetics and Evolution, 79, 199–214.
Perez Carusi, L. C., Farace, M. I., Ribicich, M. M., & Gómez Villafañe, I. E. (2009). Reproduction and parasitology of Didelphis albiventris (Didelphimorphia) in an agroecosystem landscape in central Argentina. Mammalia, 73(2), 89–97.
Porto, A., de Oliveira, F. B., Shirai, L. T., De Conto, V., & Marroig, G. (2009). The evolution of modularity in the mammalian skull I: morphological integration patterns and magnitudes. Evolutionary Biology, 36(1), 118–135.
Prevosti, F. J., Forasiepi, A., & Zimicz, N. (2013). The evolution of the Cenozoic terrestrial mammalian predator guild in South America: competition or replacement? Journal of Mammalian Evolution, 20(1), 3–21.
Reig, O.A, Kirsch, J.A.W., & Marshall, L.G. (1987). Systematic relationships of the living and neocenozoic American “opossum-like” marsupials (Suborder Didelphimorphia), with comments on the classification of these and of the Cretaceous and Paleogene New World and European metatherians. In M. Archer (ed.), Possums and opossums: studies in evolution, Vol. 1, 1–89 pp. Sydney: Surrey Beatty.
Rossi, R. V. (2005). Revisão taxonômica de Marmosa Gray, 1821 (Didelphimorphia: Didelphidae). Tese de Doutorado: Universidade de São Paulo, São Paulo.
Rossi, R.V., Voss, R.S., & Lunde, D.P. (2010). A revision of the Didelphid Marsupial Genus Marmosa. Part 1. The species in Tate’s ‘mexicana’ and ‘mitis’ sections and other closely related forms. Bulletin of the American Museum of Natural History, 1–83.
Shirai, L. T., & Marroig, G. (2010). Skull modularity in neotropical marsupials and monkeys: size variation and evolutionary constraint and flexibility. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 4(8), 663–683.
Simpson, G. G. (1972). Didelphidae from the Chapadmalal Formation in the Museo Municipal de Ciencias Naturales of Mar del Plata. Revista del Museo Municipal de Ciencias Naturales de Mar del Plata, 2, 1–40.
Slater, G. J. (2013). Phylogenetic evidence for a shift in the mode of mammalian body size evolution at the Cretaceous-Palaeogene boundary. Methods in Ecology and Evolution, 4(8), 734–744.
Smith, F. A., Lyons, S. K., Ernest, S. M., Jones, K. E., Kaufman, D. M., Dayan, T., Marquet, P. A., Brown, J. H., & Haskell, J. P. (2003). Body mass of Late Quaternary mammals: ecological archives E084-094. Ecology, 84(12), 3403.
Solari, S. (2007). New species of Monodelphis (Didelphimorphia: Didelphidae) from Peru, with notes on M. adusta (Thomas, 1897). Journal of Mammalogy, 88(2), 319–329.
Solari, S., Pacheco, V., Vivar, E., & Emmons, L. H. (2012). A new species of Monodelphis (Mammalia: Didelphimorphia: Didelphidae) from the montane forests of central Perú. Proceedings of the Biological Society of Washington, 125(3), 295–307.
Stamatakis, A., Hoover, P., & Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology, 57(5), 758–771.
Steiner, C., Tilak, M. K., Douzery, E. J., & Catzeflis, F. M. (2005). New DNA data from a transthyretin nuclear intron suggest an Oligocene to Miocene diversification of living South America opossums (Marsupialia: Didelphidae). Molecular Phylogenetics and Evolution, 35(2), 363–379.
Ter Braak, C.J.F., & Šmilauer, P. (1998). CANOCO 4. CANOCO reference manual and user’s guide to Canoco for Windows. Centre of Biometry, Wageningen.
Teta, P., D’Elía, G., Flores, D., & de La Sancha, N. (2009). Diversity and distribution of the mouse opossums of the genus Thylamys (Didelphimorphia, Didelphidae) in northeastern and central Argentina. Gayana, 73(2), 180–199.
Venditti, C., Meade, A., & Pagel, M. (2011). Multiple routes to mammalian diversity. Nature, 479(7373), 393–396.
Vieira, E. M., & Astúa de Moraes, D. (2003). Carnivory and insectivory in Neotropical marsupials. In M. E. Jones, C. R. Dickman, & M. Archer (Eds.), Predators with pouches: the biology of carnivorous marsupials (pp. 271–284). Collingwood: CSIRO Publishing.
Vilela, J. F., Oliveira, J. A., & Russo, C. A. M. (2015). The diversification of the genus Monodelphis and the chronology of Didelphidae (Didelphimorphia). Zoological Journal of the Linnean Society, 174(2), 414–427.
Voss, R.S., & Jansa, S.A. (2003). Phylogenetic studies on didelphid marsupials II. Nonmolecular data and new IRBP sequences: separate and combined analyses of didelphine relationships with denser taxon sampling. Bulletin of the American Museum of Natural History, 1–82.
Voss, R.S., & Jansa, S.A. (2009). Phylogenetic relationships and classification of didelphid marsupials, an extant radiation of New World metatherian mammals. Bulletin of the American Museum of Natural History, 1–177.
Voss, R.S., Tarifa, T., & Yensen, E. (2004). An introduction to Marmosops (Marsupialia: Didelphidae), with the description of a new species from Bolivia and notes on the taxonomy and distribution of other Bolivian forms. American Museum Novitates, 1–40.
Voss, R.S., Lunde, D.P., & Jansa, S.A. (2005). On the contents of Gracilinanus Gardner and Creighton 1989, with the description of a previously unrecognized clade of small didelphid marsupials. American Museum Novitates, 1–36.
Voss, R. S., Pine, R. H., & Solari, S. (2012). A new species of the didelphid marsupial genus Monodelphis from eastern Bolivia. American Museum Novitates, 3740, 1–14.
Voss, R. S., Lim, B. K., Díaz-Nieto, J. F., & Jansa, S. A. (2013). A new species of Marmosops (Marsupialia: Didelphidae) from the Pakaraima Highlands of Guyana, with remarks on the origin of the endemic Pantepui mammal fauna. American Museum Novitates, 3778, 1–27.
Voss, R. S., Gutiérrez, E. E., Solari, S., Rossi, R. V., & Jansa, S. A. (2014). Phylogenetic relationships of mouse opossums (Didelphidae, Marmosa) with a revised subgeneric classification and notes on sympatric diversity. American Museum Novitates, 3817, 1–27.
Acknowledgments
We thank Diego Astúa, David Flores, Sergio Solari, Pablo Teta, and Robert Voss for providing us with unpublished body mass data and Rubén Barquez and Mónica Díaz (CML) for granting access to specimens under their care. Special thanks to Santiago Catalano for his help with using his program GB-to-TNT. We acknowledge support from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina, and grant PICT 2008–1798 to NPG.
Data archiving
Data are archived in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S17654)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Amador, L.I., Giannini, N.P. Phylogeny and evolution of body mass in didelphid marsupials (Marsupialia: Didelphimorphia: Didelphidae). Org Divers Evol 16, 641–657 (2016). https://doi.org/10.1007/s13127-015-0259-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-015-0259-x