Abstract
Wolbachia is a common group of intracellular bacteria found in arthropods and filarial nematodes. Since the past decade, they have attracted considerable interest owing to their various effects on hosts, which range from reproductive manipulation to mutualism. Moreover, they can influence the mitochondrial DNA pattern which do not reflect the real evolutionary history of the target species and may be incongruent with nuclear data. Previously, Wolbachia-manipulated mitochondrial DNA (mtDNA) patterns, namely mito-nuclear discordance and deep mitochondrial splits associated with specific Wolbachia infections, have been also discovered in the genus Maculinea. Here, we present a comprehensive study on Wolbachia infestation and the genetic diversity of all Maculinea species in the Carpathian Basin. The prevalence and the pattern of the infestation highly differ among Maculinea species. Maculinea alcon and Maculinea arion are infected in 100 %, each of these species with a single strain, but the infection level of Maculinea nausithous and Maculinea teleius is much lower, additionally, they are infected with multiple strains. The genetic diversity of Maculinea species proved to be generally low, only M. nausithous showed geographic pattern based on mitochondrial sequences and allozymes. In contrast with the previous studies, we could not detect mito-nuclear discordance or find evidence for Wolbachia-induced selective sweep. Based on our results, we cannot hold only Wolbachia responsible for the restricted genetic diversity of Maculinea in the Carpathian Basin. Probably several factors shape together the level and pattern of genetic variability in Maculinea butterflies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the last half century, genetic markers have become widely used in species delimitation and in the investigation of the phylogenetic relationships among species due to the development of advanced molecular techniques. In the animal kingdom, mitochondrial DNA (mtDNA) is one of the most popular markers because of its many ideal properties (Galtier et al. 2009). Although mtDNA is proved to be suitable for barcoding animal life and highly informative in phylogenetic and phylogeographic studies, mtDNA patterns do not always reflect the real evolutionary history of the studied species and may be incongruent with nuclear data (Dupuis et al. 2012). Low intraspecific mtDNA variation can make it impossible to reveal the phylogeography of focal groups of organisms. Such reduced mtDNA diversity may be caused by an indirect selective sweep of a favoured mtDNA variant, e.g. by Wolbachia. These intracellular bacteria infect a wide variety of arthropods and filarial nematodes (Werren et al. 2008), and a recent meta-analysis estimated that more than 65 % of insect species harbour Wolbachia (Hilgenboecker et al. 2008).
Usually, Wolbachia are vertically transmitted from mother to offspring while they can manipulate the host reproduction in ways that enhance their own transmission chance to future generations. Accordingly, Wolbachia infections are associated with a variety of phenotypic effects on the hosts, such as cytoplasmic incompatibility (CI, the sperm of infected males is incapable to fertilize the eggs of females which are uninfected or infected with a different strain), male-killing (MK), feminization of genetic males and induction of parthenogenesis (PI) (Werren et al. 2008). Regardless of the specific mechanism by which they manipulate host reproduction, the spread of Wolbachia will be accompanied with the concomitant spread of all maternally inherited organelles, including mitochondria, being present in the initially infected female (Turelli and Hoffmann 1991, 1995; Hoffmann et al. 1998). This means that one particular mitotype (and its mutational derivatives) will hitchhike through the infected population in association with the selectively driven sweep of Wolbachia (Turelli and Hoffmann 1991, 1995; Turelli 1994). Consequently, Wolbachia may be expected to dramatically affect host mtDNA genome evolution.
Assuming the bacteria are transmitted with very high efficiency between generations, an infected population should have lower mtDNA diversity than an uninfected one since each Wolbachia sweep reduces the effective population size of mtDNA to one. At the same time, a single population may be infected with more than one strain which may inflate the mtDNA variability within a population given the association between the particular Wolbachia strain and the concomitant host mitotype.
Simultaneously, Wolbachia may affect levels of mtDNA differentiation. The mtDNA of infected and uninfected individuals or the individuals infected with different strains will evolve to be distinct, because there is little or no flow of mtDNA between them. Besides, the spread of Wolbachia through a single population may increase the mtDNA differentiation between populations if the bacteria are unable to invade neighbouring populations. As reported in the satyrine butterfly Coenonympha tullia (Kodandaramaiah et al. 2013), the two divergent mitochondrial clades found in North America are associated with two different Wolbachia strains leading to a ‘two barcodes—one species’ phenomenon. Neither genitalia morphology nor nuclear gene sequence supports cryptic species as an explanation, instead selective sweeps driven by Wolbachia. That is, Wolbachia can make one species appear as two due to the high intraspecific mtDNA diversity associated with possession of different bacterial strains. On the other hand, Wolbachia sweeps can result in minimal mtDNA differentiation, if there is only a very small amount of interbreeding and parallel sweeps of mtDNA variants happen in different populations. The butterfly species pair Acraea encedon and Acraea encedana bear the same male-killing Wolbachia strain which homogenizes haplotypes of the two species causing a ‘one barcode–two species’ phenomenon (Jiggins 2003). While these species are clearly distinct on morphological and nuclear DNA grounds, they appear identical on the basis of barcoding sequences of the infected individuals. The most likely explanation for this is that rare hybridization events open the door to the transfer of the male killer and associated mitotype from species to species.
As a consequence of these widespread effects, Wolbachia may interfere with the phylogenetic and phylogeographic studies of hosts. While Wolbachia influence the mtDNA variability to a large extent, patterns of variation at nuclear genes should be largely unaffected by the presence of these bacteria leading to mito-nuclear discordance (Jiggins 2003; Shoemaker et al. 2003; Kodandaramaiah et al. 2013).
Wolbachia are also present in the social parasitic Maculinea butterflies (Sielezniew et al. 2012; Patricelli et al. 2013; Ritter et al. 2013; Bereczki et al. 2014). Reduced mitochondrial diversity refers to Wolbachia-induced selective sweeps in Maculinea alcon ([Denis & Schiffermüller], 1775) and Maculinea arion (Linnaeus, 1758) (Bereczki et al. 2014; Patricelli et al. 2013; Sielezniew et al. 2012). In addition, the possibility of CI also arises in M. arion. In the Carpathian Basin, this species exists in two phenological forms (‘spring and summer types’ according to their flight periods) which co-occur in certain habitats. Bereczki et al. (2011, 2014) revealed that these forms differentiate neither on the basis of barcoding gene nor in allozymes but they are clearly distinct morphologically (based on wing and male genitalia traits), and all individuals were infected with Wolbachia. Based on the discordance between mitochondrial and morphological results, it might be assumed that Wolbachia make an effect on the speciation of M. arion causing a ‘one barcode–two species’ phenomenon (Bereczki et al. 2014). At the same time, inflated diversity and deep splits in mitochondrial sequences proved to be in correspondence with different Wolbachia strains in Maculinea nausithous (Bergsträsser, 1779) and Maculinea teleius (Bergsträsser, 1779) (Ritter et al. 2013), i.e. Wolbachia seems to have an influence on the mitochondrial DNA pattern of Maculinea hosts in different ways.
Here, we present a comprehensive study on the Wolbachia infestation and the genetic diversity of all Maculinea species in the Carpathian Basin. Our main aim is to investigate the prevalence and infestation patterns of Wolbachia in each species and reveal their possible effects on hosts’ genetic diversity and speciation using mitochondrial and nuclear gene sequences as well as allozyme loci. Therefore, our study provides new information to interpret the genetic patterns of these long-surveyed butterflies.
Materials and methods
Samples
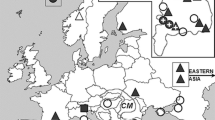
We sampled all Maculinea species in 18 geographical regions of the Carpathian Basin from 2001 to 2013 (Fig. 1a, b; Online Resource 1). The identification of the two ecotypes of M. alcon was carried out based on the initial food plants and the date of sampling. The ‘spring type’ and ‘summer type’ of M. arion were also separated phenologically. In certain habitats, two or even three Maculinea species were present. Images were collected at the end of the egg-laying period and stored at −80 °C until molecular analyses.
Sampling sites. See the abbreviations in Online Resource 1. a M. alcon and M. arion. The infection rate is 100 % in both species. b M. nausithous and M. teleius. The infection rate is indicated with the shade of symbols representing populations. Darker symbols show higher infection rate
Wolbachia screening and strain identification
DNA was extracted by homogenizing the heads or thoraxes of butterflies following the protocol in Bereczki et al. (2014). Altogether 506 individuals were screened (1–11 individuals/population) from 18 geographical regions (Online Resource 1) by the amplification of the highly conservative 16S ribosomal RNA gene with Wolbachia specific primers W-Spec of Werren and Windsor (2000). Amplification from 1 μl of DNA extracts was carried out in 25 μl final reaction volumes containing 5X PCR buffer, 0.2 mM dNTPs, 2 mM MgCl2, 0.5 mg/ml bovine serum albumin, 0.02 units/μl of Taq DNA polymerase (Phusion Hot Start II High-Fidelity, Thermo Scientific) and 0.3 μM of each primer. Amplification was carried out in an ABI Veriti thermal cycler programmed for the following: initial denaturation for 2 min at 98 °C; 40 cycles of 15 s at 98 °C, 45 s at 60 °C, 1 min 30 s at 72 °C; final elongation of 10 min at 72 °C. Amplification procedure was repeated twice in M. teleius and M. nausithous since we could obtain faint bands as a result of the first 40 cycles. We used positive (surely infected samples) and negative controls (master mix without any DNA sample) in each reaction. The success of PCRs was checked by running 2 μl of product on 1 % agarose gels stained with GelRed Nucleic Acid Stain (Biotium Inc.). The infection rates of the populations were plotted on geographic maps using QGIS v. 2.6 (2014). PCR products originating from 70 individuals (Online Resource 2) were sequenced by commercial service provider Macrogen Inc. (Seoul, South Korea). Sequences were edited and revised manually by Chromas Lite v. 2.01 and aligned by MEGA v. 6 (Tamura et al. 2013). Bayesian phylogenetic relationships were assessed in MrBayes v. 3.2.2 (Ronquist et al. 2012) as described below.
Wolbachia strain identification was carried out in M. alcon and M. arion according to Wolbachia multilocus sequence typing (MLST) system (Baldo et al. 2006) by the amplification of Wolbachia surface protein (WSP) and five conserved gene regions (gatB, coxA, hcpA, ftsZ, fbpA) following the PCR protocol mentioned above. The latter five genes determine together the sequence type (ST). WSP typing was achieved in a single individual each in M. alcon and M. arion. Wolbachia MLST was completed altogether in 32 specimens in the two species (Online Resource 2). After sequencing, we defined the strains using the reference sequences of Wolbachia MLST database (http://pubmlst.org/wolbachia/) and searched the identified sequence types in other organisms in the mentioned database.
Phylogenetic analyses of mitochondrial and nuclear DNA
The same DNA extracts were used to amplify mitochondrial and nuclear genes as those in Wolbachia studies. We sequenced the mitochondrial cytochrome c oxidase subunit I gene (COI) together with the nuclear gyceraldehyde 3-phosphate dehydrogenase (GAPDH), malate dehydrogenase (MDH) and wingless (wg) (Online Resource 2) using commercial service provider Macrogen Inc. These genes were amplified by specific primers modified at their 5′-end to include the universal sequencing primer T7promoter (Wahlberg and Wheat 2008). We followed the above amplification protocol and the guidelines of the Nymphalidae Systematics Group (http://nymphalidae.utu.fi/). After the revision and alignment of sequences, we estimated the level of genetic diversity by the following parameters: the number of variable and informative sites (V and PI), haplotype number and diversity (h and HD) (Librado and Rozas 2009), nucleotide diversity (π) (Nei and Li 1979; Nei 1987; Tajima 1983) and the number of segregating sites (θ w) (Watterson 1975; Nei 1987). In cases of nuclear genes, PHASE haplotype reconstruction option was used because of the presence of ambiguous sites (Stephens et al. 2001; Stephens and Donnelly 2003). Neutrality tests were performed on all genes to determine the departure from the neutral model of molecular evolution. Under neutrality, both Tajima’s D and Fu and Li’s D are expected to be zero. Positive D values indicate an excess of intermediate-frequency variants and can be due to the operation of natural selection. On the contrary, a value significantly less than zero indicates a higher-than-expected number of low frequency variants and might be the consequence of a recent selective sweep or processes such as background selection (Tajima 1989; Aris-Brosou and Excoffier 1996). The parameters of gene diversity and neutrality tests were calculated using the program DnaSP v. 5.10.01 (Librado and Rozas 2009).
Based on the concatenated sequences, Bayesian analyses were carried out using MrBayes 3.2.2 (Ronquist et al. 2012). The combined data were partitioned by gene and analysed as independent partitions. Several possible models of molecular evolution were sampled for each gene (both single and combined data) during the analysis using the model-jumping feature of MrBayes v. 3.2.2 applying the following sets ‘lset applyto = (all) nucmodel = 4by4 nst = mixed rates = gamma covarion = no;’. Two independent MCMC runs each with four simultaneous chains (one cold and three heated) were run for 10 million generations. The trees were sampled in every 1000 generations. Convergence of the two runs was determined by the stationary distribution plot of the log likelihood values against number of generations and confirmed by the average standard deviation of split frequencies, which were lower than 0.05 in all cases. The first 2,500,000 generations were discarded as burn-in, and trees were summarized under the 50 % majority rule method.
Haplotype networks were built in R computing environment (R Development Core Team 2014) with pegas package v. 0.6 (Paradis 2010) based on the mitochondrial COI sequences in M. nausithous and M. teleius. In this analysis, our samples (55 individuals, see in Online Resource 2: HG326644, KM517275-KM517328) and the sequences from GenBank (JX311050-JX311307, Ritter et al. 2013) were used. First, the original alignment was reduced to get the maximal overlapping sequences using MEGA v.6 (Tamura et al. 2013). Then, we deleted those specimens from the alignment which had ambiguous sites in their sequences. After this process, 101 specimens with 984 bp remained in M. nausithous and 158 specimens with 850 bp in M. teleius. Haplotype networks were created using the reduced datasets in both species.
Allozyme studies
Allozyme polymorphism was studied at 11 loci by vertical polyacrylamide gel electrophoresis altogether in 1266 individuals of four Maculinea species (Online Resource 1). Thoraxes homogenized in 300 μl of extraction buffer were used to study Gpdh, G6pgdh, Hk, Idh, Mdh, Pgi, Pgm and Sod. Abdomens homogenized in 200 μl of extraction buffer were used to analyse Acon, Aox and Est. The extraction buffer, the electrophoresis buffer systems and running conditions, together with the staining solutions, were applied as described in Bereczki et al. (2005).
Genotypes of the individuals were scored according to their enzyme pattern. Genotype and allele frequencies were calculated on the basis of banding patterns. Measures of genetic variation, i.e. average number of alleles per locus (N a), the effective number of alleles (N e), Shannon’s information index (I), average observed heterozygosity (H o) and proportion of polymorphic loci on the basis of the 95 % criterion (P%) were calculated for each species using GenAlEx v. 6.41 (Peakall and Smouse 2006).
Allele frequencies were used to estimate Cavalli-Sforza and Edwards (1967) chord distances, and a UPGMA dendrogram (Sneath and Sokal 1973) was constructed on the basis of the distance matrix using Past v. 2.17 (Hammer et al. 2001). In the case of M. alcon, a principal component analysis (PCA) was conducted using the allele frequency data of the individuals to investigate the basis of the differentiation between the two types using different food plants (‘cruciata and pneumonanthe types’). PCA was performed using Past v. 2.17 (Hammer et al. 2001).
The population genetic structure was also analysed by Bayesian-clustering method (Pritchard et al. 2000). Here, we estimated the most probable number of genetically differentiated groups (K) in our populations and assigned the individuals to these groups. Structure v. 2.3.4 was run to carry out these analyses with initial burn in 100,000 and running length 500,000. In the evaluation of the results, ΔK was computed which indicates the change in log probability between successive K values (Evanno et al. 2005). Structure Harvester Web v. 0.6.93 (Earl and vonHoldt 2012) was used to compute the ΔK values.
Results
Wolbachia infection
The prevalence of Wolbachia was 100 % both in M. alcon and M. arion irrespective of phenology or differential food plant usage while the infection level was 36.2 % in M. nausithous and 14.4 % in M. teleius (Online Resource 1; Fig. 1a, b). In Wolbachia infestation of the latter two species, we could not detect any geographical pattern (Fig. 1b).
All M. alcon individuals were infected with a single Wolbachia strain which belonged to supergroup B (Table 1; Fig. 2). The WSP allele no. 575 and the sequence type (ST) no. 235 were identified in M. alcon. Similarly, all M. arion individuals were infected with only one strain which belonged to supergroup A (Table 1; Fig. 2). A new WSP allele was identified from this species and submitted to the Wolbachia MLST database (no. 685). The sequence type (ST) no. 403 was found in M. arion.
The result of the Bayesian inference analysis with posterior probability values based on Wolbachia 16S ribosomal RNA gene sequences obtained from different Maculinea species. ALct M. alcon ‘cruciata type’, ALpt M. alcon ‘pneumonanthe type’, ARsp M. arion ‘spring type’, ARsu M. arion ‘summer type’, NA M. nausithous, TE M. teleius. See the abbreviations in Online Resource 1 and 2
M. nausithous harboured various 16S rRNA sequences belonging to both Wolbachia supergroups A and B (Fig. 2; Table 2). Additionally, highly divergent sequences occurred even in a single population of this species. Similarly, M. teleius contained different 16S rRNA sequences belonging to both supergroups A and B (Fig. 2; Table 2). The parameters of 16S rRNA gene diversity were proved to be higher in M. teleius than in M. nausithous (Table 2). Additionally, neutrality tests indicated significantly positive departure from the neutral model of molecular evolution.
The genetic diversity of Maculinea species
Mitochondrial DNA
The final COI alignment from the Carpathian Basin contained 110 sequences (Table 2, Online Resource 2) with a total length of 1369 bases out of which 60 (4.4 %) sites were variable and 45 (3.3 %) were parsimony informative. The highest number of these sites was detectable in M. nausithous. In total, 29 unique haplotypes were observed out of which 11 were included in M. teleius. At the same time, M. alcon ‘pneumonanthe type’ involved only one mitotype similarly to M. arion ‘spring type’ which also included only a single mitochondrial haplotype. Nucleotide diversity and the number of segregating sites were very low and varied from 0 to 0.00476 and from 0 to 0.00671, respectively (Table 2). The highest measures were found in M. nausithous. Neutrality tests showed negative D values almost in all cases, but significant deviation from the neutral model of molecular evolution was experienced only in M. nausithous where Fu and Li’s D was positive (Table 2).
Based on the mitochondrial COI sequences, not all Maculinea species differentiated from each other with high statistical support (Fig. 3a). Interestingly, the split of M. nausithous and M. teleius was practically without statistical support (posterior probability 0.55). In M. nausithous, strong geographical pattern was recognized. The differentiation of the three large regions (Western Hungarian, Transylvanian and Eastern Carpathian) was highly pronounced. Additionally, the populations of the Eastern Carpathian clade also diverged geographically, but reduced mitochondrial diversity was experienced in the other two regions (Western Hungarian and Transylvanian). At the same time, the resolution of COI sequences in the other three Maculinea species proved to be very low. M. nausithous and M. teleius specimens infected with Wolbachia did not form separate clades.
a The result of the Bayesian inference analysis with posterior probability values based on mitochondrial COI sequences; b consensus phylogeny from the Bayesian inference analysis with posterior probability values based on the combined dataset of three nuclear regions (GAPDH, MDH and wg). ALct M. alcon ‘cruciata type’, ALpt M. alcon ‘pneumonanthe type’, ARsp M. arion ‘spring type’, ARsu M. arion ‘summer type’, NA M. nausithous, TE M. teleius. The infected individuals are indicated with W. See the abbreviations in Online Resource 1 and 2
The mitochondrial haplotype network which included also the sequences of Ritter et al. (2013) showed five main haplotype groups in M. nausithous (Online Resource 5a). The central group involved mostly Asian individuals and specimens from Poland and Bulgaria. The Hungarian, Slovenian and Transylvanian individuals as well as the majority of Croatian specimens formed a distinct haplotype group in the Carpathian Basin. The Western European individuals constituted two independent groups separated from each other by numerous mutation steps. One of these groups involved the majority of Western German specimens and certain French individuals (Western Europe 2 in Online Resource 5a) linked to the so-called Wolbachia clade although they were differentiated from it by numerous mutations, and Ritter et al. (2013) could not detect current infection in this group. Wolbachia clade included mostly but not exclusively infected individuals from Croatia, Russia and the Eastern Carpathian region of Romania but none of the specimens from Hungary and Slovenia despite the fact that these populations are mostly infected with Wolbachia (Fig. 1b). In the case of M. teleius, there is a central haplotype group which contained the majority of individuals both from Europe and Asia (Online Resource 5b). Certain individuals from various geographical regions were separated from this central group by one or two mutation steps (and three in a single case). Only the Chinese and the majority of Japanese specimens differed from the central set of haplotypes by numerous mutations. A Wolbachia-dominated clade which involved Asian specimens was also recognizable in this species although several infected individuals can be found in the central group as well.
Nuclear DNA
Altogether 1680 bases were studied from three different gene regions (GAPDH, MDH and wingless) in 58 individuals (Table 2, Online Resource 2). In general, the diversity of these DNA fragments was lower than that of mitochondrial sequences. The highest variability was detected in M. arion and the lowest in M. teleius. Neutrality tests did not indicate significant departure from the neutral model in any of the species.
Based on the concatenated nuclear sequences, all Maculinea species differed from each other perfectly (Fig. 3b). Some pattern was experienced only in M. nausithous but it did not correspond with geographical regions. The infected individuals clustered into separate clades neither in this case. The resolution of the different nuclear regions did not differ from each other considerably although wingless gene seemed to be the less suitable for the reconstruction of phylogeography (Online Resource 3a, b, c).
Allozymes
The allozyme variability of Maculinea species was generally low. M. arion exhibited the highest level of polymorphism while M. teleius did the lowest level (Table 3).
All Maculinea species clearly differed from each other based on allozyme loci as well (Fig. 4a). Allozymes showed highly homogeneous pattern in M. arion and M. teleius (Online Resource 4). At the same time, both the dendrogram and the Bayesian analysis of the genetic structure indicated strong differentiation among the large geographical regions in M. nausithous similarly to mitochondrial sequences (Fig. 4a, Online Resource 4). The pattern emerging in M. alcon corresponded with different food plant usage but this correspondence was not entirely clear (certain ‘pneumonanthe type’ populations, i.e. ALpt-Dra and ALpt-Gyk clustered into the ‘cruciata type’ populations, see in Fig. 4a). Simultaneously, principal component analysis clearly showed that the ‘cruciata and pneumonanthe type’ populations were separated along the first axis which explained 46.9 % of the total variance, and the locus esterase showed the largest loading in this axis (Fig. 4b).
a UPGMA dendrogram based on allozymes using Cavalli-Sforza and Edwards’ chord distances. b The results of PCA in M. alcon. Each symbol represents one sample in the reduced space by variances. The first two axes explain 66.6 % of the total variance. The ‘cruciata and pneumonanthe types’ of M. alcon clearly differ along the first axis determined by the alleles 2 and 3 of esterase locus. ALct M. alcon ‘cruciata type’, ALpt M. alcon ‘pneumonanthe type’, ARsp M. arion ‘spring type’, ARsu M. arion ‘summer type’, NA M. nausithous, TE M. teleius. See the abbreviations in Online Resource 1
Discussion
In this paper, we provide a comprehensive view about Wolbachia infection and the genetic diversity of Maculinea butterflies in the Carpathian Basin. The prevalence and the pattern of the infestation highly differ among Maculinea species. M. alcon and M. arion are infected in 100 %, each of these species with a single strain. Similarly, Sielezniew et al. (2012) reported a 100 % prevalence of a single strain in the Polish and Lithuanian populations of M. alcon. Simultaneously, all studied individuals in the Polish and Italian populations of M. arion proved to be infected with one strain (Sielezniew et al. 2012; Patricelli et al. 2013). The Wolbachia 16S rRNA gene sequences were identical in all studied European populations in M. alcon and in M. arion, separately, but the two types of strains harboured by these species did not mix with each other in any particular case probably because there is cytoplasmic incompatibility between them. Interestingly, 19 populations of Euphydryas aurinia in the UK (Smee 2011) proved to be also infected in 100 % with the same strain as M. alcon in our study. Nevertheless, the phenotype of this Wolbachia strain has remained unknown in both host species.
The establishment of the Wolbachia’s effects on the hosts strongly requires experimental confirmation since one particular strain may cause the same (Dyson et al. 2002) or different (Fujii et al. 2001) reproductive alterations in different hosts. Namely, the phenotype seems to be dependent on interactions between the bacteria and the genomic make-up of the individual host. Without any experimental confirmation, we can only hypothesize the potential phenotypes of the strains based on the infection rate and the genetic diversity of host species. The infestation pattern, i.e. 100 % prevalence of a single strain each in M. alcon and M. arion refers to the perfect vertical transmission of Wolbachia in these hosts. Vertically transmitted endosymbionts are generally considered to be evolved towards mutualism because their evolutionary fate is closely linked to that of their hosts (Fine 1975; Yamamura 1993; Zug and Hammerstein 2014). Probably these particular strains were able to overcome these hosts’ immune system and rule out the other strains competitively through cytoplasmic incompatibility.
The effects of Wolbachia on these hosts’ gene diversity seem to be variable in the different European regions. The Polish and Lithuanian populations of M. alcon have a complete lack of mitochondrial sequence variation. Moreover, barcoding gene sequences proved to be identical from Western Europe to Kazakhstan and Kyrgyzstan (Sielezniew et al. 2012). The Polish populations of M. arion also have restricted mtDNA variation. However, mitochondrial haplotype diversity varies according to geographical regions in Italy. Generally, Italian populations share variable mitochondrial haplotypes apart from some Northern and Central Italian populations which have reduced diversity (Patricelli et al. 2013). In both species, the nuclear elongation factor 1α characterized by low substitution rate showed higher variability than the mitochondrial sequences whose evolutionary speed is commonly known to be high. On the contrary, in the Carpathian Basin, nuclear variation based on 1680 bases from three different gene regions (GAPDH, MDH and wingless) was lower even than the restricted mitochondrial diversity, thus the nuclear data were uninformative to reconstruct the phylogeography of these species. In addition, neutrality tests did not result in significantly negative values in any of these species. Therefore, we could not find evidence for Wolbachia-induced selective sweeps which arose in the previous studies in spite of the presence of a single strain and the highly reduced mitochondrial variation.
Additionally, it seems that not Wolbachia are responsible for the controversial patterns of mitochondrial and morphological variability of the different forms of M. arion. The results of the present study do not support the ‘one barcode–two species’ hypothesis. On the one hand, the allozymes do not show differentiation between the phenological forms. Although allozyme diversity of Maculinea species is generally low compared with other lycaenids (Schmitt et al. 2003; Schmitt and Hewitt 2004; Aagaard et al. 2002), it is suitable for species separation in this genus (Pecsenye et al. 2007). On the other hand, although these forms harbour the same strain, we could not detect mito-nuclear discordance and significant deviation from the neutral model of molecular evolution. One further possibility to explain the differentiation between the ‘spring and summer types’ of M. arion is their differential host ant usage, but we do not possess any data about it in the Carpathian Basin. Thus, there is an urgent need of information about host ants on this geographic scale. The two forms of M. alcon seem to differentiate only based on their initial host plant usage since the ‘cruciata and pneumonanthe type’ populations show a strong trend to differ on the basis of esterase locus which is known to be related to adaptation to host plants. In Colias butterflies, esterase D allozymes co-vary with food plant use, suggesting a role in detoxifying plants’ chemical defences (Burns 1975).
The infection level of M. nausithous and M. teleius is much lower than in M. alcon and M. arion. On the one hand, this means lower infection rate in both species. On the other hand, probably fewer Wolbachia are present in one individual because we could amplify the highly conservative 16S rRNA gene only in double PCRs. Additionally, these hosts—even in a single population of M. nausithous—are infected with multiple strains belonging to supergroup A or B. This infestation pattern refers to incomplete vertical transmission of Wolbachia in these host species and extensive horizontal transfer in/between them. Although several potential horizontal transfer routes can exist in all Maculinea species, e.g. transfer by Myrmica host ants being in regular physical contact with caterpillars (ant workers feed them by regurgitation or caterpillars feed on the ant brood) or transfer by parasitoids (in certain cases, the host’s immune system may neutralize the parasitoid larva, see Davies and Vinson 1986; Hu et al. 2003), the lifecycle of M. teleius and M. nausithous is closely related to each other in many respects. Females in both species lay their eggs on Sanguisorba officinalis. Besides, they can use the same host ants and even the same ant nest may harbour M. teleius and M. nausithous individuals since they often occur in the same habitat (Tartally 2008). Additionally, caterpillars can be parasitized by the same wasp species, e.g. Neotypus melanocephalus which infects the young larvae on the food plant (Shaw et al. 2009). Namely, extensive horizontal transfer routes can exist to mediate Wolbachia in both species (and between them) which corresponds with the presumably incomplete vertical transmission. In the conventional view, horizontal transmission favours parasitism (Anderson and May 1982), but we also need experimental studies to identify the phenotype of Wolbachia in these hosts.
As it is indicated by the significantly positive values of the neutrality tests in the case of Wolbachia 16S rRNA gene in these host species, there is likely to be strong selection on Wolbachia themselves, possibly exerted by host defensive mechanisms. Wolbachia strains can differ in their ability to transfect different hosts and not all host species are equally permissive (Werren et al. 2008). M. teleius and M. nausithous seem to be more resistant to Wolbachia than M. alcon and M. arion which is inferred from their much lower infection rate and the attempted invasion of multiple strains into these hosts. Probably different strains try to colonize these hosts and they could compete with each other.
Previously, cryptic speciation has been hypothesized for M. teleius and M. nausithous, based on deep mitochondrial split in each of these species (Ugelvig et al. 2011; Pech et al. 2004; Als et al. 2004). Ritter et al. (2013) tested the theory of cryptic speciation on a comprehensive sample across the Palaearctic ranges and revealed that deep mitochondrial divergence did not correspond with microsatellite data but was concordant with Wolbachia infection in both species. Haplotypes previously attributed to cryptic species were part of Wolbachia-infected clades thus deep intraspecific divergences found in DNA barcode studies coincide with specific infection patterns. In contrast with these results, in the Carpathian Basin, mitochondrial haplotypes of specimens infected with Wolbachia formed separate clades neither in M. nausithous nor in M. teleius. Although the genetic diversity was the highest in M. nausithous, the phylogenetic and allozyme patterns mainly reflect the biogeographical history of the species since the great part of the variability arises from the differentiation of the large geographic regions which coincides the disjunct distribution of M. nausithous. Namely, M. nausithous only occurs on the Western and the Eastern edges of the Carpathian Basin with a large distributional gap in the centre where the species is missing. Interestingly, we could not detect this pattern in the nuclear genome maybe because of data deficiency (we could not amplify nuclear sequences from the Eastern Carpathian clade at all because of DNA degradation). The structured haplotype network reflects complex phylogeographical pattern in M. nausithous. Beside the various Asian haplotypes, the European populations are also divided to distinct haplogroups which suggest that the species survived during Pleistocene ice ages within different European glacial refugia. At the same time, one of the Western European haplogroups is interposed between the Asian and Wolbachia-dominated clade. Although, Ritter et al. (2013) could not detect current infection in this group (Western Europe 2), it is possible that the pronounced genetic distance between the two Western European haplogroups is the result of a historical infection. Therefore, it seems that biogeographical factors and Wolbachia infestation could shape together the genetic structure of M. nausithous. Simultaneously, the diversity of M. teleius was very low on the basis of all the studied genetic marker sets and neutrality tests did not lead to significantly negative results. The haplotype network that includes specimens from the whole Palearctic shows the higher diversity of the species in Eastern Asia. This elevated genetic variability refers to isolated refugial areas in this region owing to the complex topography. Simultaneously, the considerable differentiation of Wolbachia-dominated clade indicates the effect of the infestation on the genetic diversity of M. teleius although further studies need to discover the exact effects of Wolbachia on this host.
In summary, our research provides a comprehensive description of the Wolbachia infestation and the genetic diversity of Maculinea butterflies in the Carpathian Basin. Based on our results, we cannot hold Wolbachia responsible for the highly reduced genetic diversity of the target species exclusively. Probably several factors shape together the level and pattern of genetic variability in Maculinea butterflies, e.g. biogeography and/or population dynamics. Wolbachia are only one of these factors but their importance is very high not only in terms of evolutionary but also conservation biology. A case study about North American Lycaeides butterfly species emphasizes that Wolbachia has to be considered before introducing individuals from one population to another because they may cause failure of introduction attempts due to CI (Nice et al. 2009). Introducing individuals from a source population infected with Wolbachia to another population infected with another strain can destroy the population that we wanted to save originally. Maculinea species are endangered and their reintroduction has been also achieved several times (Wynhoff 1998; Andersen et al. 2014; Thomas et al. 2009). Therefore, we cannot work out and execute reintroduction action plans responsibly without considering Wolbachia infection.
References
Aagaard, K., Hindar, K., Pullin, A. S., James, C. H., Hammarstedt, O., Balstad, T., et al. (2002). Phylogenetic relationships in brown argus butterflies (Lepidoptera: Lycaenidae: Aricia) from northwestern Europe. Biological Journal of the Linnean Society, 75(1), 27–37. doi:10.1046/j.1095-8312.2002.00004.x.
Als, T. D., Vila, R., Kandul, N. P., Nash, D. R., Yen, S.-H., Hsu, Y.-F., et al. (2004). The evolution of alternative parasitic life histories in large blue butterflies. Nature, 432(7015), 386–390. doi:10.1038/nature03020. http://www.nature.com/nature/journal/v432/n7015/suppinfo/nature03020_S1.html.
Andersen, A., Simcox, D. J., Thomas, J. A., & Nash, D. R. (2014). Assessing reintroduction schemes by comparing genetic diversity of reintroduced and source populations: a case study of the globally threatened Large Blue butterfly (Maculinea arion). Biological Conservation, 175, 34–41. doi:10.1016/j.biocon.2014.04.009.
Anderson, R. M., & May, R. M. (1982). Coevolution of hosts and parasites. Parasitology, 85(02), 411–426. doi:10.1017/S0031182000055360.
Aris-Brosou, S., & Excoffier, L. (1996). The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism. Molecular Biology and Evolution, 13(3), 494–504.
Baldo, L., Dunning Hotopp, J. C., Jolley, K. A., Bordenstein, S. R., Biber, S. A., Choudhury, R. R., et al. (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Applied and Environmental Microbiology, 72(11), 7098–7110. doi:10.1128/aem.00731-06.
Bereczki, J., Pecsenye, K., Peregovits, L., & Varga, Z. (2005). Pattern of genetic differentiation in the Maculinea alcon species group (Lepidoptera, Lycaenidae) in Central Europe. Journal of Zoological Systematics and Evolutionary Research, 43(2), 157–165. doi:10.1111/j.1439-0469.2005.00305.157-165.
Bereczki, J., Tóth, J. P., Tóth, A., Bátori, E., Pecsenye, K. & Varga, Z. (2011). Genetic structure of phenologically differentiated Large Blue (Maculinea arion) populations (Lepidoptera: Lycaenidae) in the Carpathian Basin. European Journal of Entomology, 108(4), 519−527.
Bereczki, J., Tóth, J. P., Sramkó, G., & Varga, Z. (2014). Multilevel studies on the two phenological forms of large blue (Maculinea arion) (Lepidoptera: Lycaenidae). Journal of Zoological Systematics and Evolutionary Research, 52(1), 32–43. doi:10.1111/jzs.12034.
Burns, J. M. (1975). Isozymes in evolutionary systematics. In C. L. Markert (Ed.), Isozymes IV: genetics and evolution (pp. 49–62). New York: Academic.
Cavalli-Sforza, L. L., & Edwards, A. W. F. (1967). Phylogenetic analysis: models and estimation procedures. Evolution, 21, 550–570.
Davies, H. D., & Vinson, S. B. (1986). Passive evasion by eggs of braconid parasitoid Cardiochiles nigriceps of encapsulation in vitro by haemocytes of host Heliothis virescens. Possible role for fibrous layer in immunity. Journal of Insect Physiology, 32(12), 1003–1010. doi:10.1016/0022-1910(86)90119-8.
Dupuis, J. R., Roe, A. D., & Sperling, F. A. H. (2012). Multi-locus species delimitation in closely related animals and fungi: one marker is not enough. Molecular Ecology, 21(18), 4422–4436. doi:10.1111/j.1365-294X.2012.05642.x.
Dyson, E. A., Kamath, M. K., & Hurst, G. D. D. (2002). Wolbachia infection associated with all-female broods in Hypolimnas bolina (Lepidoptera: Nymphalidae): evidence for horizontal transmission of a butterfly male killer. Heredity, 88(3), 166–171.
Earl, D., & vonHoldt, B. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361. doi:10.1007/s12686-011-9548-7.
Evanno, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology, 14(8), 2611–2620. doi:10.1111/j.1365-294X.2005.02553.x.
Fine, P. E. M. (1975). Vectors and vertical transmission: an epidemiologic perspective. Annals of the New York Academy of Sciences, 266(1), 173–194. doi:10.1111/j.1749-6632.1975.tb35099.x.
Fujii, Y., Kageyama, D., Hoshizaki, S., Ishikawa, H., & Sasaki, T. (2001). Transfection of Wolbachia in Lepidoptera: the feminizer of the adzuki bean borer Ostrinia scapulalis causes male killing in the Mediterranean flour moth Ephestia kuehniella. Proceedings of the Royal Society of London, Series B: Biological Sciences, 268(1469), 855–859. doi:10.1098/rspb.2001.1593.
Galtier, N., Nabholz, B., Glémin, S., & Hurst, G. D. D. (2009). Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Molecular Ecology, 18(22), 4541–4550. doi:10.1111/j.1365-294X.2009.04380.x.
Hammer, Ø., Harper, D. A. T., & Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Paleontologica Electronica, 4(1), 9.
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., & Werren, J. H. (2008). How many species are infected with Wolbachia? – a statistical analysis of current data. FEMS Microbiology Letters, 281(2), 215–220. doi:10.1111/j.1574-6968.2008.01110.x.
Hoffmann, A. A., Hercus, M., & Dagher, H. (1998). Population dynamics of the Wolbachia infection causing cytoplasmic incompatibility in Drosophila melanogaster. Genetics, 148(1), 221–231.
Hu, J., Zhu, X.-X., & Fu, W.-J. (2003). Passive evasion of encapsulation in Macrocentrus cingulum Brischke (Hymenoptera: Braconidae), a polyembryonic parasitoid of Ostrinia furnacalis Guenée (Lepidoptera: Pyralidae). Journal of Insect Physiology, 49(4), 367–375. doi:10.1016/S0022-1910(03)00021-0.
Jiggins, F. M. (2003). Male-killing Wolbachia and mitochondrial DNA: selective sweeps, hybrid introgression and parasite population dynamics. Genetics, 164(1), 5–12.
Kodandaramaiah, U., Simonsen, T. J., Bromilow, S., Wahlberg, N., & Sperling, F. (2013). Deceptive single-locus taxonomy and phylogeography: Wolbachia-associated divergence in mitochondrial DNA is not reflected in morphology and nuclear markers in a butterfly species. Ecology and Evolution, 3(16), 5167–5176. doi:10.1002/ece3.886.
Librado, P., & Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25(11), 1451–1452. doi:10.1093/bioinformatics/btp187.
Nei, M. (1987). Molecular evolutionary genetics. New York: Columbia University Press.
Nei, M., & Li, W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences, 76(10), 5269–5273.
Nice, C. C., Gompert, Z., Forister, M. L., & Fordyce, J. A. (2009). An unseen foe in arthropod conservation efforts: the case of Wolbachia infections in the Karner blue butterfly. Biological Conservation, 142(12), 3137–3146. doi:10.1016/j.biocon.2009.08.020.
Paradis, E. (2010). PEGAS: an R package for population genetics with an integrated-modular approach. Bioinformatics, 26(3), 419–420. doi:10.1093/bioinformatics/btp696.
Patricelli, D., Sielezniew, M., Ponikwicka-Tyszko, D., Ratkiewicz, M., Bonelli, S., Barbero, F., et al. (2013). Contrasting genetic structure of rear edge and continuous range populations of a parasitic butterfly infected by Wolbachia. BMC Evolutionary Biology, 13(1), 14.
Peakall, R. O. D., & Smouse, P. E. (2006). Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6(1), 288–295. doi:10.1111/j.1471-8286.2005.01155.x.
Pech, P., Fric, Z., Konvička, M., & Zrzavý, J. (2004). Phylogeny of Maculinea blues (Lepidoptera: Lycaenidae) based on morphological and ecological characters: evolution of parasitic myrmecophily. Cladistics, 20(4), 362–375. doi:10.1111/j.1096-0031.2004.00031.x.
Pecsenye, K., Bereczki, J., Tihanyi, B., Tóth, A., Peregovits, L., & Varga, Z. (2007). Genetic differentiation among the Maculinea species (Lepidoptera: Lycaenidae) in eastern Central Europe. Biological Journal of the Linnean Society, 91(1), 11–21.
Pritchard, J. K., Stephens, M., & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155(2), 945–959.
QGIS Development Team (2014). QGIS Geographic Information System. Open Source Geospatial Foundation Project.
R Development Core Team. (2014). R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Ritter, S., Michalski, S. G., Settele, J., Wiemers, M., Fric, Z. F., Sielezniew, M., et al. (2013). Wolbachia infections mimic cryptic speciation in two parasitic butterfly species, Phengaris teleius and P. nausithous (Lepidoptera: Lycaenidae). Plos One, 8(11), e78107. doi:10.1371/journal.pone.0078107.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61(3), 539–542. doi:10.1093/sysbio/sys029.
Schmitt, T., & Hewitt, G. M. (2004). The genetic pattern of population threat and loss: a case study of butterflies. Molecular Ecology, 13(1), 21–31. doi:10.1046/j.1365-294X.2004.02020.x.
Schmitt, T., Giessl, A., & Seitz, A. (2003). Did Polyommatus icarus (Lepidoptera: Lycaenidae) have distinct glacial refugia in southern Europe? Evidence from population genetics. Biological Journal of the Linnean Society, 80(3), 529–538. doi:10.1046/j.1095-8312.2003.00261.x.
Shaw, M. R., Stefanescu, C., & Van Nouhuys, S. (2009). Parasitoids of European butterflies. In J. Settele, T. G. Shreeve, M. Konvicka, & H. Van Dyck (Eds.), Ecology of butterflies in Europe (pp. 130–156). Cambridge: Cambridge University Press.
Shoemaker, D., Keller, G., & Ross, K. G. (2003). Effects of Wolbachia on mtDNA variation in two fire ant species. Molecular Ecology, 12(7), 1757–1771. doi:10.1046/j.1365-294X.2003.01864.x.
Sielezniew, M., Rutkowski, R., Ponikwicka-Tyszko, D., Ratkiewicz, M., Dziekańska, I., & Švitra, G. (2012). Differences in genetic variability between two ecotypes of the endangered myrmecophilous butterfly Phengaris (=Maculinea) alcon– the setting of conservation priorities. Insect Conservation and Diversity, 5(3), 223–236. doi:10.1111/j.1752-4598.2011.00163.x.
Smee, M. R. (2011). Population ecology and genetics of the marsh fritillary butterfly Euphydryas aurinia. Exeter: University of Exeter.
Sneath, P. H., & Sokal, R. R. (1973). Numerical taxonomy. San Francisco: W. H. Freeman.
Stephens, M., & Donnelly, P. (2003). A comparison of Bayesian methods for haplotype reconstruction from population genotype data. The American Journal of Human Genetics, 73(5), 1162–1169. doi:10.1086/379378.
Stephens, M., Smith, N. J., & Donnelly, P. (2001). A new statistical method for haplotype reconstruction from population data. The American Journal of Human Genetics, 68(4), 978–989. doi:10.1086/319501.
Tajima, F. (1983). Evolutionary relationship of DNA sequences in finite populations. Genetics, 105(2), 437–460.
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123(3), 585–595.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12), 2725–2729. doi:10.1093/molbev/mst197.
Tartally, A. (2008). Myrmecophily of Maculinea butterflies in the Carpathian Basin (Lepidoptera: Lycaenidae). PhD, University of Debrecen
Thomas, J. A., Simcox, D. J., & Clarke, R. T. (2009). Successful conservation of a threatened Maculinea butterfly. Science, 325(5936), 80–83. doi:10.1126/science.1175726.
Turelli, M. (1994). Evolution of incompatibility-inducing microbes and their hosts. Evolution, 48(5), 1500–1513. doi:10.2307/2410244.
Turelli, M., & Hoffmann, A. A. (1991). Rapid spread of an inherited incompatibility factor in California Drosophila. Nature, 353(6343), 440–442. doi:10.1038/353440a0.
Turelli, M., & Hoffmann, A. A. (1995). Cytoplasmic incompatibility in Drosophila simulans: dynamics and parameter estimates from natural populations. Genetics, 140(4), 1319–1338.
Ugelvig, L. V., Vila, R., Pierce, N. E., & Nash, D. R. (2011). A phylogenetic revision of the Glaucopsyche section (Lepidoptera: Lycaenidae), with special focus on the Phengaris–Maculinea clade. Molecular Phylogenetics and Evolution, 61(1), 237–243. doi:10.1016/j.ympev.2011.05.016.
Wahlberg, N., & Wheat, C. W. (2008). Genomic outposts serve the phylogenomic pioneers: designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Systematic Biology, 57(2), 231–242. doi:10.1080/10635150802033006.
Watterson, G. A. (1975). On the number of segregating sites in genetical models without recombination. Theoretical Population Biology, 7(2), 256–276. doi:10.1016/0040-5809(75)90020-9.
Werren, J. H., & Windsor, D. M. (2000). Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proceedings of the Royal Society of London, Series B: Biological Sciences, 267(1450), 1277–1285. doi:10.1098/rspb.2000.1139.
Werren, J. H., Baldo, L., & Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nature Reviews Microbiology, 6(10), 741–751. doi:10.1038/nrmicro1969.
Wynhoff, I. (1998). Lessons from the reintroduction of Maculinea teleius and M. nausithous in the Netherlands. Journal of Insect Conservation, 2(1), 47–57. doi:10.1023/A:1009692723056.
Yamamura, N. (1993). Vertical transmission and evolution of mutualism from parasitism. Theoretical Population Biology, 44(1), 95–109. doi:10.1006/tpbi.1993.1020.
Zug, R., & Hammerstein, P. (2014). Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biological Reviews. doi:10.1111/brv.12098.
Acknowledgments
The study was supported by the Hungarian Scientific Research Fund (OTKA K109223 and K84071), János Bolyai Scholarship of the Hungarian Academy of Sciences, EU EESD-ENV-2OO2-NAS (Mac Man) project and Social Renewal Operational Programme (TÁMOP-4.2.2/B-10/1-2010-0024). Grateful acknowledgments are due to S. Szabó, A. Tartally, T. Korompai, Z. Ilonczay, A. Ambrus, L. Peregovits, L. Rákosy, F. Rebousek and R. Verovnik for collecting samples. The technical assistance of V. Mester, T. Tóth, K. Kecsmár, H. Boros and B. Prokaj in the molecular work is very much respected. The support of the Nature Conservation Authorities is also greatly appreciated.
Data accessibility
DNA sequences: GenBank accessions: HG326619-HG326646, KM517249-KM517565
Sampling locations and data uploaded as online Supporting Information.
Allozyme genotypes: Dryad doi: 10.5061/dryad.v8q8r
Wolbachia WSP and MLST sequence information uploaded into Wolbachia MLST database.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Additional supporting information may be found in the online version of this article at the publisher’s website.
Online Resource 1
Sampling sites. Abb. – abbreviations of localities. Region – the geographic region from which the samples originate. The table includes sampling time, the number of Maculinea individuals tested for Wolbachia in parentheses and the number of individuals used in allozyme studies in bold. (PDF 238 kb)
Online Resource 2
GenBank accession numbers of the sequenced mitochondrial and nuclear genes of Maculinea butterflies as well as 16S rRNA genes of Wolbachia found in Maculinea species. ID – individual identifiers of Maculinea specimens. The infected individuals are indicated with W. Specimens from which Wolbachia strain identification was carried out are indicated with ‘+’. WSP – Wolbachia surface protein, MLST – Multilocus Sequence Typing based on five genes (gatB, coxA, hcpA, ftsZ, fbpA). (PDF 63 kb)
Online Resource 3
Results of Bayesian inference analyses of single nuclear gene datasets in Maculinea. (a) GAPDH, (b) MDH, (c) wg. (PDF 31 kb)
Online Resource 4
Results of the Bayesian-clustering Structure analysis based on 11 allozyme loci. The most probable K values are indicated in the case of each Maculinea species. See the abbreviations of populations in Online Resource 1. M. alcon: ‘pneumonanthe type’ populations are in bold. M. arion: ‘spring type’ populations are in bold. Populations belonging to the same geographic region are joined together on the upper part of each barplot. (PDF 209 kb)
Online Resource 5
COI haplotype networks for Maculinea nausithous (a) and M. teleius (b) including the sequences of Ritter et al. (2013). Colours show the geographic regions from where haplotypes originate. Circle size is proportional to haplotypes frequency. Dots on lines linking haplotypes indicate the number of mutations. A separate Wolbachia clade is present in both species but there are numerous infected individuals outside this clade. (PDF 98 kb)
Rights and permissions
About this article
Cite this article
Bereczki, J., Rácz, R., Varga, Z. et al. Controversial patterns of Wolbachia infestation in the social parasitic Maculinea butterflies (Lepidoptera: Lycaenidae). Org Divers Evol 15, 591–607 (2015). https://doi.org/10.1007/s13127-015-0217-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-015-0217-7