Abstract
Cervical cancer is the third most common gynecologic cancer in the world. Exploration of the molecular mechanism underlying cervical cancer pathogenesis will provide new insights into the development of novel therapies. In this study, we were aimed to characterize a novel miRNA in cervical cancer tumorigenesis. First, we measured the expressional change of miR-144-3p in clinical tissues and cancer cells. Second, we employed cell proliferation, cell migration, and invasion assays to understand its functional role in cervical cancer. Then, we confirmed in vitro findings in xenograft cancer model. Last, we mapped out a downstream target of miR-144-3p and validated its functional role in cancer cells. In the results, miR-144-3p was found significantly downregulated in cervical cancer cells and tissues. Over-expressing miR-144-3p suppressed cancer cells growth and metastasis. Consistent with in vitro results, over-expressing miR-144-3p led to tumor growth inhibition in vivo. Further on, MAPK6 was identified as an endogenous target of miR-144-3p in cervical cancer. Knocking down MAPK6 inhibited cervical cancer cells proliferation, migration, and invasion potential. Our investigation was the first time to report miR-144-3p as a tumor suppressive miRNA in cervical cancer. It inhibited tumor growth by targeting MAKP6. The newly identified signalling axis may serve as novel therapeutic targets to manage cervical cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervical cancer is one of the major causes of cancer death among the women worldwide. About 90% of the deaths caused by cervical cancer were reported from developing countries. It is estimated about 530,000 new cases diagnosed annually. The average age as being diagnosed is 47 years [23]. Most cervical cancers are caused by human papilloma virus (HPV) infection. Viral DNA can integrate into the host genome DNA to activate oncogenic transformation. There are more than 100 subtypes of HPV. Among them, HPV16 and HPV18 are most commonly associated with the occurrence of cervical cancer. Other than HPV infection, age, smoking, using oral contraceptives, and exposure to diethylstilboestrol may be associated with increased risk of developing cervical cancer [5, 12].
It takes 10–15 years before cervical intraepithelial neoplasia turns into invasive cancer. Detecting the cervical neoplasia at the early stage usually lead to a better management to prevent and treat the development of cervical cancer. The discovery of the link between HPV infection and cervical cancer leads the invention of prophylactic vaccination. Since then, the prophylactic vaccination is becoming the primary prevention approach to reduce the morbidity and mortality of cervical cancer [12, 20]. The standard treatments for cervical cancer patients (FIGO stage IA-IB1) include lymph node dissection, radical hysterectomy, and radiation alone or with chemotherapy. For patients with cervical cancer at advanced stage, the most effective approaches are external beam radiotherapy in combination with cisplatin-based chemotherapy. However, in many developing countries, the current 3-year to 5-year survival rate in cervical cancer patients, for all stages combined, is still less than 50%. To further understand the biological mechanisms regulating the tumorigenesis of the cervical tissue will inspire more prevention or treatment strategies to improve the cure rate of cervical cancer in all stages [20, 23].
The key HPV proteins that induce the oncogenesis of host cells are E6 an E7. The expression of E6 and E7 in host cells inactivates p53 and pRB pathways to initiate the tumorigenesis. In addition to these classic pathological mechanisms, mounting evidence has suggested epigenetic regulation also plays a key role cervical carcinogenesis. These oncogenic events at epigenetic level include DNA methylation or histone modification that alter the transcriptional regulation of oncogenic or tumor suppressive genes or non-coding RNAs including lncRNAs or miRNAs that regulate post-transcriptionally the key signalling molecules in controlling the growth or development of tumors in cervical tissue [9]. MiRNAs are small non-coding RNA molecules containing about 22 nucleotides. They function in RNA silencing and post-transcriptional regulation. The dysregulated miRNA expression has been reported frequently as being associated with tumorigenesis mechanism in various cancers [11, 21]. Many newly identified miRNAs in cancers have been proposed as potential biomarkers or intervention targets. In cervical cancer, aberrant miRNA expression profiles have been revealed in multiple studies [6]. Functional studies have confirmed many miRNAs such as miR-10a, miR-21, miR-196a, miR-125a, or miR-183 can act as either oncogenic or tumor-suppressive factors to control cervical carcinogenesis [7, 8, 13, 18, 27]. In addition, HPV infection may also module the expression of miRNAs in host cells that forms an epigenetic mechanism for tumor transformation [22].
In our study, we investigated miR-144-3p, as a new epigenetic factor in regulating tumorigenesis of cervical cancer. The expressional level of the miRNA in cervical cancer was first evaluated in cells and patient tissues. Functional characterization of the miRNA was done with cancer cell lines and in vivo xenograft model. We also proposed a novel endogenous target of miR-144-3p here. The functional significance of this novel factor in cervical cancer was established in the cancer cell model.
Methods
Cell culture
Human cervical cancer lines Hela, C33A, HT-3, ME-180, HCC94, and MS751 were obtained from ATCC. They were grown in DMEM supplemented with 10% fetal bovine serum at 5% CO2/37 °C. Primary normal cervical squamous cells (NCSC) were isolated from adjacent non-tumor sites of cervical tissues from the patients with informed consent. They were grown in serum-free keratinocyte growth medium supplemented with bovine pituitary extract, epithelial growth factor, and antibiotics (Invitrogen, Waltham, MA, USA). The cells were passaged every 2 to 3 days. Short-tandem repeat profiling of human cell lines was performed to confirm the identity of the cells. No cross-contaminated or genetically drifted cells were found.
Clinical cervical cancer tissue specimens
Clinical evaluation of miR-144-3p expression in human cervical tumor tissues was approved by the Ethical Committee of Heze Municipal Hospital. A total of 23 cancer patients were enrolled and the informed consent was obtained. The tumor and the adjacent non-tumor tissues were harvested for the pair comparison.
Real-time PCR analysis
Total RNA was extracted using Trizol method, and cDNA library was made using high capacity cDNA archive kit from Applied Biosystems according the standard protocol. Transcript expression of miR-144-3p (Forward: 5′-ACACTCCAGCTGGGTACAGTA-3′; Reverse, 5′-CTCAACTGGTGTCGTG-5′) and U6 (Forward: 5′-CTCGCTTCGGCAGCACA-3′; Reverse: 5′-AACGCTTCACGAATTTGCGT-3′) was measured by SYBR green real-time PCR. The real-time PCR was performed on QuantStudio real-time PCR platform (Applied Biosystems, Waltham, MA, USA) following manufacturer’s instruction.
miRNA mimic and siRNAs
miRNA-144-3p mimic and the scramble control were ordered from mirVana™ Libraries (Thermo Fisher Scientific, Waltham, MA, USA). miRNAs were transfected into cells with Lipofectamine 2000 reagent following the manufacturer’s manual. siRNA against MAPK6 and scramble control were synthesized and validated by Dharmacon. The siRNA was transfected with Lipofectamine® RNAiMAX reagent according to manufacturer’s instruction.
Cell proliferation assay
The cell growth rate was monitored with Cell Counting Kit-8 following manufacturer’s instruction. Briefly, 6000 cells were plated into a single well of a 96-well plate. CCK-8 reagent was added to the well 3 h before the measurement. The cell proliferation curves were plotted by determining the absorbance at 450 nm in a plate reader at the indicated time point.
Cell migration assay
The assay was performed in CytoSelect™ 24-Well Cell Migration kit from CellBiolabs following the manufacturer’s instruction. Briefly, cells were placed at 25,000 cells per well in the top chamber. Cell growth medium–supplemented 10% FBS was used as a chemoattractant in the bottom chamber. Twenty-four hours later, migratory cells passing through polycarbonate membrane into the lower well were quantified using CyQuant® GR fluorescent dye in a plate reader.
Cell invasion assay
The assay was performed in CytoSelect™ 24-Well Cell Invasion kit from CellBiolabs following the manufacturer’s instruction. The upper surface of the insert membrane is coated with a uniform layer of dried Bovine Type I Collagen matrix. Briefly, cells were placed at 25,000 cells per well in the top chamber. Cell growth medium–supplemented 10% FBS was used as a chemoattractant in the bottom chamber. Twenty-four hours later, the invaded cells into the insert membrane were stained and quantified in a plate reader at OD 560 nm.
Luciferase assay
We cloned the 3′-UTR of MAPK6 containing predicted miR-144-3p targeting site (Fig. 3a) to the downstream of the firefly luciferase gene in the vector (pMIR-REPORT) as denoted WT. And we mutated the miR-144-3p targeting sequence in WT to create MUT vector (Fig. 3a).
miR-144-3p mimics or scramble control were transfected into Hela cells expressing either WT or MUT and Renilla Luciferase vector (pRL-SV40). The luciferase activities were quantified by the Dual-Luciferase® Reporter (DLR™) Assay System in a microtiter reader according to standard protocol.
Western-blotting analysis
Protein expression was analyzed by western-blotting based on standard method. Briefly, cells were collected in RIPA buffer supplemented with proteases inhibitors cocktail. Protein concentration was determined by BCA assay. Forty micrograms of protein lysate was denatured and heated in LDS sample buffer (Life Technologies, Pleasanton, CA, USA) and fractionated in 12% SDS/PAGE. The fractionated protein was then transferred to a nitrocellulose membrane. The membranes were probed with the primary antibodies in 1:1000 dilution. The target protein level was measured with SuperSignal West Pico PLUS chemiluminescent kit. MAPK6 and ACTINB antibodies were obtained from Cell Signaling.
Xenograft experiment in mice
The experimental procedure was described in Tang et al. [24]. Briefly, Hela cells were transfected with either miR-144-3p or its scramble. Twenty-four hours later, the cells were harvested and counted. After washed with PBS, the cells were suspended at a concentration of 2 × 107 cells/ml. Rag (−/−) mice aged 6 weeks were used for our xenograft study. The cells of 50 μl were injected to either flank of the subject subcutaneously: one side for miR-144-3p transfected cells and the other side for the control. The mice were fed and monitored for 3 weeks before sacrificed. The tumors were harvested and measured by weight. The animal studies complied with Guide for the Care and Use of Laboratory Animals (8th edition, NIH) and was approved by the Animal Study Ethics Committee of Heze Municipal Hospital.
Statistical analysis
The data were shown as the means ± SDs. The differences between the groups were analyzed using Student’s t test. If more than two experimental groups were involved, the experimental differences were analyzed by one-way ANOVA analysis. Only p < 0.05 was considered significant.
Results
miRNA-144-3p was downregulated in cervical cancer cells and tumor tissues
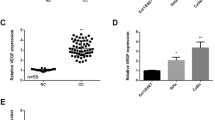
To examine the level of miR-144-3p in cervical cancer, we firstly performed real-time PCR analysis of miR-144-3p in six common human cervical cancer cell lines: Hela, C33A, HT-3, ME-180, HCC94, and MS751. We also included three primary normal cervical squamous cells (NCSC) to establish the expressional baseline of miR-144-3p. As shown (Fig. 1a), miR-144-3p was downregulated significantly in all six cancer cell lines. In Hela and HCC94 cells, miR-144-3p was reduced by more than twofold. To validate downward trend of miR-144-3p expression in cervical cancer, we performed tissue analysis on 23 human cervical cancer samples. The change was determined by comparing the miRNA expression at the tumor sites with that from the adjacent non-tumor tissues. As shown (Fig. 1b), the normalized expression of miR-144-3p showed a significant reduction in tumor tissues. The studies in cells and cancer tissues both indicated a potential role of miR-144-3p in tumorigenesis of cervical cancer.
miRNA-144-3p is suppressed in cervical cancer cells and tumor tissues. a The comparison of miRNA-144-3p expression levels in cervical cancer lines (Hela, HT-3, C33A, ME-180, HCC94, and MS751) and primary normal cervical squamous cells (NCSC) from three donors. The representative result from three independent experiments was shown. b The expressional levels of miR-144-3p in 23 cervical tumor tissues and the matched non-tumor tissues were quantified by real-time PCR. (**p < 0.01, significantly difference from the control in a pairwise Student’s t test)
miRNA-144-3p inhibited cervical cancer in vitro
Next, we proceeded to characterize the function of miR-144-3p in cervical cancer. Since miR-144-3p expressed at lowest levels in Hela and HCC94, we chose these two lines as the experimental models and restore the expression of miR-144-3p by transfecting miR-144-3p mimic. First, we examined the cell growth through 72 h. As shown (Fig. 2a), the expression of miR-144-3p markedly reduced the proliferating rates of Hela and HCC94. Second, we examined their migratory potential in a trans-well system without matrix layer. As shown (Fig. 2b), the migratory capabilities of Hela and HCC94 were greatly reduced by miR-144-3p over-expression. Last, we also measured the invasive properties of the transfected cells. Since collagens are the most constituents in cervical ECM tissue, we chose type I collagen-based matrix to evaluate the cells invasive potential. As shown (Fig. 2c), the percentages of number of invasive cells in the matrix were significantly decreased in both lines transfected with miR-144-3p. The representative staining figures were shown in Fig. 2d. In summary, these results indicated miR-144-3p inhibited the growth and metastatic potential of cervical tumor cells.
miRNA-144-3p inhibits cervical cancer in vitro. a Cell proliferation study of Hela or HCC94 transfected with either miRNA-144-3p or miRNA scramble control. Forty-eight hours after transfection, the growth rate of cells was monitored using CCK-8 kit for 72 h. b Cell migration study of Hela or HCC94 transfected with either miRNA-144-3p or miRNA scramble control. The cell migration potential was measured with CytoSelect™ 24-Well Cell Migration assay. The migrated cells were stained with CyQuant® GR dye and quantified in a fluorescence plate reader. c Cell invasion potential of transfected cells was measured with CytoSelect™ 24-well cell invasion assay with type I collagen-based matrix. The cells in the membrane were stained and quantified at OD 560 nm. d The representative image of invasive cells at the bottom of the matrix membrane. The representative results from at least three independent experiments were shown. The data were presented as means ± S.D. of at least three independent experiments. (*p < 0.05, **p < 0.01, ***p < 0.001 as compared with control)
miRNA-144-3p inhibited cervical cancer in vivo
To study the anti-tumor effect of miR-144-3p in vivo, we sought to use xenograft model. Hela cells were transfected with either control miRNA or miR-144-3p. The transfected cells were then inoculated in nude mice. Three weeks later, the tumor were collected and weighted. As shown (Fig. 3a), average tumor size from the sites inoculated with Hela cells expressing miR-144-3p was markedly smaller than that inoculated with cells transfected with miRNA control. The representative image comparing the tumors from two different inoculation conditions is shown in Fig. 3b. Therefore, in vivo experiment further supported the tumor suppressive function of miR-144-3p in cervical cancer model.
miRNA-144-3p inhibits cervical cancer in vivo. a Statistic analysis of tumor growth of Hela xenograft in mice. Hela cells transfected with miR-144-3p or miRNA scramble control were inoculated on either flank of the same nude mice (n = 6). Three weeks later, xenografts were excised and weighted. b The comparison of tumors excised from xenograft models. The representative image was shown. The data were presented as means ± S.D. of at least three independent experiments. (*p < 0.05, **p < 0.01, ***p < 0.001 as compared with control)
MAPK6 was identified as downstream target of miR-144-3p
Next, we sought to elucidate the downstream target of miR-144-3p in cancer cells. The prediction result from TargetScan [1] suggested 3′UTR of MAPK6 carries a conserved miR-144-3p binding site (Fig. 4a). To test there is a direct binding of miR-144-3p to 3′UTR of MAPK6, we cloned the 3′UTR of MAPK6 containing the miRNA targeting site to the downstream of luciferase reporter gene–denoted WT vector. In comparison, the miRNA targeting site was mutated in MUT vector. We co-transfected Hela cells with the luciferase gene reporters (WT or MUT) with either scramble or miR-144-3p. As shown (Fig. 4b), luciferase gene activity from WT vector was suppressed greatly by miR-144-3p but not scramble miRNAs, suggesting miRNA-144-3p bound directly to 3′UTR of MAPK6. In MUT vector, miR-144-3p failed to inhibit the luciferase activity, suggesting the binding of miRNA to 3′UTR was interrupted by the mutation. In the next experiment, we proceeded to examine the expressional regulation of miRNA-144-3p on MAPK6 at protein level. Hela cells were transfected with miR-144-3p or control miRNA. The MAPK6 expression was then measured by western blotting. Agreeing with the result from luciferase assay, miR-144-3p downregulated protein expression of MAPK6 in cells (Fig. 4c). Taken together, we confirmed MAPK6 was an endogenous target of miRNA-144-3p.
MAPK6 is an endogenous target of miR-144-3p in cervical cancer. a Schematic diagram of a conserved putative miR-144-3p targeting site in 3′UTR of MAPK6 (WT). The site mutations were introduced to miR-144-3p targeting site of 3′UTR (MUT). b Luciferase reporter assay study of the interaction between miR-144-3p and 3′UTR of MAPK6. Hela cells were first transfected with luciferase reporter genes conjugated with either wild-type (WT) or mutant (MUT) 3′UTR of MAPK6. Twenty-four hours later, the cells were transfected further with either miR-144-3p mimic or miRNA scramble control. Transfected cells were cultured for 48 h and then harvested for the measurement of luciferase activities in a plate reader. c Western blotting analysis of MAPK6 protein expression in cells transfected with miR-144-3p or miRNA scramble control. Beta-actin was used as loading control. The normalized densitometry analysis of MAPK6 expression was also shown. The data were presented as means ± S.D. of at least three independent experiments: (*p < 0.05, **p < 0.01, ***p < 0.001 as compared with control)
Last, we wanted to confirm MAPK6 also carried a functional role in tumorigenesis of cervical cancer. We transfected HCC94 cells with MAPK6 siRNA or scramble control. The knockdown efficiency was high as shown in Fig. 5a. The transfected cells were first monitored on their proliferation rates. As shown (Fig. 5b), knocking down MAPK6 reduced the proliferative potential of HCC94. Second, we proceeded to measure cell migration capacity. As shown (Fig. 5c), cells transfected with HCC94 showed reduced migration potential. Last but not least, knocking down MAPK6 also dampened cell invasion rate in the type I collagen–based matrix of the transwell system (Fig. 5d). In summary, these results suggested MAPK6 was an oncogenic factor in cervical cancer cells.
Knocking down MAPK6 inhibits cervical cancer cells. a Western blotting analysis of MAPK6 protein expression in cells transfected with MAPK6 siRNA or scramble control. Beta-actin was used as loading control. The normalized densitometry analysis of MAPK6 expression was also shown. b Cell proliferation study of HCC94 transfected with either MAPK6 siRNA or the scramble control. The growth rate of cells was measured using CCK-8 kit for 72 h. c Cell migration study of HCC94 transfected with either MAPK6 siRNA or the scramble control. The cell migration potential was measured with CytoSelect™ 24-Well Cell Migration assay. The migrated cells in the lower chamber were stained with CyQuant® GR dye and quantified in a fluorescence plate reader. d Cell invasion potential analysis of transfected cells was done with CytoSelect™ 24-well cell invasion assay with type I collagen-based matrix. The cells in the membrane were stained and quantified at OD 560 nm. The representative results from three independent experiments were shown. The data were presented as means ± S.D. of at least three independent experiments. (*p < 0.05, **p < 0.01, ***p < 0.001 as compared with control)
Discussion
In this report, we have characterized the novel function of miR-144-3p in cervical cancer. Both cell culture and animal studies suggest miR-144-3p could act as a tumor suppressor in the pathogenesis of cervical cancer. Over-expression of miR-144-3p inhibits cancer cells in culture and xenograft model. The suppression of miR-144-3p is also detected in both cancer lines and patients’ sample tissues. Further evaluation reveals MAPK6 is an endogenous target of miR-144-3p and may contribute to tumor cells migration and invasion. This novel signalling axis identified in cervical cancer provides a new insight from tumorigenesis to therapy.
The most characterized causative factor of cervical cancer is HPV infection. Mounting evidences also reveal the genetic and epigenetic changes in the host can also promote the cancer progression or initiate the tumorigenesis process. Aberrant miRNA expression has been associated with cervical cancer in multiple studies [3, 10]. Wang X et al. reported an aberrant miRNA expression profile containing and proposed a list of tumor-suppressive and oncogenic miRNA candidates in cervical cancer development [25]. In the study, miRNAs differentially expressed in cervical cancer were analyzed by microarray assay in five pairs of age-matched cancerous and normal cervical tissues. Among 455 miRNAs probed, 33 miRNAs that were reported changed significantly in the cancer tissues. Further on, the authors have validated candidates in cancer cell growth assay. For instance, miR-143 and miR-145 were found downregulated in cancer tissues and their over-expression suppressed cancer cell growth. In contrast, miR-146a was upregulated in tumor samples and its over-expression promoted cancer cell proliferation. Another study has profiled miRNA expression signature associated with cervical cancer prognosis [14]. The study was aimed to identify miRNAs as prognostic markers to predict cancer survival score. They used PCR-based miRNA assay to analyze 102 cervical tumor samples. In the study, miR-9 and miR-200a were identified to carry the potential predictive value for patient survival. In particular, over-expressing miR-200 reduced cell motility in transwell system, suggesting its regulatory role in cervical cancer metastasis. There is another comprehensive study to profile miRNA expression in invasive squamous cell carcinomas in early stage using TaqMan real-time PCR [15]. About 70 miRNAs showing significant differences were reported. Among the ten most significant miRNAs, miR-127 expressional change was implicated with lymph node metastasis. Subsequent functional characterization revealed inhibiting miR-199a-suppressed cancer cell proliferation. Taken together, miRNA deregulation may play a significant role through the stages of cervical tissue malignant transformation. The miRNA profiling studies provide a myriad of promising candidates to exploit as prognostic or therapeutic targets to treat cervical cancer.
We are the first time to report miR-144-3p carrying a significant function in cervical cancer formation and progression. This is consistent with the past studies that proposed miR-144-3p as an anti-tumor factor in various cancers. But the downstream target of the miRNA is cancer type specific suggesting a complexity in the downstream regulatory network of miRNA-144-3p. In pancreatic cancer, miR-144-3p induces cancer cells growth arrest in G1/S phase and activates apoptotic program. The endogenous target of miR-144-3p in this model is centrosomal protein of 55 (CEP55) [29]. In gastric cancer study, miR-144-3p is found to suppress cancer cell progression. The expressional level of miR-144-3p is correlated with tumor size, lymph node metastasis, TNM stage, and depth of invasion. The endogenous target of miR-144-3p in gastric cancer has been identified as pre-leukemia transcription factor 3 (PBX3) [16]. The inhibition of PBX3 by miRNA is accountable for the suppression of epithelial-to-mesenchymal transition (EMT) in cancer cells. In laryngeal squamous cell carcinoma, miR-144-3p acts as tumor suppressive factor as well [28]. In this model, miR-144-3p can also suppress EMT transition process as in gastric cancer. However, E26 transformation specific-1 (ETS-1), as a different downstream target, was proposed in this study. Given EMT as a critical step in cervical cancer progression, it will be interesting to investigate if miR-144-3p can regulate this process through targeting MAPK6 in cervical cancer model. However, there are some studies suggesting miR-144-3p can be oncogenic factor in certain types of cancers. Xiao W et al. have reported the function of miR-144-3p in oncogenic process of clear cell renal cell carcinoma [26]. Conversely, in this model, miR-144-3p activates cancer cell growth, metastasis, and chemo-resistance. AT-rich interactive domain 1A (ARID1A) was identified as an endogenous target of miR-144-3p. Similarly, another study also reported miR-144-3p induces tumor growth and invasion in papillary thyroid carcinoma [17]. The associated mechanism was proposed through targeting aired box gene 8 (PAX8). In addition, blocking the miRNA enhances the tumor suppression effect delivered by X-ray exposure or paclitaxel in papillary thyroid carcinoma. Therefore, the oncogenic effect of miR-144-3p could be cancer specific and depends on its downstream targets in the particular types of cells or tissues.
MAPK6, also called ERK3, is an atypical MAPK. It was firstly identified in 1991 but unlike ERK1/2 its function is less characterized [4]. Recently, a study has reported p21 protein activated kinases (PAKs) are downstream factors of MAPK6 [2]. PAKs are known to play a critical role in regulating cell adhesion and migration through regulating actin cytoskeletal dynamic. Over-expression of MAPK6 increases cell migratory behavior through activating PAKs in breast cancer cells. In other cancer types, MAPK6 has been proposed to carry a similar function in promoting cancer invasion. For instance, in lung cancer cells, MAPK6 interacts with steroid receptor coactivator 3 (SRC-3) that subsequently leads to the activation of ETS transcription factor PEA3 to promote MMP expression. This regulatory mechanism by MAPK6 is implicated with the proinvasive activity of lung cancer [19]. Our study model also supports MAPK6 as oncogenic factor particularly in promoting cell migration and invasion. Therefore, to dissect out the mechanistic role of MAPK6 in cervical cancer, it will be important to elucidate its downstream effectors. As suggested by the past studies, MAPK6 may regulate the pathways in cytoskeletal dynamic and ECM remodelling gene expression.
Conclusion
We are the first time to report miR-144-3p as novel tumor suppressor in cervical cancer. Mechanistic characterization also reveals MAPK6 is directly targeted by miR-144-3p to inhibit cervical cancer. Further study on cervical cancer mouse model with MAPK6 siRNA or kinase inhibitor will be important to test the therapeutic relevance of this signalling axis in treating cervical cancer. Nevertheless, our study supports MAPK6 as promising target to control cancer metastasis or provide an alternative for treating cervical cancer.
References
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4. doi:https://doi.org/10.7554/eLife.05005
Al-Mahdi R, Babteen N, Thillai K, Holt M, Johansen B, Wetting HL, Seternes OM, Wells CM (2015) A novel role for atypical MAPK kinase ERK3 in regulating breast cancer cell morphology and migration. Cell Adhes Migr 9:483–494. https://doi.org/10.1080/19336918.2015.1112485
Banno K, Iida M, Yanokura M, Kisu I, Iwata T, Tominaga E, Tanaka K, Aoki D (2014) MicroRNA in cervical cancer: OncomiRs and tumor suppressor miRs in diagnosis and treatment. TheScientificWorldJournal 2014:178075. https://doi.org/10.1155/2014/178075
Boulton TG, Nye SH, Robbins DJ, Ip NY, Radziejewska E, Morgenbesser SD, DePinho RA, Panayotatos N, Cobb MH, Yancopoulos GD (1991) ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65:663–675
Burd EM (2003) Human papillomavirus and cervical cancer. Clin Microbiol Rev 16:1–17
Duenas-Gonzalez A, Lizano M, Candelaria M, Cetina L, Arce C, Cervera E (2005) Epigenetics of cervical cancer. An overview and therapeutic perspectives. Mol Cancer 4:38. https://doi.org/10.1186/1476-4598-4-38
Fan Z, Cui H, Xu X, Lin Z, Zhang X, Kang L, Han B, Meng J, Yan Z, Yan X, Jiao S (2015) MiR-125a suppresses tumor growth, invasion and metastasis in cervical cancer by targeting STAT3. Oncotarget 6:25266–25280. https://doi.org/10.18632/oncotarget.4457
Fan D, Wang Y, Qi P, Chen Y, Xu P, Yang X, Jin X, Tian X (2016) MicroRNA-183 functions as the tumor suppressor via inhibiting cellular invasion and metastasis by targeting MMP-9 in cervical cancer. Gynecol Oncol 141:166–174. https://doi.org/10.1016/j.ygyno.2016.02.006
Fang J, Zhang H, Jin S (2014) Epigenetics and cervical cancer: from pathogenesis to therapy. Tumour Biol 35:5083–5093. https://doi.org/10.1007/s13277-014-1737-z
Gao C, Zhou C, Zhuang J, Liu L, Liu C, Li H, Liu G, Wei J, Sun C (2018) MicroRNA expression in cervical cancer: novel diagnostic and prognostic biomarkers. J Cell Biochem 119:7080–7090. https://doi.org/10.1002/jcb.27029
Hayes J, Peruzzi PP, Lawler S (2014) MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 20:460–469. https://doi.org/10.1016/j.molmed.2014.06.005
Hillemanns P, Soergel P, Hertel H, Jentschke M (2016) Epidemiology and early detection of cervical cancer. Oncol Res Treat 39:501–506. https://doi.org/10.1159/000448385
Hou T, Ou J, Zhao X, Huang X, Huang Y, Zhang Y (2014) MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. Br J Cancer 110:1260–1268. https://doi.org/10.1038/bjc.2013.829
Hu X, Schwarz JK, Lewis JS Jr, Huettner PC, Rader JS, Deasy JO, Grigsby PW, Wang X (2010) A microRNA expression signature for cervical cancer prognosis. Cancer Res 70:1441–1448. https://doi.org/10.1158/0008-5472.CAN-09-3289
Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG, Bae DS (2008) Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res 14:2535–2542. https://doi.org/10.1158/1078-0432.CCR-07-1231
Li B, Zhang S, Shen H, Li C (2017) MicroRNA-144-3p suppresses gastric cancer progression by inhibiting epithelial-to-mesenchymal transition through targeting PBX3. Biochem Biophys Res Commun 484:241–247. https://doi.org/10.1016/j.bbrc.2017.01.084
Liu C, Su C, Chen Y, Li G (2018) MiR-144-3p promotes the tumor growth and metastasis of papillary thyroid carcinoma by targeting paired box gene 8. Cancer Cell Int 18:54. https://doi.org/10.1186/s12935-018-0550-y
Long MJ, Wu FX, Li P, Liu M, Li X, Tang H (2012a) MicroRNA-10a targets CHL1 and promotes cell growth, migration and invasion in human cervical cancer cells. Cancer Lett 324:186–196. https://doi.org/10.1016/j.canlet.2012.05.022
Long W, Foulds CE, Qin J, Liu J, Ding C, Lonard DM, Solis LM, Wistuba II, Qin J, Tsai SY, Tsai MJ, O’Malley BW (2012b) ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion. J Clin Invest 122:1869–1880. https://doi.org/10.1172/JCI61492
Pimple S, Mishra G, Shastri S (2016) Global strategies for cervical cancer prevention. Curr Opin Obstet Gynecol 28:4–10. https://doi.org/10.1097/GCO.0000000000000241
Rupaimoole R, Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16:203–222. https://doi.org/10.1038/nrd.2016.246
Shishodia G, Verma G, Das BC, Bharti AC (2018) miRNA as viral transcription tuners in HPV-mediated cervical carcinogenesis. Front Biosci 10:21–47
Small W Jr, Bacon MA, Bajaj A, Chuang LT, Fisher BJ, Harkenrider MM, Jhingran A, Kitchener HC, Mileshkin LR, Viswanathan AN, Gaffney DK (2017) Cervical cancer: a global health crisis. Cancer 123:2404–2412. https://doi.org/10.1002/cncr.30667
Tang T, Wong HK, Gu W, Yu MY, To KF, Wang CC, Wong YF, Cheung TH, Chung TK, Choy KW (2013) MicroRNA-182 plays an onco-miRNA role in cervical cancer. Gynecol Oncol 129:199–208. https://doi.org/10.1016/j.ygyno.2012.12.043
Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM (2008) Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One 3:e2557. https://doi.org/10.1371/journal.pone.0002557
Xiao W, Lou N, Ruan H, Bao L, Xiong Z, Yuan C, Tong J, Xu G, Zhou Y, Qu Y, Hu W, Gao Y, Ru Z, Liu L, Xiao H, Chen K, Yang H, Zhang X (2017) Mir-144-3p promotes cell proliferation, metastasis, sunitinib resistance in clear cell renal cell carcinoma by downregulating ARID1A. Cell Physiol Biochem 43:2420–2433. https://doi.org/10.1159/000484395
Yao Q, Xu H, Zhang QQ, Zhou H, Qu LH (2009) MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun 388:539–542. https://doi.org/10.1016/j.bbrc.2009.08.044
Zhang P, Cao L, Fan P, Mei Y, Wu M (2016) LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep 17:1204–1220. https://doi.org/10.15252/embr.201642067
Zheng H, Guo Z, Zheng X, Cheng W, Huang X (2018) MicroRNA-144-3p inhibits cell proliferation and induces cell apoptosis in prostate cancer by targeting CEP55. Am J Transl Res 10:2457–2468
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Clinical evaluation of miR-144-3p expression in human cervical tumor tissues was approved by the Ethical Committee of Heze Municipal Hospital. The animal studies complied with Guide for the Care and Use of Laboratory Animals (8th edition, NIH) and was approved by the Animal Study Ethics Committee of Heze Municipal Hospital.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, J., Zhao, Y., Li, F. et al. MiR-144-3p: a novel tumor suppressor targeting MAPK6 in cervical cancer. J Physiol Biochem 75, 143–152 (2019). https://doi.org/10.1007/s13105-019-00681-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13105-019-00681-9