Abstract
The density peaks (DP) clustering approach, a novel density-based clustering algorithm, detects clusters with arbitrary shape. However, this method uses a crisp neighborhood relation to calculate local density. It cannot identify the different values of the neighborhood membership degrees of the points with respect to different distances from core point. The proposed FN-DP (fuzzy neighborhood density peaks) clustering algorithm uses fuzzy neighborhood relation to define the local density in FJP (fuzzy joint points) algorithm. The proposed algorithm integrates the speed of DP clustering algorithm with the robustness of FJP algorithm. The experimental results illustrate the superior performance of our algorithm compared with the DP clustering approach.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
In unsupervised learning, as one of the most important techniques, the clustering can be used in many fields data mining, pattern recognition, document retrieval, image segmentation and so on [1]. Clustering methods are generally divided into three groups: hierarchical clustering, density/neighborhood-based clustering and partitioning/centroid-based clustering. Main procedures of agglomerative hierarchical clustering are as follow. In the first step, closer objects are merged in a cluster, and then, objects a little bit far away from the previous ones are merged in the same cluster and so forth. In centroid-based methods, however, clusters are represented by centroids which have common features of some certain clusters and may not necessarily be members of the data set. In the next step the objects are assigned to these clusters according to the similarity degrees to the centroids. In hierarchical clustering, the remoteness of objects from each other is considered, while in centroid-based methods their remoteness from the centroids is considered. Partitioning-based clustering is represented by k-means [2]. Other methods of partitioning-based clustering include k-modes and fuzzy c-means (FCM) [3–5]. Density-based clustering is based on the idea that a cluster in a data space is a contiguous region of high point density, separated from other such clusters by contiguous regions of low point density. Neither do these methods require the number of clusters as input parameters, nor do they make assumptions about the underlying density or the variance within the groups that may exist in the data. To our knowledge, density-based clustering is probably introduced for the first time by Wishart [6]. DBSCAN [7] algorithm introduces density-based clustering independently to the Computer Science Community, also proposing the use of spatial index structures to achieve a scalable clustering algorithm. To address one weakness of DBSCAN: the problem of detecting meaningful clusters in data of varying density, Ankerst et al. [8] propose a cluster analysis method based on the OPTICS algorithm computing the augmented cluster-ordering of the database objects. DENCLUE [9] proposes a notion of density-based clusters using a kernel density estimation. An algorithmic framework, called GDBSCAN [10], which generalizes the topological properties of density-based clusters, can be found in Sander et al. Some of density-based clustering methods are similar to hierarchical clustering, the main difference lying in their respective linkage criterion.
Density peaks (DP) clustering [11], a density-based algorithm, is proposed by Rodriguez and Laio. Unlike traditional density-based clustering methods, this algorithm can be considered as a combination of density-based and centroid-based. It starts by determining the centroids of clusters according to two important quantities: \(\rho\) and \(\delta\). The second step is to determine which objects to merge in a cluster. Unlike traditional density-based clustering methods, it is based on the local density of objects. All objects are in descending order according to the local density. An unclassified object is assigned to the cluster that contains a certain classified object satisfying a condition. It is the nearest of all the classified objects to the unclassified object. Similarly to other density-based methods, DP clustering algorithm is able to recognize clusters with arbitrary shape. The computation speed of this algorithm is more advantageous than traditional density-based clustering methods. In order to overcome the problem that the global structure of data is not considered, Du et al. [12] propose a density peaks clustering based on k nearest neighbors (DPC-KNN). In addition, the DP clustering performs not well when it finds some pseudo cluster centers. In order to overcome this difficulty, Liang et al. propose the 3DC clustering [13] based on the divide-and-conquer strategy and the density-reachable concept.
In real life, the boundary between clusters could not be precisely defined such that some of the objects could belong to more than one cluster with different positive degrees of membership. A classic example is the fuzzy clustering. In these methods, the Fuzzy c-Means (FCM) algorithm is perhaps the most important and widely used method. The vast majority of the research work [14–21] is based on this method. These methods suppose the fuzziness of clustering with respect to the possibility of the membership of some objects into various clusters. Nasibov and Ulutagay propose a different approach of fuzziness based on a new Fuzzy Joint Points (FJP) method [22] which perceives the neighborhood concept from a level-based viewpoint which means that the objects are considered in how much detail in construction of homogenous classes. It means that the fuzzier the objects, the more similar they are. Based on this approach, many clustering methods [23–25] are proposed.
In the case that number of clusters are known, the fuzzy clustering methods show excellent performance in specifying datasets with sphere-like shape. In these methods, FJP algorithm is robust since it uses fuzzy relation in neighborhood analysis. However, it performs poorly in terms of computation time. On the other hand, DP clustering is able to detect clusters in any shape without specifying the number of clusters. In order to be able to run correctly in a wide range of change interval could be more advantageous, in this study, the fuzzy neighborhood- density peaks(FN-DP) clustering which integrates the speed of DP clustering algorithm with the robustness of FJP algorithm is proposed.
The rest of this paper is organized as follows. In Sect. 2, the DP clustering method is mentioned and some concepts about the FJP are defined. In Sect. 3, a detailed description of FN-DP is given. In Sect. 4, experimental results are presented on synthetic data sets and real world data sets. Finally, some conclusions and the intending work are given in the last section.
2 Related work
2.1 Density peaks clustering
Let \(\mathbf{X}=\left\{ {{\mathbf{x}}_{1}},{{\mathbf{x}}_{2}},\ldots ,{{\mathbf{x}}_{n}} \right\}\) denote a dataset of n data objects. Each object \({{\mathbf{x}}_{i}},1\le i\le n\) has m attributes. Thus, for each \(i,1\le i\le n\), and for \(j,1\le j\le m\), let \({{x}_{i,j}}\) be the j-th attribute of \({{\mathbf{x}}_{i}}\). The Euclidean distance \(d\left( {{\mathbf{x}}_{i}},{{\mathbf{x}}_{j}} \right)\) between any points \({{\mathbf{x}}_{i}},{{\mathbf{x}}_{j}}\in \mathbf{X}\) can be determined as follows:
Unlike DBSCAN, the DP clustering finds the cluster centers before data points are assigned. Determining the cluster centers is of vital importance to guarantee good clustering results. Because it determines the number of the clusters, and affects the assignation indirectly. In the following, we will describe the calculation of \(\rho\) and \(\delta\) in much more detail.
DP represents data objects as points in a space and adopts a distance metric, such as Eq. (1), as a dissimilarity between objects. Let \(D=\left\{ {{d}_{1}},{{d}_{2}},\ldots ,{{d}_{{{N}_{d}}}} \right\}\) a set of all the distances between every two points in data set, where all the distances from smallest to greatest. \({N_d} = \left( \begin{array}{l} n\\ 2 \end{array} \right)\), where N is the number of points in the dataset. Unlike DBSCAN, the neighborhood radius is determined not by the direct value, but by the percentage. \({{d}_{c}}\) indicates a percentage and is the only input parameter, which is called a cutoff distance. The neighborhood radius \(\varepsilon\) is defined as:
where \(\left\lceil \cdot \right\rceil\) is the ceiling function and \({{d}^{\max }}={{d}_{{{N}_{d}}}}=\max d\left( {{\mathbf{x}}_{i}},{{\mathbf{x}}_{j}} \right)\). The method takes one parameter\({{d}_{c}}\) which is a percentage.
The neighborhood set of point \({{\mathbf{x}}_{i}}\in \mathbf{X}\) with parameter \(\varepsilon\) (\(\varepsilon\)-neighborhood set) is as follows:
\({{\mu }_{{{\mathbf{x}}_{i}}}}\left( {{\mathbf{x}}_{j}} \right)\) denote the membership degree of the point \({{\mathbf{x}}_{j}}\) to the neighborhood set of the point \({{\mathbf{x}}_{i}}\), as follows:
The local density \({{\rho }_{i}}\) [7] of a point \({{\mathbf{x}}_{i}}\) is defined as:
The calculation of the delta value [7], again, is quite simple. The minimum distance between the point of \({{\mathbf{x}}_{i}}\) and any other points with higher density, denoted by \({{\delta }_{i}}\) is defined as
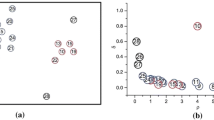
When the local density and delta values for each point have been calculated, this method identifies the cluster centers by anomalously large \({{\rho }_{i}}\) and \({{\delta }_{i}}\). On the basis of this idea, cluster centers always appear on the upper-right corner of the decision graph. After cluster centers have been found, the DP clustering assigns remaining points to the same cluster as its nearest neighbors with higher density.
2.2 Basic concepts about the fuzzy joint points method
Let \(F\left( {{\Re }^{m}} \right)\) denote the set of m-dimensional fuzzy sets of the space \({{\Re }^{m}}\). \({{{\mu }'}_{A}}:{{\Re }^{m}}\to \left[ 0,1 \right]\) denotes the membership function of the fuzzy set \(A\in F\left( {{\Re }^{m}} \right)\).
Definition 2
A conical fuzzy point \(A=\left( a,R \right)\in F\left( {{\Re }^{m}} \right)\) of the space \({{\Re }^{m}}\) is a fuzzy set with membership function (Fig. 1) [22].
where \(a\in {{\Re }^{m}}\) is the center of fuzzy point A, and R is the radius of its support supp A, where
The \(\alpha\)-level set of conical fuzzy point \(A=\left( a,R \right)\) is calculated as
In this study, the short term “fuzzy point” is used, instead of conical fuzzy point defined in Eq. (7).
3 The proposed algorithm
Before introducing the proposed method, the fuzzy neighborhood relation is defined.
3.1 Fuzzy neighborhood relation
Points \({{x}_{1}}\) and \({{x}_{2}}\) have the same number of neighbors within \(\varepsilon \le {{d}^{\max }}\) radius (Fig. 2) [24]. There is an obvious difference between these points. It is obvious that the points \({{x}_{1}}\) and \({{x}_{2}}\) in Fig. 2 are the same according to the crisp neighborhood relation used in the DP clustering method. Because in classical case there is no difference with respect to membership degrees between points within the same neighborhood radius of core point (Fig. 3) [24]. In other words, points \({{y}_{1}}\) and \({{y}_{2}}\) have the same neighborhood membership degrees to the point x. As it is seen from Fig. 3, the membership degrees of \({{y}_{1}}\) and \({{y}_{2}}\) are both equal to 1. This study expands the neighborhood set determined in Eq. (4) to the fuzzy neighborhood case. Utilizing fuzzy neighborhood function provides an advantage that there are different values of the neighborhood membership degrees of the points with respect to different distances from core point.
Note that, in order to form a fuzzy relation \(\mu :X\times X\to \left[ 0,1 \right]\), the idea of the membership function of the fuzzy set defined in Eq. (4) is introduced. In order to be consistent with the parameter of the original DP clustering method, the radius of the considered fuzzy points is calculated as Eq. (2) in the proposed algorithm.
Thus, such a neighborhood membership function [24] is defined as
In the previous example, if fuzzy neighborhood function is used, point \({{x}_{1}}\) will have a higher membership degree of being a core point than that of point \({{x}_{2}}\).
In neighborhood relation determined by Eq. (4), neighborhood degrees of points with varying distances to the core point will be different from each other (Fig. 3).
As it is seen from Fig. 4 [24], points \({{y}_{1}}\) and \({{y}_{2}}\) have different neighborhood membership degrees to the point x. Hence, the membership degree of \({{y}_{1}}\), i.e. \({{\alpha }_{1}}\), is higher than the membership degree of \({{y}_{2}}\), i.e. \({{\alpha }_{2}}\).
3.2 FN-DP
Based on the definitions given above, a new local density is redefined for fuzzy logic approach, as follows:
where \({\mu }'\) denotes the fuzzy neighborhood function defined by Eq. (10).
The main advantage of transformation of the DP clustering algorithm to the FN-DP clustering algorithm and using fuzzy sets theory is that the fuzzy neighborhood function that make local density more sensitive can be utilized. So the FN-DP clustering method could be more robust to the datasets various shapes and densities.
The idea of Sect. 3.1 is introduced into the DP clustering algorithm. The following algorithm is a summary of the proposed algorithm based on fuzzy neighborhood relation.
Algorithm. The proposed algorithm. |
Inputs: The samples\(\mathbf{X}\in {{\Re }^{n\times m}}\) The parameter\({{d}_{c}}\) Outputs: The label vector of cluster index:\(\mathbf{y}\in {{\Re }^{n\times 1}}\) Method: Step 1: Calculate distance matrix according to Eq. (1) Step 2: Calculate \({{\rho }_{i}}\) for point \({{\mathbf{x}}_{i}}\) according to Eq. (11) Step 3: Calculate \({{\delta }_{i}}\) for point \({{\mathbf{x}}_{i}}\) according to Eq. (6) Step 4: Plot decision graph and select cluster centers Step 5: Assign each remaining point to the cluster Step 6: Return y |
Now, the time complexity of the proposed algorithm is given. Assume that N is the number of objects in the data set. The computational complexity of the similarity matrix is \(O\left( {{N}^{2}} \right)\). This also need \(O\left( {{N}^{2}} \right)\) to compute the new local density. In addition, the cost of the sorting process with quick sort \(O\left( N\log N \right)\). As the complexity in the assignment procedure is \(O\left( N \right)\), the total time cost of the proposed algorithm is \(O\left( {{N}^{2}} \right)+O\left( {{N}^{2}} \right)+O\left( N\log N \right)+O\left( N \right)\tilde{\ }O\left( {{N}^{2}} \right)\).
4 Experiments and results
In order to demonstrate the feasibility of the proposed algorithm, 12 synthetic datasets with various shapes and densities are used. To further compare FN-DP clustering algorithm based on fuzzy neighborhood relation with the original algorithm based on crisp neighborhood relation, 5 synthetic datasets are used. Moreover, by experiments on real-world datasets, the proposed method is compared with the original algorithm in terms of clustering accuracy (ACC), normalized mutual information (NMI) and adjusted Rand index (ARI) [26–31].
We conduct experiments in a desktop computer with a core i7 DMI2-Intel 3.6 GHz processor and 16 GB RAM running MATLAB 2013A. The cutoff distance \({{d}_{c}}\) used in the DP clustering algorithm and the FN-DP clustering algorithm is given from 0.1 to 100% at an increment 0.1% for all 5 synthetic datasets. On the other hand, on real-world datasets, The parameter \({{d}_{c}}\) is selected from the sequence {0.1% 0.5% 1% 2% 3% 4% 5% 6% 7% 8% 9% 10%} based on the clustering performance. In order to better demonstrate the feasibility of the proposed algorithm, the best results only are presented in terms of clustering accuracy.
4.1 Experiments on synthetic datasets
As shown in Fig. 5a–d, the proposed method does an excellent job in clustering datasets with spherical or ellipsoidal shape. Among them, A1, A2 and A3 datasets are large datasets with varying number of clusters. The experiment results demonstrate the robustness of the FN-DP clustering method in terms of the quantity. As shown in Fig. 5e–g, the proposed algorithm gets extraordinarily favorable performance to these three datasets with different sizes and shapes. As shown in Fig. 5h–k, the datasets S1 to S4 are two-dimensional sets with varying complexity in terms of spatial data distributions. The data sets have 5000 points around 15 clusters with a varying degrees of overlap. The performance of the proposed algorithm is perfect for data sets with varying complexity. As these experiments illustrate our algorithm is very effective in finding clusters of arbitrary shape, density, distribution and number.
4.2 Experiments for robustness
To evaluate the performances of the algorithms, “correct range percent (CRP)’’ is used as an indicator to indicates the percentage of correct result range of \({{d}_{c}}\) parameter to the whole [0%, 100%] interval. So the CRP criteria is calculated as follows:
where \(CR{{P}_{i,j}}=\left( {{d}_{c}} \right)_{i,j}^{U}-\left( {{d}_{c}} \right)_{i,j}^{L}\) is the j-th continuous interval of the parameter \({{d}_{c}}\), in which the algorithm can give correct results in the i-th dataset, where \(\left( {{d}_{c}} \right)_{i,j}^{U}\) is the low bound and \(\left( {{d}_{c}} \right)_{i,j}^{L}\) is the upper bound in the j-th continuous interval.
In addition, the strict criteria will be loosened, because even experts cannot find absolutely correct clustering results visually in some datasets. For Aggregation and R15 datasets, all cases that the accuracy is more than 99% is considered acceptable. Because of that D31 dataset has more clusters, the benchmark of the clustering accuracy is revised to 96%.
In order to show these results visually, comparisons are given as histogram (Fig. 6). The CRP values for the clusterings obtained with FN-DP are, in all the cases superior to the one obtained by DP. Flame, Twospirals and Aggregation have clusters with non-spherical shape. It is obvious that FN-DP results are significantly better than those obtained by the original algorithm for these three datasets. On R15 and D31 datasets with spherical shape, in comparisons with the DP clustering method, our method always shows a small advantage in terms of the CRP criteria. It’s tempting to conclude that our algorithm does an excellent job compared with DP, when the dataset has some cluster with non-spherical shape. On the other hand, when a dataset present the non-spherical distribution, the CRP values of the clusters formed by the two methods are close. However, the proposed method performs slightly better.
4.3 Experiments on real-world datasets
4.3.1 Real-world datasets
The real-world datasets used in the experiment also are taken from the UCI Machine Learning Repository, including Iris Plants Database (Iris), Wine Recognition Database (Wine), the Heart Disease database (Heart), Ionosphere database (Ionosphere), Wisconsin Diagnostic Breast Cancer (WDBC), Waveform Database (Waveform), Ringnorm data set (Ringnorm) and Pen-Based Recognition of Handwritten Digits (Penbased). The details of these datasets are listed in Table 1.
4.3.2 Performance on real-world datasets
Table 2 lists the clustering accuracy of our proposed algorithm and the original algorithm. In the following tables, the numbers highlighted in bold indicate that the corresponding algorithm has the best performance in terms of its corresponding evaluation. Tables 2, 3 and 4 show comparisons against DP and DBSCAN in terms of the different quality measures (ACC, NMI and ARI). In these tables, the symbol - means that DBSCAN detects outliers on the corresponding data set. Thus, in these cases, we cannot evaluate the performance of DBSCAN using these quality measures. In addition, on Iris, Wine, Waveform and Ringnorm data sets, numbers of clusters are found accurately by DBSCAN. It only detects two clusters, one cluster, one cluster and one cluster, respectively. By comparison with the DP clustering algorithm and DBSCAN, our method obtains better clustering performance in terms of the all quality measures (ACC, NMI and ARI) on all datasets, as shown in Tables 2, 3 and 4. In the following tables, the numbers highlighted in bold indicate that the corresponding algorithm has the best performance in terms of its corresponding evaluation.
To explain further how to choose the parameter \({{d}_{c}}\) of DP and FN-DP, we use the Heart data set as an example. Figure 7 shows the ACC values obtained by the two methods with different \({{d}_{c}}\) on the Heart data set. The horizontal axis represents the parameter \({{d}_{c}}\). As explained earlier (in the beginning of Sect. 4), the parameter \({{d}_{c}}\) is selected from the sequence {0.1% 0.5% 1% 2% 3% 4% 5% 6% 7% 8% 9% 10%}. The vertical axis represents the ACC value. Obviously, when we set parameter \({{d}_{c}}\) to 1%, FN-DP obtains the best result (0.8111) on this data set. Nevertheless, when parameter is set to 6%, DP obtains the best result (0.8074) on the Heart data set. In this paper, the parameter \({{d}_{c}}\) is selected based on the clustering performance. Thus, on the Heart data set, the parameter \({{d}_{c}}\) of FN-DP is set to 1%, whereas the parameter \({{d}_{c}}\) of DP is set to 6. As a result, the parameter \({{d}_{c}}\)of the two algorithms is not the same. On other data sets, we present the best results in terms of clustering accuracy. Thus the parameter of the two algorithms may be different on each data set. Similar strategy on parameter selection has been used in some literatures [32–35]. It is interesting to note that if the parameter \({{d}_{c}}\) of the two algorithms is set to 1%, the ACC value of the proposed method is slightly higher than that of DP. By contrast, if the parameter \({{d}_{c}}\) of the two algorithms is set to 6%, the ACC value of the two algorithms is the same. In addition, if and only if the parameter \({{d}_{c}}\) of the two algorithms is set to 7%, the ACC value of DP is slightly higher than that of FN-DP as shown in Fig. 7. In most cases, FN-DP results are significantly better than those obtained by the original algorithm on this data set. This again proves that FN-DP obtains more robust performance than the original algorithm.
Figure 8 shows the running time spent on clustering using FN-DP and DP methods. We run every algorithm 20 times on each data set and get the average. It is clear the running times of the two methods are close, within a difference of 0.0001, on the first five data sets. However, the running times of FN-DP are slightly smaller than those of DP on some larger data sets (Waveform, Ringnorm and Penbased). The reason for this result may be that MATLAB has a very strong processing capability for matrix manipulations. The code of the original algorithm does not use matrix manipulations to compute the local density. In all cases, the two methods are comparable in terms of efficiency. Our proposed method brings a boost of performance without loss of efficiency.
4.4 Experiments on image data
We test the quality of this algorithm on image data. Figure 9 shows the original images from the Berkeley database and the segmentation evaluation database [36–38]. Figure 10 shows image segmentation results. Unlike K-means, the number of groups is specified in advance. The number of groups may depend on the result of decision graph. Thus, FN-DP can be separated from all the other objects in the background, as shown in Fig. 10. In addition, it is also insensitive to the choice of \({{d}_{c}}\) in this test. Because the same results are obtained within [0.1%, 10%] the range of \({{d}_{c}}\) over these images. Experimental results show that FN-DP can be a feasible preprocessing method for image segmentation.
5 Conclusions
A new fuzzy neighborhood function is introduced into this paper and the FN-DP clustering algorithm based on this function is proposed. Experiments show that our algorithm is very effective in finding clusters with arbitrary shape, density, distribution and number. The proposed algorithm combines the speed of the DP clustering method and robustness of FJP algorithm. It is observed that our algorithm is more robust than the original algorithm to datasets with various shapes and densities. However, in this stage, the aim is not to find the optimal values of the parameters, but to prove that one can get more realistic and robust results by using fuzzy neighborhood relation in the proposed algorithm instead of using crisp neighborhood relation utilized in the original algorithm. The experimental results on real-world dataset illustrate the superior performance of our algorithm compared with the DP clustering approach.
The combination of our proposed algorithm and the \(\gamma\)-graph displays a possibility that an automatic cluster centroid selection method is developed. FN-DP costs much time in the calculation of the similarity matrix, thus we will try to introduce the idea of the grid into our method. The cost is only associated with the number of cells. And the number of cells K is far less than the number of objects N.
References
Yu Z, Li L, Liu J, Zhang J, Han G (2015) Adaptive noise immune cluster ensemble using affinity propagation. IEEE Trans Knowl Data Eng 27(12):3176–3189
MacQueen JB (1967) Some methods for classification and analysis of multivariate observation. In: Proceedings of the fifth Berkeley symposium on mathematical statistics probability, vol 1, No. 14, pp 281–297
Huang Z (1998) Extensions to the k-means algorithm for clustering large data sets with categorical values. Data Min Knowl Disc 2(3):283–304
Backer E, Jain AK (1981) A clustering performance measure based on fuzzy set decomposition. IEEE Trans Pattern Anal Mach Intell 1:66–75
Pal NR, Bezdek JC (1995) On cluster validity for the fuzzy c-means model. IEEE Trans Fuzzy Syst 3(3):370–379
Wishart D (1969) Mode analysis: a generalization of nearest neighbour which reduces chaining effects. In: Proceedings of the colloquium in numerical taxonomy
Ester M, Kriegel HP, Sander J, Xu XW (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. In: Proceedings of second international conference on knowledge discovery data mining, vol 96, No. 34, pp 226–231
Ankerst M, Breunig MM, Kriegel HP, Sander J (1999) OPTICS: ordering points to identify the clustering structure. In: Proceedings of ACM SIGMOD Conference, vol 28, No. 2, pp 49–60
Hinneburg A, Keim DA (1998) An efficient approach to clustering in large multimedia databases with noise. In: Proceedings of the fourth international conference on knowledge discovery and data mining
Sander J, Ester M, Kriegel HP, Xu X (1998) Density-based clustering in spatial databases: the algorithm DBSCAN and its applications. Data Mining Knowl Discov 2(2):169–194
Rodriguez A, Laio A (2014) Clustering by fast search and find of density peak. Science 344(6191):1492–1496
Du M, Ding S, Jia H (2016) Study on density peaks clustering based on k-nearest neighbors and principal component analysis. Knowl-Based Syst 99:135–145
Liang Z, Chen P (2016) Delta-density based clustering with a divide-and-conquer strategy: 3DC clustering. Pattern Recognit Lett 73:52–59
Škrjanc I (2015) Evolving fuzzy-model-based design of experiments with supervised hierarchical clustering. IEEE Trans Fuzzy Syst 23(4):861–871
Yu Z, Chen H, You J, Liu J, Wong HS, Hana G, Li L (2015) Adaptive fuzzy consensus clustering framework for clustering analysis of cancer data. IEEE/ACM Trans Comput Biol Bioinform 12(4):887–901
Wu CH, Ouyang CS, Chen LW, Lu LW (2015) A new fuzzy clustering validity index with a median factor for centroid-based clustering. IEEE Trans Fuzzy Syst 23(3):701–718
Hu L, Chan KC (2016) Fuzzy clustering in a complex network based on content relevance and link structures. IEEE Trans Fuzzy Syst 24(2):456–470
Wang Y, Chen L, Mei JP (2014) Incremental fuzzy clustering with multiple medoids for large data. IEEE Trans Fuzzy Syst 22(6):1557–1568
Zheng Y, Jeon B, Xu D, Wu QM, Zhang H (2015) Image segmentation by generalized hierarchical fuzzy C-means algorithm. J Intell Fuzzy Syst 28(2):961–973
Klawonn F, Kruse R, Winkler R (2015) Fuzzy clustering: more than just fuzzification. Fuzzy Sets Syst 281:272–279
He YL, Wang XZ, Huang JZ (2016) Fuzzy nonlinear regression analysis using a random weight network. Inf Sci 364:222–240
Nasibov EN, Ulutagay G (2007) A new unsupervised approach for fuzzy clustering. Fuzzy Sets Syst 158:2118–2133
Nasibov EN (2008) A robust algorithm for solution of the fuzzy clustering problem on the basis of the fuzzy joint points method. Cybern Syst Anal 44(1):7–17
Nasibov EN, Ulutagay G (2009) Robustness of density-based clustering methods with various neighborhood relations. Fuzzy Sets Syst 160:3601–3615
Ulutagay G, Nasibov EN (2010) Influence of transitive closure complexity in FJP-based clustering algorithms. Turkish J Fuzzy Syst 1(1):3–20
Wen X, Shao L, Xue Y, Fang W (2015) A rapid learning algorithm for vehicle classification. Inf Sci 295(1):395–406
Xia Z, Wang X, Sun X, Liu Q, Xiong N (2016) Steganalysis of LSB matching using differences between nonadjacent pixels. Multimed Tools Appl 75(4):1947–1962
Yu Z, Zhu X, Wong HS, You J, Zhang J, Han G (2016) Distribution-based cluster structure selection. IEEE Trans Cybern. doi:10.1109/TCYB.2016.2569529
Zhang Y, Sun X, Wang B (2016) Efficient Algorithm for K-barrier coverage based on integer linear programming. China Commu 13(7):16–23
Gu B, Sheng VS, Wang Z, Ho D, Osman S, Li S (2015) Incremental learning for ν-support vector regression. Neural Netw 67:140–150
Yu Z, Luo P, You J, Wong HS, Leung H, Wu S, Zhang J, Han G (2016) Incremental semi-supervised clustering ensemble for high dimensional data clustering. IEEE Trans Knowl Data Eng 28(3):701–714
Dong CR, Ng WWY, Wang XZ, Chan PPK, Yeung DS (2014) An improved differential evolution and its application to determining feature weights in similarity-based clustering. Neurocomputing 146:95–103
Huang G, Song S, Gupta JND, Wu C (2014) Semi-supervised and unsupervised extreme learning machines. IEEE Trans Cybern 44(12):2405–2417
Ashfaq RAR, Wang XZ, Huang JZ, Abbas H, He YL (2017) Fuzziness based semi-supervised learning approach for intrusion detection system. Inf Sci 378:484–497
Wang XZ, Wang YD, Wang LJ (2004) Improving fuzzy c-means clustering based on feature-weight learning. Pattern Recogn Lett 25(10):1123–1132
Zeng S, Yang X, Gou J, Wen J (2016) Integrating absolute distances in collaborative representation for robust image classification. CAAI Trans Intell Technol 1(2):189–196
Alpert S, Galun M, Brandt A, Basri R (2012) Image segmentation by probabilistic bottom-up aggregation and cue integration. IEEE Trans Pattern Anal Mach Intell 34(2):315–327
Ma Z, Liu Q, Sun K, Zhan S (2016) A syncretic representation for image classification and face recognition. CAAI Trans Intell Technol 1(2):173–178
Acknowledgements
The work is supported by the National Natural Science Foundation of China under Grant No. 61379101 and No. 61672522, the National Basic Research Program of China under Grant No. 2013CB329502, the Priority Academic Program Development of Jiangsu Higer Education Institutions (PAPD), and the Jiangsu Collaborative Innovation Center on Atmospheric Environment and Equipment Technology (CICAEET).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Du, M., Ding, S. & Xue, Y. A robust density peaks clustering algorithm using fuzzy neighborhood. Int. J. Mach. Learn. & Cyber. 9, 1131–1140 (2018). https://doi.org/10.1007/s13042-017-0636-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-017-0636-1