Abstract
The complete mitochondrial genome sequence of the hybrid of Acipenser dabryanus (♀) × A. schrenckii (♂) was first determined by using the next-generation sequencing in this study. The circular mitochondrial genome was 16,439 bp in length, which contained 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, one displacement loop locus and an origin of replication on the light-strand, showed a typical vertebrate pattern. All of the genes were encoded on the heavy strain except for ND6 and eight tRNA genes. The overall nucleotide composition was 30.19% A, 23.74% T, 29.65% C, 16.42% G, with 53.93% AT, respectively. Compared with the complete mitogenome of the parents, the results indicated that the mitochondria of hybrid sturgeon could be consistent with a maternal inheritance. Phylogenetic analyses using concatenated nucleotide sequences of the 13 protein-coding genes with two different methods (maximum likelihood and neighbor-joining analysis) both highly supported that A. dabryanus (♀) × A. schrenckii (♂) showed a close relationship with A. dabryanus and A. sinensis. These data provide useful information for a better understanding of the mitogenomic diversities and evolution in fish as well as novel genetic markers for studying population genetics and species identification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Acipenser dabryanus and A. schrenckii are two species of Acipenseriformes, which belong to the genus of Acipenser. A. dabryanus, a critically endangered and endemic species, is also called “the Yangtze River sturgeon”, which has been distributed in the upper and middle sections of the Yangtze River (Liu et al. 2017a). The natural production of A. dabryanus have declined sharply in the past decades due to dam construction, overfishing, pollution, etc., in the Yangtze River (Liu et al. 2017a). A. schrenckii of the Amur River basin and its tributaries is one of the most important economically valuable fish in Russia and in China (Liu et al. 2017b; Li et al. 2016). However, growth depression, late sexual maturity, and disease susceptibility restrict the culture of population of A. schrenckii (Dong et al. 2014; Wei et al. 2011). Because of heterosis, hybrid sturgeon has been an important species in sturgeon aquaculture in China (Gao et al. 2017; Shen et al. 2014).
In the present study, we first determined the complete mitogenome sequence of the hybrid sturgeon (A × A). The specimen of A × A used in this study were obtained by artificial hybridization from the Conservation and Utilization of Fishes resources in the Upper Reaches of the Yangtze River Key Laboratory of Sichuan Province, Neijiang Normal University. A 30–40 mg fin clip was collected and preserved in 95% ethanol at 4 °C. Total genomic DNA was extracted from these caudal fins by a Tissue DNA Kit (OMEGA E.Z.N.A.) following the manufacturer’s protocol. Subsequently, the genomic DNA was sequenced using the next-generation sequencing, and then the mitogenome was assembled using A. dabryanus as reference.
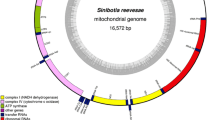
The complete mitochondrial genome of the A × A was determined to be 16,439 bp in length, including 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, a displacement loop (D-loop) locus and an origin of replication on the light-strand (OL) (Table 1; Fig. 1). The overall nucleotide composition was 30.19% A, 23.74% T, 29.65% C, 16.42% G, with 53.93% AT, respectively. Most of coding genes were encoded on the heavy strain (H-strand) except for ND6 and eight tRNA genes, which were encoded on the light strain. The common initiation codon was ATG in the 13 protein-coding genes, except COX1, which used GTG. 9 protein-coding genes stop with the complete termination codon TAG (ND1, COXI, ND3, ND6) or TAA (ATP8, ATP6, COXIII, ND4L, ND5), while the rest ends with an incomplete termination codon T(ND2, COXII, ND4, Cyt b). Moreover, all the 22 tRNA genes, length ranging from 67 to 75 bp, and the tRNA Cys was the shortest while tRNA Leu was the longest. The 12S rRNA and 16S rRNA were 960 and 1701 bp, respectively. Additionally, an 32 bp origin of replication on the light-strand (OL) was found in the A × A mitogenome between tRNA Asn and tRNA Cys. Furthermore, the displacement loop (D-loop) was located between tRNA Pro and tRNA Phe, with 727 bp in length (Fig. 1; Table 1). The genomic sequence has been deposited in GenBank with an accession number MF958972.
The gene map of the complete mitochondrial genome of the hybrid of Acipenser dabryanus (♀) × A. schrenckii (♂). Arrows indicate the direction of transcription. tRNA genes are named using single-letter amino acid abbreviations. Genes encoded on the H-strand and L-strand are shown outside and inside the circular map of the mitogenome, respectively. The innermost circle indicates the GC content graph
To confirm the phylogenetic relationships between A × A and other Acipenserinae subfamily fishes, phylogenetic analyses were performed on the concatenated dataset of 13 PCGs at nucleotide level with neighbor-joining (NJ) and maximum likelihood (ML) methods (Zou et al. 2017). The tree topologies produced by NJ and ML analyses were equivalent (Fig. 2). The 15 taxa all belong to Acipenseridae, Acipenserinae except Culter erythropterus, which belongs to Cyprinidae, Culterinae. So C. erythropterus was used as an outgroup. The other 14 species from 2 genus were divided into two clades. Species of Huso huso, A. stellatus, A. ruthenus, A. baerii and, A. gueldenstaedtii were clustered into clade B, and the rest of species were clustered into clade A. The A × A and its female parent (A. dabryanus) aggregated into a clade at the first. The situation also showed in H. dauricus × A. schrenckii (Liu et al. 2017b). This phenomenon is inseparable from the maternal inheritance characteristics of the mitochondria. The A × A has a closer relationship with A. sinensis than its male parent (A. schrenckii), due to its female (A. dabryanus) has a close relationship with A. sinensis (Li et al. 2016). Although H. dauricus and H. huso both pertain to Huso but located in different branches, which is slightly different from conventional morphology-based classification of Acipenserinae species (Li et al. 2016; Birstein et al. 1999). The difference may be owing to the limited availability of mitogenomes from Acipenserinae species (Wei et al. 2011).
The phylogenetic analyses investigated using neighbor-joining (NJ) and maximum likelihood (ML) analysis indicated evolutionary relationships among 15 taxa based on nucleotide sequences of 13 concatenated protein-coding genes. NJ posterior probability (blue number) and ML bootstrap support values (black number) are shown on the nodes. Culter erythropterus (GenBank: NC 024749) was used as an outgroup. (Color figure online)
In summary, the present study first reported the complete mitochondrial genome of the hybrid sturgeon of A × A, The circular molecule was 16,439 bp long and showed a typical vertebrate mitogenome structure. Phylogenetic analyses showed that mitochondrial genomes of A × A remain maternally inherited, which was consistent with the mitochondrial inheritance mechanism. The complete mitochondrial genome sequence of the A × A provided an important dataset for a better understanding of the mitogenomic diversities and evolution in fish as well as novel genetic markers for studying population genetics and species identification.
References
Birstein VJ, Doukakis P, DeSalle R (1999) Molecular phylogeny of Acipenserinae and black caviar species identification. J Appl Ichthyol 15:12–16
Dong XL, Yu J, Zhang Y, Sun DJ (2014) Complete mitochondrial genome of Acipenser schrenckii (Acipenseriformes, Acipenseridae). Mitochondrial DNA 25:350
Gao FT, Wei M, Zhu Y, Guo H, Chen SL, Yang GP (2017) Characterization of the complete mitochondrial genome of the hybrid Epinephelus moara♀ × Epinephelus lanceolatus♂, and phylogenetic analysis in subfamily epinephelinae. J Ocean Univ China 16:555–563
Li S, Guo WT, Liu XQ, Qu HT, Guan M, Su W (2016) Complete mitochondrial genome of the hybrid of Acipenser schrenckii (♀) × Huso dauricus (♂). Mitochondrial DNA 27:7–8
Liu JJ, Tan C, Xiao K, Wang BZ, Liu XQ, Du HJ (2017a) The complete mitochondrial genome of Dabry’s sturgeon (Acipenser dabryanus). Mitochondrial DNA B 2:54–55
Liu XQ, Du HJ, Xiao K, Hu YC, Liu JJ, Zhao X (2017b) The complete mitochondrial genome of the hybrid sturgeon of Huso dauricus (♀) × Acipenser schrenckii (♂). Mitochondrial DNA B 2:11–12
Shen L, Shi Y, Zou YC, Zhou XH, Wei QW (2014) Sturgeon aquaculture in China: status, challenge and proposals based on nation-wide surveys of 2010–2012. J Appl Ichthyol 30:1547–1551
Wei QW, Zou Y, Li P, Li L (2011) Sturgeon aquaculture in China: progress, strategies and prospects assessed on the basis of nation-wide surveys (2007–2009). J Appl Ichthyol 27:162–168
Zou YC, Xie BW, Qin CJ, Wang YM, Yuan DY, Li R, Wen ZY (2017) The complete mitochondrial genome of a threatened loach (Sinibotia reevesae) and its phylogeny. Genes Genomics 39:767–778
Acknowledgements
This work was supported by the National Natural Science Funds (No. 31402305), the Educational Commission of Sichuan Province of China (No. 15ZA0285), the department of science and technology of Sichuan Province of China (No. 2015JY0262) and the scientific research project of Neijiang Normal University (No. 16JC08).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zou, Y., Zhang, J., Wen, Z. et al. The complete mitochondrial genome of the hybrid of Acipenser dabryanus (♀) × A. schrenckii (♂) and its phylogeny. Conservation Genet Resour 10, 881–884 (2018). https://doi.org/10.1007/s12686-017-0887-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-017-0887-x