Abstract
Castration-resistant prostate cancer (CRPC) represents the final stage of prostate cancer (PCa). Cabazitaxel, a taxane chemotherapy drug, is used in treating CRPC. However, patients with CRPC eventually develop resistance to cabazitaxel, and the underlying mechanism remains unclear. Here, we aimed to investigate potential genetic alterations that may play a role in CRPC resistance to cabazitaxel. Using microarray data from the GSE158494 dataset, we identified ten critical genes (CXCL8, ITGB8, CLIP4, MAP1B, WIPI1, MMP13, CXCL1, C1S, GOLGA8B, and CXCL6) associated with CRPC cell resistance to cabazitaxel. The potential function of these key genes in PCa progression was analyzed using different databases, including Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA), and Chinese Prostate Cancer Genome and Epigenome Atlas (CPGEA). Our findings revealed altered expression of these genes in the development of PCa. Furthermore, CXCL1 and GOLGA8B were found to influence the disease-free survival (DFS) status of patients with PCa, with GOLGA8B affecting the overall prognosis in patients with PCa. Additionally, GOLGA8B expression was associated with the infiltration of various immune cells in PCa, and it was upregulated in clinical PCa and CRPC samples. Through CCK-8 assays, we established that GOLGA8B could influence the sensitivity of CRPC cells to cabazitaxel and docetaxel. In conclusion, we identified GOLGA8B as a crucial gene that influences PCa progression and contributes to CRPC resistance to cabazitaxel.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Prostate cancer (PCa) is a prevalent malignancy, primarily affecting older men, with an incidence rate exceeding 20%. It ranks as the second most frequent cause of cancer-related death in men in the United States of America [1]. China is experiencing a rapid increase in PCa incidence and mortality rates [2]. Currently, a range of effective treatments is available for PCa, with androgen deprivation therapy (ADT) being the preferred first-line treatment [3]. However, after undergoing ADT, these patients ultimately develop castration-resistant prostate cancer (CRPC) [4]. CRPC can be managed through various therapeutic modalities, including chemotherapy [5]. Taxane-based chemotherapeutic agents such as docetaxel have been used in CRPC treatment [6]. However, patients with CRPC treated with docetaxel eventually develop resistance to the drug, necessitating the use of alternative taxane drugs to address docetaxel-resistant CRPC.
Cabazitaxel belongs to the taxane family of drugs. It can bind tubulin and promote microtubule assembly by stabilizing them and preventing their depolymerization, which consequently interferes with cell division and results in cell death [7, 8]. Cabazitaxel has been shown to exert cytotoxic activity and retard the growth of several docetaxel-resistant tumor cells [9]. Furthermore, the roles of cabazitaxel in inhibiting human xenograft tumor growth [10] and in treating CRPC [11, 12] have been reported. Despite the efficacy of cabazitaxel in treating CRPC, patients ultimately become resistant to cabazitaxel, but the mechanism underlying this resistance remains unclear.
Genetic alterations have emerged as significant contributors to resistance to chemotherapeutic drugs in PCa. Studies have shown the pivotal role of genetic alterations as a fundamental factor underlying chemotherapy resistance in PCa, particularly for drugs like docetaxel and vinblastine [13,14,15]. Furthermore, research has highlighted that certain gene mutations can cause resistance to cabazitaxel in PCa [16, 17]. Consequently, investigating alterations in gene expression becomes crucial for elucidating the mechanisms underlying resistance to cabazitaxel in CRPC. In the present study, the GSE158494 dataset retrieved from the Gene Expression Omnibus (GEO) database was utilized to determine the upregulated genes in cabazitaxel-resistant CRPC cells. We identified ten upregulated hub genes that may be important for CRPC cells resistant to cabazitaxel. Furthermore, we singled out one of the ten hub genes, GOLGA8B, which can affect PCa occurrence, progression, and prognosis. We hypothesized that GOLGA8B can influence PCa development and cabazitaxel resistance simultaneously. Thus, we selected GOLGA8B for further study. GOLGA8B was identified as being upregulated in clinical PCa and CRPC specimens, demonstrating its effect on the sensitivity of CRPC cells to both cabazitaxel and docetaxel. This investigation reinforces the pivotal role of GOLGA8B in the development of PCa and resistance of CRPC to cabazitaxel.
2 Materials and methods
2.1 Data collection
This study involved the analysis of multiple datasets retrieved from various sources. We utilized the GSE158494 dataset, containing microarray data derived from cabazitaxel-sensitive and -resistant DU145 and PC-3 CRPC cells. Genes showing differential expression between cabazitaxel-sensitive and -resistant PC-3 and DU145 cells were considered hub genes crucial for the occurrence of cabazitaxel resistance. Additionally, two more datasets, GSE33455 and GSE36135, were acquired, both providing microarray data of docetaxel-resistant CRPC cells. These datasets were sourced from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Furthermore, we analyzed the GSE35988 dataset, encompassing information on patients diagnosed with PCa and CRPC. In addition to these datasets, patient data from two distinct databases were examined: The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) and Chinese Prostate Cancer Genome and Epigenome Atlas (CPGEA; http://www.cpgea.com).
2.2 Data handling
Raw microarray data retrieved from various databases underwent normalization using the “limma” package in the R software. During this process, the probe ID was replaced with the corresponding gene ID. Genes meeting the criteria of |log2FC > 1| and P < 0.05 were classified as cabazitaxel resistance-related genes. Additionally, clinical data were obtained from diverse databases.
2.3 Pathway analysis
Pathway analysis was conducted through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. The online tool Metascape, accessible at http://metascape.org/, was used for this analysis, and a bubble map was generated using the “ggplot2” R package.
2.4 Survival analysis
The online web tool “GEPIA” facilitated the exploration of the correlation between cabazitaxel resistance-related genes and the disease-free survival (DFS) status in PCa patients. Univariate and multivariate Cox regression analyses were performed to determine the variables necessary to develop the nomogram. According to the median gene expression of each gene in PCa patients from TCGA database, patients were divided into high and low gene expression groups. Additionally, the “forest plot” package in the R software was employed to display P-values, hazard ratios (HRs), and 95% confidence intervals (CIs) for each variable. Subsequently, a nomogram was constructed based on the outcomes of the multivariate Cox proportional hazards analysis, serving as a predictive tool for overall recurrence.
2.5 Immune infiltration analysis
Gene Set Cancer Analysis (GSCA; http://bioinfo.life.hust.edu.cn/GSCA/#/) is a public online web tool used to download the immune cell infiltration information of PCa patients from TCGA database. We first downloaded this immune infiltration information from GSCA. Using data from TCGA database, Spearman correlation analysis was conducted to evaluate the relationship between the expression of cabazitaxel resistance-related genes and immune cell infiltration (Additional file 1).
2.6 Clinical specimen collection
Specimens were obtained from individuals diagnosed with PCa and CRPC at Tongji Hospital, School of Medicine, Tongji University. The collection procedure was approved by the Ethics Committee of Tongji Hospital, School of Medicine, Tongji University (SBKT-2021-220). Comprehensive information about the study was provided to all participating patients, and they provided informed consent for the utilization of their samples in the research.
2.7 Cell culture and drug treatment
PCa cell lines were obtained from the Chinese Academy of Science Cell Bank (Shanghai, China). The human CRPC cell lines PC-3 and DU145 were cultured in Roswell Park Memorial Institute (RPMI) 1640 media (Sigma, Darmstadt, Germany; Catalog No. R8758) containing 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, Waltham, MA, USA, Catalog No. 10091). Cell cultures were maintained at 37 ℃ in a humidified atmosphere with 5% CO2 and 95% air. Docetaxel and cabazitaxel were acquired for the study (SelleckChem, Houston, TX, USA, Catalog No. S1148 and S3022, respectively). The CRPC cells were exposed to docetaxel or cabazitaxel at a dose of 2 nmol/L for 24 h. Furthermore, both PC-3 and DU145 cells were treated with docetaxel (2 nmol/L) for 2 weeks to induce resistance. Upon observing that docetaxel did not affect cell growth, we concluded that these cells were resistant to docetaxel (Additional file 2).
2.8 Cell transfection and lentivirus production
Cell transfection experiments were conducted using Lipofectamine 2000 (Thermo Fisher Scientific, Catalog No. 11668019). Specifically, shGOLGA8B lentivirus was designed to facilitate specific gene knockdown. Gene-specific shRNAs and a control lentivirus (shControl) were procured from Youze Biotechnology Company (Hunan, China).
2.9 RNA extraction and qRT-PCR
Total RNA was isolated from clinical samples and CRPC cell lines using the TRIzol reagent (Sigma-Aldrich, St. Louis, MO, USA, Catalog No. T9424). Subsequently, cDNA synthesis was performed using a reverse transcription kit (Advantage® RT-for-PCR Kit, Takara Bio Inc., Kusatsu, Japan, Catalog No. 639505). Gene expression analysis was conducted using a real-time PCR kit (TB Green® Premix Ex Taq™ II, Takara Bio Inc., Catalog No. RR420A) following the manufacturer’s instructions. The PCR primer sequences for GOLGA8B and GAPDH (reference gene) are listed in Table 1. mRNA expression was quantified using the 2−ΔΔCt technique.
2.10 Antibodies
Rabbit polyclonal anti-GOLGA8B antibody (Catalog No. ab155806) and rabbit monoclonal anti-GAPDH antibody (Catalog No. ab9485) were purchased from Abcam UK (Cambridge, UK).
2.11 Western blotting
Proteins were extracted from tissues and cell lines using RIPA lysis buffer. Following extraction, protein samples were processed with treated with Dual Color Protein Loading Buffer (Thermo Fisher Scientific). Protein separation was achieved through SDS-PAGE using a 10% gel, followed by protein transfer onto nitrocellulose membranes sourced from Merck KGaA (Darmstadt, Germany). The Protein-Free Rapid Blocking Buffer (Thermo Fisher Scientific) was used to block the membranes. The membranes were exposed to primary antibodies against GOLGA8B (dilution 1:1000) and GAPDH (dilution 1:1000; Abcam UK) overnight at 4 ℃. The following day, the membranes were washed thrice using 1 × TBST (10 min/cycle) and then incubated at room temperature for 1.5 h with the corresponding secondary antibody [Catalog No. A0208, HRP-labeled Goat Anti-Rabbit IgG (H + L), obtained from Beyotime Biotechnology, Shanghai, China]. Finally, after exposing the membranes to X-ray film, protein identification was conducted.
2.12 Cell proliferation assay
The Cell Counting Kit 8 (CCK 8; Dojindo, Japan) was used to assess the cell proliferation capacity. Briefly, cells were placed in 96-well plates at a density of 3000 cells/well. They were then cultivated for specific lengths of time (0, 24, 48, or 72 h) in 200 µL of RPMI 1640 supplemented with 10% FBS. Subsequently, cell viability was determined using the CCK8 assay following the provided protocol. A spectrophotometer (LD942, Beijing, China) was used to measure the absorbance at 450 nm.
2.13 Statistical analysis
R v4.0.3 (Institute for Statistics and Mathematics, Vienna, Austria; https://www.r-project.org) was used to analyze the matrix data. The Wilcoxon test was applied for comparisons between two groups, and the Kruskal–Wallis test was utilized for comparisons involving more than two groups. Statistical metrics included P values, 95% CIs, and HRs. Statistical significance was set at P < 0.05 (two-tailed).
3 Results
3.1 Identification of ten upregulated hub genes in cabazitaxel-resistant CRPC cells
To uncover the critical genes involved in the development of CRPC resistance to cabazitaxel, we conducted an analysis using the GSE158494 dataset. This dataset comprised microarray data from both cabazitaxel-sensitive and -resistant DU145 and PC-3 CRPC cells. The generation of volcano plots facilitated the visualization of differential gene expression between cabazitaxel-sensitive and -resistant CRPC cells (Fig. 1A, B). This analysis revealed the upregulation of ten hub genes in cabazitaxel-resistant cells compared with that in the sensitive cells, with no detection of downregulated genes (Fig. 1C).
Ten cabazitaxel resistance-related genes were identified from the GSE158494 dataset. A Volcano map reflects the differently expressed genes between cabazitaxel-sensitive and -resistant DU145 cells from the GSE158494 dataset. B Volcano map reflects the differently expressed genes between cabazitaxel-sensitive and -resistant PC-3 cells from the GSE158494 dataset. C Ten genes associated with cabazitaxel resistance from both DU145 and PC-3 cells
3.2 Pathways enriched in association with cabazitaxel resistance-related hub genes
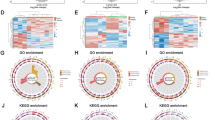
We focused on identifying pathways associated with cabazitaxel resistance-related genes, using GO and KEGG pathway analyses. Bubble maps were constructed to illustrate the pathways exhibiting enrichment. We defined a pathway with the greatest number of enriched genes and the minimum P-value as the most important one. In GO analysis, notable enrichment was observed in the humoral immune response pathway (Fig. 2A). Furthermore, KEGG pathway analysis revealed a predominant enrichment in the IL-17 signaling pathway (Fig. 2B). These findings strongly imply that the hub genes may play a role in cabazitaxel resistance by regulating the immune response.
3.3 Expression analysis of cabazitaxel resistance-related genes in PCa patients
To examine whether the identified cabazitaxel resistance-related genes from CRPC cells alter their expression upon the onset of PCa, we assessed the data acquired from TCGA, comparing the expression profiles of PCa patients and healthy individuals. Among the ten identified hub genes, GOLGA8B was found to be upregulated (P < 0.0001) in PCa tissues, whereas ITGB1, CLIP4, MAP1B, CXCL1, C1S, and CXCL6 exhibited downregulation upon PCa onset (P < 0.05; Fig. 3A). To account for population diversity, since TCGA database primarily includes data from the Western population, the analysis was extended to the CPGEA database, which includes data from Chinese PCa patients. In this dataset, WIPI1 and GOLGA8B (P < 0.0001) were upregulated in Chinese PCa patients, whereas the other genes were downregulated (P < 0.0001; Fig. 3B). Furthermore, any alteration in the expression of these genes during CRPC development was assessed by analyzing the GSE35988 dataset from the GEO database, which contains data from both PCa and CRPC patients. This analysis indicated that CXCL8, MMP13, and GOLGA8B were upregulated in CRPC samples (P < 0.05), whereas the other genes exhibited downregulation (P < 0.01; Fig. 3C). These findings collectively suggest that these cabazitaxel resistance-related genes may play pivotal roles in the onset and progression of both PCa and CRPC.
Expression of cabazitaxel resistance-related genes in PCa and CRPC patients from different databases. Expression of ten cabazitaxel resistance-related genes in PCa patients from A TCGA and B CPGEA databases. C Expression of cabazitaxel resistance-related genes in PCa and CRPC patients from the GSE35988 dataset. “–” represents no statistical differences. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
3.4 Impact of CXCL1 and GOLGA8B on the survival status of PCa patient
We investigated the potential impact of these hub genes on the survival outcomes of PCa patients. Using the online tool GEPIA, the correlation between the expression levels of the ten hub genes and the DFS status of PCa patients was assessed. The expression levels of CXCL1 (P = 0.031) and GOLGA8B (P = 0.00077) exhibited correlations with the DFS status of PCa patients (Fig. 4).
3.5 Impact of GOLGA8B on the prognosis of PCa patients
To ascertain the potential influence of these ten hub genes on PCa patient prognosis, survival clinical data from TCGA database were utilized. Single and multiple Cox regression models were developed to assess their impact on patient prognosis. In the single Cox regression model, CXCL1, CXCL6, CXCL8, and GOLGA8B were determined as genes influencing patient prognosis (P < 0.05; Fig. 5A). However, the multiple Cox regression model identified only GOLGA8B as a risk factor influencing patient prognosis (P < 0.05; Fig. 5B). Furthermore, a nomogram developed to predict the role of these cabazitaxel resistance-related genes in patient prognosis revealed that GOLGA8B was the only gene influencing prognosis (P < 0.001; Fig. 5C). Finally, a calibration curve was constructed to validate these results (Fig. 5D), reaffirming that GOLGA8B can impact the survival and prognosis of PCa patients.
Cox regression reflects the risk of cabazitaxel resistance-related genes in influencing the DFS status of patients. A Single-factor and B muti-factor Cox regression analyses depict the role of cabazitaxel resistance-related genes in influencing the DFS status of patients. C Nomogram illustrates the function of cabazitaxel resistance-related genes in affecting the DFS status of patients. D Calibration curve of the nomogram
3.6 Association between GOLGA8B expression and immune cell infiltration in PCa
Infiltration of immune cells has been established to play a pivotal role in PCa progression [18]. Furthermore, cabazitaxel can affect breast cancer development by influencing immune cell infiltration [19]. Therefore, we hypothesized that cabazitaxel resistance-related genes are correlated with immune cell infiltration in PCa, contributing to its progression. Accordingly, we analyzed the correlation between cabazitaxel resistance-related genes and immune cell infiltration in PCa depending on the data from TCGA. The previous findings collectively emphasize the potential significance of GOLGA8B in PCa development and cabazitaxel resistance. Thus, we conducted an analysis to identify the correlation between some immune cells and GOLGA8B in PCa. Utilizing data from TCGA, we examined the association between GOLGA8B expression and various immune cell types within PCa. The analysis revealed a positive correlation between GOLGA8B expression and CD4+ T cells and neutrophils, whereas it demonstrated a negative correlation with B cells, CD8+ T cells, macrophages, and myeloid dendritic cells (Fig. 6). These results suggest that GOLGA8B might potentially contribute to PCa development and cabazitaxel resistance through its influence on immune cell infiltration.
3.7 Expression of GOLGA8B in clinical PCa and CRPC samples
We verified whether the expression of GOLGA8B changes upon the onset of PCa in clinical patients. For this purpose, we collected 16 paired tumor and pan-cancer normal tissues from clinical PCa patients, including seven CRPC patients. The clinical information is presented in Table support 1 (Table S1). In this study, we collected normal para-cancer and tumor tissues to assess GOLGA8B expression at both the mRNA and protein levels. The analysis revealed an increase in GOLGA8B expression levels (P < 0.01) in PCa tissues compared with that in normal tissues (Fig. 7A, B). Using clinical data of the patients involved, seven paired CRPC cases were selected for a subsequent evaluation of GOLGA8B expression. The results demonstrated an upregulation of GOLGA8B in CRPC tissues at both the mRNA and protein levels (P < 0.05; Fig. 7C, D). These findings underscore the pivotal role of GOLGA8B in the onset and progression of both PCa and CRPC.
3.8 Impact of GOLGA8B on cabazitaxel sensitivity in CRPC cells
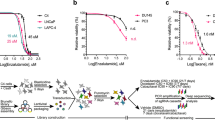
As GOLGA8B exhibited upregulation in cabazitaxel-resistant CRPC cells, it implied a potential role of this gene in CRPC resistance to cabazitaxel. Consequently, we evaluated how GOLGA8B affects the sensitivity of CRPC cells to cabazitaxel. After administering cabazitaxel to the DU145 and PC-3 cell cultures, an increase in the mRNA and protein expression levels of GOLGA8B was observed after a 24 h treatment period (P < 0.05; Fig. 8A–C). A GOLGA8B knockdown (shGOLGA8B) lentivirus was then constructed and administered to CRPC cells. The analysis demonstrated that shGOLGA8B lentivirus effectively reduced the expression of GOLGA8B in CRPC cells (P < 0.01; Fig. 8D–F). Next, we used a CCK-8 assay to detect the function of GOLGA8B in affecting the sensitivity of CRPC cells to cabazitaxel. We transfected the shGOLGA8B or shControl lentivirus into both DU145 and PC-3 cells and then treated them with cabazitaxel. We found that cabazitaxel decreased the proliferation of CRPC cells (P < 0.001; Fig. 8G, H). Additionally, we observed that CRPC cells transfected with shGOLGA8B lentivirus exhibited a decrease in cell proliferation ability (P < 0.05; Fig. 8G, H). We also found that cells with GOLGA8B knockdown were more sensitive to cabazitaxel (P < 0.0001; Fig. 8G, H). These findings underscore the involvement of GOLGA8B in modulating the sensitivity of CRPC cells to cabazitaxel, and cabazitaxel can effectively inhibit CRPC cell proliferation.
GOLGA8B is upregulated in cabazitaxel-resistant CRPC cells and affects the sensitivity of CRPC cells to cabazitaxel. A, B mRNA levels of GOLGA8B after a 24 h treatment of DU145 (A) and PC-3 (B) cells. C Protein levels of GOLGA8B after 24 h treatment of CRPC cells with cabazitaxel. D–F mRNA levels of GOLGA8B after transfection of DU145 (D) and PC-3 (E) cells with GOLGA8B knockdown (shGOLGA8B) lentivirus. F Protein levels of GOLGA8B after transfection of CRPC cells with shGOLGA8B lentivirus. G, H Cell proliferation ability of DU145 (G) and PC-3 (H) cells was assessed after transfection with shGOLGA8B lentivirus, or lack thereof, followed by treatment with cabazitaxel. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. GADPH was used as an inner control in both qRT-PCR and western blotting. Cab: cabazitaxel
3.9 Impact of GOLGA8B on docetaxel sensitivity in CRPC cells
In the context of the primary use of cabazitaxel for treating docetaxel-resistant tumors, we investigated the impact of GOLGA8B on the sensitivity of CRPC cells to docetaxel. To achieve this, we utilized another dataset, GSE33455, containing microarray data of docetaxel-sensitive and -resistant CRPC cells. Analysis of this dataset revealed an upregulation of GOLGA8B in docetaxel-resistant CRPC cells compared with that in their docetaxel-sensitive counterparts (P < 0.01; Fig. 9A, B). Analysis of the GSE158494 dataset, containing data on docetaxel-resistant CRPC cells, revealed an increased expression level of GOLGA8B in these cells compared with that in the control (P < 0.05; Fig. 9C, D). Subsequently, we induced resistance to docetaxel in CRPC cells PC-3 and DU145 by treating them with docetaxel for 2 weeks. We observed that upon developing resistance to docetaxel, both PC-3 and DU145 cells exhibited an increase in GOLGA8B expression at both the mRNA and protein levels (P < 0.01; Fig. 9E–H). Furthermore, we transfected DU145 and PC-3 cells with shGOLGA8B or shControl lentivirus, treated them with docetaxel, and assessed their proliferation ability in the presence of docetaxel. The results indicated that the proliferation ability of CRPC cells decreased upon treatment with docetaxel (P < 0.01; Fig. 9I, J). Additionally, the data demonstrated that the knockdown of GOLGA8B rendered both DU145 and PC-3 cells highly sensitive to docetaxel (P < 0.001; Fig. 9I, J). Collectively, these findings highlight that, in addition to influencing sensitivity to cabazitaxel, GOLGA8B can impact the sensitivity of CRPC cells to docetaxel.
GOLGA8B is upregulated in docetaxel-resistant CRPC cells and affects the sensitivity of CRPC cells to docetaxel. A, B Expression of GOLGA8B in docetaxel-sensitive and -resistant DU145 (A) and PC-3 (B) cells from the GSE33455 dataset. C, D Expression of GOLGA8B in docetaxel-sensitive and -resistant DU145 C and PC-3 D cells from the GSE158494 dataset. E, F mRNA levels of GOLGA8B after 24 h treatment of DU145 (E) and PC-3 (F) cells with docetaxel. G, H Protein levels of GOLGA8B after 24 h treatment of DU145 (G) and PC-3 (H) cells with docetaxel. I, J Cell proliferation ability of DU145 (I) and PC-3 (J) cells treated by docetaxel after transfection with shGOLGA8B lentivirus, or lack thereof. *P < 0.05, **P < 0.01. GADPH was used as an inner control in both qRT-PCR and western blotting. Doc: docetaxel, DocR: docetaxel resistance
4 Discussion
Due to the rapid increase in morbidity and mortality rates, PCa poses a significant threat to the health of older men worldwide [1]. Consequently, more effective treatments to enhance the survival of affected individuals are an immediate necessity. Various effective approaches have been employed for PCa treatment, with ADT being the primary option. Nevertheless, individuals undergoing ADT ultimately develop CRPC, and those entering this stage typically exhibit a median survival duration of less than 20 months [20].
Chemotherapy has demonstrated its effectiveness in treating CRPC. Drugs such as docetaxel and vinblastine have been employed in CRPC treatment for improving patient survival rates [13]. Docetaxel, as an efficacious therapeutic medication, can extend survival durations and enhance the efficacy of ADT in CRPC treatment [21, 22]. Nevertheless, individuals receiving docetaxel treatment may eventually develop resistance, necessitating the use of alternative second-line chemotherapy drugs.
Cabazitaxel, classified as a third-generation, semisynthetic taxane drug that binds to tubulin, is effective in treating tumors that have developed resistance to other taxanes [23]. A phase III open-label clinical trial in 2010 demonstrated the capacity of cabazitaxel to reduce the mortality risk in patients with CRPC following the development of docetaxel resistance [11]. Additionally, another study reported that cabazitaxel can delay tumor progression and enhance the survival rates of CRPC patients [12]. These findings highlight cabazitaxel as an effective treatment option for CRPC, despite potential adverse effects in carcinoma treatment [24]. Nevertheless, resistance to cabazitaxel can also emerge over time in patients previously responsive to the drug. Thus, investigating the potential mechanisms underlying CRPC resistance to cabazitaxel holds promise for improving CRPC management and treatment.
Genetic alterations have been established as pivotal contributors to PCa resistance to chemotherapy drugs. Several studies have indicated that genetic alterations represent a crucial factor in PCa resistance to chemotherapeutic agents, such as docetaxel and vinblastine [13,14,15]. Furthermore, additional research has revealed genetic alterations as an underlying cause of PCa resistance to cabazitaxel. The upregulation of TUBB3 in PCa cells reportedly leads to resistance to cabazitaxel [16]. Furthermore, the amplification of ABCB1 can cause CRPC cells to become resistant to cabazitaxel [17]. These results indicate that genetic changes are crucial for PCa resistance to cabazitaxel. However, until now, systematic studies on hub genes important for CRPC resistance to cabazitaxel are lacking.
In 2020, a study utilized next-generation RNA sequencing to comprehensively compare genes with differential expression between cabazitaxel-sensitive and -resistant CRPC cells [25]. Through the analysis of this study, hub genes were identified as potentially significant contributors to cabazitaxel resistance in CRPC. Furthermore, an examination of their roles in influencing PCa progression was performed. Within the analysis, the upregulation of ten hub genes in cabazitaxel-resistant CRPC cells was observed. Additionally, these genes, associated with cabazitaxel resistance, demonstrated an impact on the onset and progression of both PCa and CRPC. A pivotal gene, GOLGA8B, was identified as capable of influencing the survival and prognosis of patients with PCa. Lastly, upregulation of GOLGA8B was observed in patients with PCa or CRPC, and it was found to affect the sensitivity of CRPC cells to both cabazitaxel and docetaxel.
GOLGA8B is associated with various diseases, and alterations in GOLGA8B expression may contribute to the development of conditions such as dementia and coronary atherosclerosis [26, 27]. Additionally, GOLGA8B expression has been linked to the occurrence of multiple types of tumors. For instance, one study revealed that GOLGA8B can promote lung squamous cell carcinoma by suppressing the expression of STAT3 [28]. Furthermore, certain long non-coding RNAs play a role in the development of clear cell renal cell carcinoma by regulating GOLGA8B expression levels [29]. Another study, utilizing bioinformatics approaches, highlighted the potential significance of GOLGA8B in PCa [30]. In our study, we observed that GOLGA8B not only influences PCa progression and prognosis but also serves as a key gene in conferring resistance against both cabazitaxel and docetaxel in CRPC.
Nevertheless, our study has certain limitations. First, the data on cabazitaxel-resistant CRPC were obtained solely from cell lines within the GSE158494 dataset rather than from clinical samples. The information derived from cell lines alone may substantially differ from clinical data, and relying on a single dataset may introduce bias. However, collecting clinical cabazitaxel-resistant CRPC samples is extremely challenging, limiting our analysis to cell lines. Consequently, a possibility of incompleteness exists in identifying cabazitaxel resistance-related genes, and bias could be present. Second, while assessing GOLGA8B expression in clinical PCa and CRPC samples, the sample size was small. Therefore, the findings may not be fully representative, and the results require validation with a larger sample size. Third, although GOLGA8B was ascertained to play a pivotal role in CRPC resistance to cabazitaxel, the precise underlying mechanism remains unclear. Consequently, future research should prioritize elucidating the mechanism via which GOLGA8B promotes CRPC resistance to cabazitaxel.
5 Conclusion
In this study, we identified ten hub genes that may play important roles in cabazitaxel resistance in CRPC. Furthermore, GOLGA8B was demonstrated to play a pivotal role in the onset and progression of CRPC and serve as a key regulator of cabazitaxel resistance in CRPC.
Data availability
The datasets supporting the conclusions of this article are available in the public databases such as GEO, TCGA and CPGEA. Other data can get from the correspondence author.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. https://doi.org/10.3322/caac.21654.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. https://doi.org/10.3322/caac.21660.
Chen X, Wu Y, Wang X, Xu C, Wang L, Jian J, Wu D, Wu G. CDK6 is upregulated and may be a potential therapeutic target in enzalutamide-resistant castration-resistant prostate cancer. Eur J Med Res. 2022;27:105. https://doi.org/10.1186/s40001-022-00730-y.
Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T, Mason M, Matveev V, Wiegel T, Zattoni F, et al. EAU guidelines on prostate cancer. part II: treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur Urol. 2014;65:467–79. https://doi.org/10.1016/j.eururo.2013.11.002.
Olson KB, Pienta KJ. Recent advances in chemotherapy for advanced prostate cancer. Curr Urol Rep. 2000;1:48–56. https://doi.org/10.1007/s11934-000-0035-z.
Arsov C, Winter C, Rabenalt R, Albers P. Current second-line treatment options for patients with castration resistant prostate cancer (CRPC) resistant to docetaxel. Urol Oncol. 2012;30:762–71. https://doi.org/10.1016/j.urolonc.2010.02.001.
Oberlies NH, Kroll DJ. Camptothecin and taxol: historic achievements in natural products research. J Nat Prod. 2004;67:129–35. https://doi.org/10.1021/np030498t.
Attard G, Greystoke A, Kaye S, De Bono J. Update on tubulin-binding agents. Pathol Biol. 2006;54:72–84. https://doi.org/10.1016/j.patbio.2005.03.003.
Vrignaud P, Semiond D, Benning V, Beys E, Bouchard H, Gupta S. Preclinical profile of cabazitaxel. Drug Des Devel Ther. 2014;8:1851–67. https://doi.org/10.2147/DDDT.S64940.
Cisternino S, Bourasset F, Archimbaud Y, Semiond D, Sanderink G, Scherrmann JM. Nonlinear accumulation in the brain of the new taxoid TXD258 following saturation of P-glycoprotein at the blood-brain barrier in mice and rats. Br J Pharmacol. 2003;138:1367–75. https://doi.org/10.1038/sj.bjp.0705150.
de Bono JS, Oudard S, Ozguroglu M, Hansen S, Machiels JP, Kocak I, Gravis G, Bodrogi I, Mackenzie MJ, Shen L, et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet. 2010;376:1147–54. https://doi.org/10.1016/S0140-6736(10)61389-X.
Oudard S. TROPIC: phase III trial of cabazitaxel for the treatment of metastatic castration-resistant prostate cancer. Futur Oncol. 2011;7:497–506. https://doi.org/10.2217/fon.11.23.
Chen X, Ma J, Wang X, Zi T, Qian D, Li C, Xu C. CCNB1 and AURKA are critical genes for prostate cancer progression and castration-resistant prostate cancer resistant to vinblastine. Front Endocrinol. 2022;13:1106175. https://doi.org/10.3389/fendo.2022.1106175.
Deng L, Gu X, Zeng T, Xu F, Dong Z, Liu C, Chao H. Identification and characterization of biomarkers and their functions for docetaxel-resistant prostate cancer cells. Oncol Lett. 2019;18:3236–48. https://doi.org/10.3892/ol.2019.10623.
Liu RJ, Li SY, Liu LQ, Xu B, Chen M. Identification of biomarkers, pathways and potential therapeutic target for docetaxel resistant prostate cancer. Bioengineered. 2021;12:2377–88. https://doi.org/10.1080/21655979.2021.1936831.
Sekino Y, Han X, Kawaguchi T, Babasaki T, Goto K, Inoue S, Hayashi T, Teishima J, Shiota M, Yasui W, et al. TUBB3 reverses resistance to docetaxel and cabazitaxel in prostate cancer. Int J Mol Sci. 2019. https://doi.org/10.3390/ijms20163936.
Lombard AP, Lou W, Armstrong CM, D’Abronzo LS, Ning S, Evans CP, Gao AC. Activation of the ABCB1 amplicon in docetaxel- and cabazitaxel-resistant prostate cancer cells. Mol Cancer Ther. 2021;20:2061–70. https://doi.org/10.1158/1535-7163.MCT-20-0983.
Strasner A, Karin M. Immune infiltration and prostate cancer. Front Oncol. 2015;5:128. https://doi.org/10.3389/fonc.2015.00128.
Cao X, Li B, Chen J, Dang J, Chen S, Gunes EG, Xu B, Tian L, Muend S, Raoof M, et al. Effect of cabazitaxel on macrophages improves CD47-targeted immunotherapy for triple-negative breast cancer. J Immunother Cancer. 2021. https://doi.org/10.1136/jitc-2020-002022.
Cornford P, van den Bergh RCN, Briers E, Van den Broeck T, Cumberbatch MG, De Santis M, Fanti S, Fossati N, Gandaglia G, Gillessen S, et al. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate cancer. part II-2020 update treatment of relapsing and metastatic prostate cancer. Eur Urol. 2021;79:263–82. https://doi.org/10.1016/j.eururo.2020.09.046.
Sweeney CJ, Chen YH, Carducci M, Liu G, Jarrard DF, Eisenberger M, Wong YN, Hahn N, Kohli M, Cooney MM, et al. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer. N Engl J Med. 2015;373:737–46. https://doi.org/10.1056/NEJMoa1503747.
Vale CL, Burdett S, Rydzewska LHM, Albiges L, Clarke NW, Fisher D, Fizazi K, Gravis G, James ND, Mason MD, et al. Addition of docetaxel or bisphosphonates to standard of care in men with localised or metastatic, hormone-sensitive prostate cancer: a systematic review and meta-analyses of aggregate data. Lancet Oncol. 2016;17:243–56. https://doi.org/10.1016/S1470-2045(15)00489-1.
Mita AC, Denis LJ, Rowinsky EK, Debono JS, Goetz AD, Ochoa L, Forouzesh B, Beeram M, Patnaik A, Molpus K, et al. Phase I and pharmacokinetic study of XRP6258 (RPR 116258A), a novel taxane, administered as a 1 hour infusion every 3 weeks in patients with advanced solid tumors. Clin Cancer Res. 2009;15:723–30. https://doi.org/10.1158/1078-0432.CCR-08-0596.
Nader R, El Amm J, Aragon-Ching JB. Role of chemotherapy in prostate cancer. Asian J Androl. 2018;20:221–9. https://doi.org/10.4103/aja.aja_40_17.
Jimenez N, Reig O, Montalbo R, Mila-Guasch M, Nadal-Dieste L, Castellano G, Lozano JJ, Victoria I, Font A, Rodriguez-Vida A, et al. Cell plasticity-related phenotypes and taxanes resistance in castration-resistant prostate cancer. Front Oncol. 2020;10: 594023. https://doi.org/10.3389/fonc.2020.594023.
Chen H, Wang Y, Sun B, Bao X, Tang Y, Huang F, Zhu S, Xu J. Negative correlation between endoglin levels and coronary atherosclerosis. Lipids Health Dis. 2021;20:127. https://doi.org/10.1186/s12944-021-01545-2.
Blue EE, Thornton TA, Kooperberg C, Liu S, Wactawski-Wende J, Manson J, Kuller L, Hayden K, Reiner AP. Non-coding variants in MYH11, FZD3, and SORCS3 are associated with dementia in women. Alzheimers Dement. 2021;17:215–25. https://doi.org/10.1002/alz.12181.
Li Z, Li Y, Li N, Shen L, Liu A. Silencing GOLGA8B inhibits cell invasion and metastasis by suppressing STAT3 signaling pathway in lung squamous cell carcinoma. Clin Sci (Lond). 2022;136:895–909. https://doi.org/10.1042/CS20220128.
Wang J, Zhang C, He W, Gou X. Construction and comprehensive analysis of dysregulated long non-coding RNA-associated competing endogenous RNA network in clear cell renal cell carcinoma. J Cell Biochem. 2019;120:2576–93. https://doi.org/10.1002/jcb.27557.
Cheng Y, Li L, Qin Z, Li X, Qi F. Identification of castration-resistant prostate cancer-reAlated hub genes using weighted gene co-expression network analysis. J Cell Mol Med. 2020;24:8006–17. https://doi.org/10.1111/jcmm.15432.
Acknowledgements
We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Funding
This study was supported by Natural Science Foundation of Shanghai Municipal Science and Technology Committee (NO.22ZR1456800), National Natural Science Foundation of China (NO.82001610) and Key Discipline of Tongji Hospital in Shanghai (ZD24-MN).
Author information
Authors and Affiliations
Contributions
HPL and XAW designed the study, performed the experiments and analyzed the data. HPL drafted the manuscript. CDX and MHZ performed the experiments and assisted with data analysis. XC revised the manuscript. All authors have read and approved the final manuscript. HPL and XAW confirmed the authenticity of the data.
Corresponding authors
Ethics declarations
Ethic approval and consent participate
The study was approved by the ethic committee of Tongji Hospital, School of Medicine, Tongji University (SBKT-2021-220). Each participate volunteered to join and signed the informed consent form. The study conformed to the provisions of the Declaration of Helsinki.
Competing interests
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, H., Wang, X., Zhai, M. et al. Exploration of the influence of GOLGA8B on prostate cancer progression and the resistance of castration-resistant prostate cancer to cabazitaxel. Discov Onc 15, 152 (2024). https://doi.org/10.1007/s12672-024-00973-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-024-00973-7