Abstract
Silver nanoparticles (NPs) have been demonstrated as a promising antibacterial candidate to fight against resistant pathogens. In this study, different shapes of silver nanostructures (i.e., sphere, rod, and cube) were synthesized by green methods. Morphology, size, and crystalline structure of the produced structures were characterized by UV–visible spectroscopy, scanning electron microscopy (SEM), and X-ray diffraction (XRD). For evaluation of antibacterial activity of silver nanostructures with various shapes, measurement of minimum inhibitory concentrations (MICs) was carried out against Gram-positive (Staphylococcus aureus and Bacillus subtilis) and Gram-negative (Pseudomonas aeruginosa and Escherichia coli) bacteria. The results showed that the concentration of silver nanostructures that prevents bacteria growth is different for each shape, the cubic and rod shape (with sharp edge and vertex) in lower concentrations being more effective than spherical nanoparticles. MTT assay to assess the toxicity of silver nanoparticles showed a concentration and shape-dependent decrease in cell viability in cancer human cells (MCF-7), signifying shape- and dose-dependent toxicity. In addition, the interaction of different nanostructures with serum albumin was evaluated. According to these results, AgNPs with sharper geometry resulted in protein degradation and higher toxicity as compared with smooth or spherical geometries. The results showed that the geometry of silver nanostructures can have quite a significant role in the definition of biological and antibacterial efficacy of NPs, which has significant implications in the design of NPs for various antibacterial applications and will require more consideration in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The antibiotic resistance among microbes is one of the biggest challenges in many fields such as medicine, healthcare settings, and wastewater treatments. Therefore, development of antimicrobial compounds is urgent and a priority to improve bactericidal potential. In this regard, the nanomaterials due to their physical and chemical properties are considered as an effective and potential antimicrobial drugs. It has been understood that metal particles in the nanometer size range exhibit properties that are different from both the ion and the bulk material. Among nanomaterials, silver nanoparticles (NPs) are emerged as a promising candidate to fight against pathogens because of their high surface area-to-volume ratio and the unique chemical and physical properties [1, 2]. It was proposed that the high specific surface area and high fraction of surface atoms of silver NPs will lead to high antimicrobial activity [3]. Experimental evidences suggest that silver ion react with proteins by combining the thiol (SH) groups, which leads to the inactivation of the proteins and bacterial inactivation [3]. However, the detailed antimicrobial effects of silver NPs were not fully investigated.
Chemical reduction is the most applied method for the preparation of silver NPs. However, this method involves using or generation of highly toxic and hazardous materials, which can limit the use of silver NPs for biomedical applications. Consequently, to avoid the use of organic solvents and toxic reducing agent, using biological compounds or green synthesis of nanoparticles has been investigated as a biocompatible method for the synthesis of such particles. The green synthesis of silver NPs involves three main steps, which must be evaluated based on green chemistry perspectives, including (1) selection of solvent, (2) selection of environmentally reducing agent, and (3) selection of non-toxic substances for the silver NPs stability [4]. Most of these methods were carried out in water, an environmentally-friendly solvent. In polysaccharide method [5], silver NPs are prepared using water as an environmentally solvent and polysaccharides as a capping agent or in some cases polysaccharides serve as both a reducing and a capping agent. Eco-friendly bio-organisms including plant extracts and microorganism such as proteins of fungi, which act as both reducing and capping agents forming stable and shape-controlled silver NPs. We have recently developed green methods for synthesis of silver NPs. In our previous studies, green synthesis of silver NPs was accomplished by using fungal species including Fusarium oxysporum [6,7,8] and Neurospora intermedia [9].
It is recognized that antibacterial properties of silver NPs are dependent on their physicochemical properties such as composition, size, and shape. Therefore, synthesis of silver nanostructures with suitable shapes and size is required. To the best of our knowledge, there is no evidence on the synthesis of different shapes of nanosilvers by green methods. In this study, different shapes of silver nanostructures were synthesized by using green methods. The produced structures were characterized by UV–visible spectroscopy, SEM, and XRD analysis. The silver nanostructures of known shapes were further used for studying the antibacterial effects against Gram-positive and Gram-negative bacteria. In addition, the biological activity and toxicity on human cell were investigated.
2 Materials and Methods
2.1 Materials
Chitosan with medium molecular weight, starch, silver nitrate, and β-d-glucose were purchased from Sigma-Aldrich. Bacterial strains and the fungus F. oxysporum were obtained from Iran Zistdaru-Danesh and Persian Type Culture Collective, respectively.
2.2 Synthesis of Silver Nanostructures with Different Shapes
2.2.1 Synthesis of Cubes
Chitosan was dissolved in 1% acetic acid solution [10]. A mixture of 4 ml of AgNO3 (0.2 M) and 4 ml of glucose (0.2 M) was added to 10 ml of chitosan solution (3.46 mg/ml) under stirring at room temperature. Then, 20 μl of HCl (2 M) was added and, subsequently, the reaction mixture was autoclaved (121 °C, 15 psi) for 1 h. Finally, the reaction mixture was centrifuged (at 15000 rpm for 10 min) and the resulted NPs were washed two times with distilled water to remove the excess amounts of chitosan and AgNO3. The resulting solution was dark green in color, indicating the formation of silver cubic structures.
2.2.2 Synthesis of Spheres
For the synthesis of sphere nanoparticles, a modified green method was applied by using the fungus F. oxysporum and starch [8]. Briefly, 1 ml of a starch solution (1%, w/w) was added to 1 ml of an aqueous solution of AgNO3 (0.08 M), and the final pH was adjusted to 6.8 using a phosphate buffer. Then, the culture supernatant of F. oxysporum was added to the above solution. Finally, the reaction mixture was incubated at 50 °C and agitated at 200 rpm for 24 h to complete the biosynthesis of the nanoparticles. After incubation time, the mixture was centrifuged (at 15,000 rpm for 10 min) and the resulted AgNPs washed two times with distilled water. The resulting solution was yellow in color indicating the formation of silver spherical nanoparticles.
2.2.3 Synthesis of Nanorices
The experimental procedure of silver nanorice resembled the synthesis of sphere nanoparticles except for the pH of solution, concentration of AgNO3, incubation time, and temperature. The concentration of AgNO3 was 1 M, while the final pH of solution was adjusted to 3 using HCl. Keeping the ratio between culture supernatant and AgNO3 unchanged, the reaction mixture was incubated at 30 °C and agitated at 200 rpm for 3 days to complete the biosynthesis of the nanoparticles.
2.2.4 Synthesis of Nanorods (Blunt Ends)
The experimental procedure resembled the synthesis of nanorices except for the concentration of AgNO3. In this experiment, the concentration of AgNO3 was increased to 1.2 M. The orange silver nanorice particles turned to red, indicating the formation of silver nanorod structures.
2.2.5 Synthesis of Nanorods (Sharp Ends)
The blunted end nanorods prepared at a previous stage were incubated with the same condition for 10 days. After 10 days of aging, the solution of nanorods changed to brown.
2.3 Characterization of Silver Nanoparticles
2.3.1 UV–Visible Spectral Analysis
The color changes of reaction mixtures were used as evidence for the formation of different shapes of silver nanostructures. Therefore, 1-ml samples were withdrawn at various intervals, and the absorbance was measured by a double beam UV–visible spectrophotometer (Cary 100, Varian) at a resolution of 1 nm in the range 200–800 nm.
2.3.2 XRD
X-ray diffraction was carried out using a Philips PW-1730 System (Philips, the Netherlands) operating at the Co Kα wavelength of 1.7889 A, 30 mA, and 40 kV. The diffractograms were recorded over 2θ range from 20° to 80°. Samples were prepared by casting silver solutions on a silicon substrate.
2.3.3 SEM
The size and morphology of the synthesized silver nanostructures were accomplished by SEM (ΣIGMA/VP). The SEM images were recorded at 15.00 kV. Thin film was prepared by drop coating of purified green synthesized nanosilver from all prepared solutions onto carbon-coated copper SEM grids.
2.4 Antibacterial Activity of Different Shape Silver Nanostructures
The minimum inhibitory concentration (MIC) values for Gram-positive (Staphylococcus aureus and Bacillus subtilis) and Gram-negative (Escherichia coli and Pseudomonas aeruginosa) were determined by a standard microdilution method in liquid medium (Mueller–Hinton broth), using 96-well plates. The inocula of these bacteria were adjusted to 0.5 MacFarland densities. Stock solutions of nanosilvers (1 mg/ml) were prepared in sterile distilled water and diluted to the required concentrations using Mueller–Hinton broth. Appropriate concentrations of nanosilvers were added to the bacterial cultures in each well to obtain respective concentration (2 to 100 μg/ml) of silver NPs and incubated for 48 h. The lowest concentration that completely inhibited the growth of the microorganism as compared with the growth in the culture control well was taken as the MIC of silver NPs.
2.5 Cytotoxicity Assay
The cellular cytotoxicity of silver nanostructures was measured using MTT assays against MCF-7 cell line. MCF-7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) cell culture medium supplemented with penicillin (100 μg ml−1), streptomycin (100 U ml−1), and 10% heat-inactivated FBS. The cells were kept in at 37 °C humidified atmosphere with 5% CO2. Cells (105) were seeded into each well of 96-well culture plates for 24 h. The cells were exposed to different shapes of silver nanostructures, all at concentrations of 5, 10, 25, 35, 50, and 75 μg ml−1 in DMEM supplemented with 10% FBS incubated for 24 h at 37 °C and 5% CO2. Following incubation, the test medium was removed and the culture wells were washed twice with 1× PBS buffer. The cell viability was determined using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. Viable cells are capable of metabolizing MTT while dead cells are not. The cells were incubated with 100 μl of MTT solution (0.5 mg ml−1 MTT diluted in PBS) for 4 h at 37 °C and 5% CO2. Subsequently, the MTT solution was discarded and DMSO (100 μl) was added to every well. The optical density (OD) was measured at 600 nm. The cell viability for each treatment was calculated as the ratio of the mean OD of separate wells (n = 3) relative to that of the control, where only cell culture medium was added. Values are calculated as mean ± standard deviation.
2.6 Evaluation of Protein Interaction
In order to define the interaction of various shapes of silver nanostructures with proteins, SDS–PAGE (12%) was employed. To this end, 0.9 ml of human serum albumin (HSA) (0.1 mg/ml) was prepared and 0.1 ml of various silver samples with the same concentration (30 μg/ml) were added and incubated at 37 °C for 1 h. The samples were centrifuged at 16,000 rpm for 20 min to separate the protein-coated nanosilver. The protein-coated nanosilver was resuspended in 500 μl of phosphate-buffered saline (PBS) pH 7.4 and centrifuged again to remove not adsorbed HSA. Finally protein-coated nanosilver dispersed in 40 μl of PBS followed by the addition of 6 μl of loading buffer, containing β-mercaptoethanol, and the samples were loaded to SDS–polyacrylamide gel electrophoresis (12%) for analysis [11].
3 Results and Discussion
3.1 UV–Visible Analysis of Silver Nanostructures
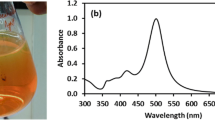
UV–visible spectroscopy was used for evaluation of shape related optical properties of synthesized structures. Several studies revealed that optical properties of silver NPs are strongly dependent on both size and shape. Overall appearance and compartment of optical absorption spectrum of silver NPs including wavelength region of surface plasmon absorption peak, number of absorption peaks, and spectrum broadening are predominantly related to the shape of nanoparticle; however, details of the optical absorption curve including fine position of surface plasmon peak are dependent upon the size of the nanoparticle. Figure 1 demonstrates the dependency of absorption spectrum on the symmetry of silver NPs morphology, so spherical silver NPs exhibits absorption spectrum with only one absorption peak at wavelength 447 nm with symmetrical absorption intensity around peak position. In addition, the absorption intensity at long wavelengths is imperceptible. Generally this is the optical spectrum characteristic of silver spherical nanoparticles. For non-spherical structures, optical absorption demonstrates a broad spectrum with main absorption peaks at 470, 436, 391, and 406 nm for nanorice, nanorod with sharp ends, nanorod with blunt ends, and cubic nanostructures, respectively. All absorption spectra in comparison with optical absorption spectrum of spherical silver NPs demonstrated non-symmetrical absorption intensity around surface plasmon absorption peak position. Due to the broad size distribution of non-spherical nanostructures, the quadrupole modes and longitudinal plasmon peaks for different nanoparticles are located at different wavelengths near each other; therefore, the second plasmon peak has been disappeared. This is the main reason of spectrum broadening. However, some morphologies such as monodisperse nanorod with sharp ends exhibits longitudinal and transverse plasmon resonance peaks at 436 and 551 nm, respectively. In addition, nanorod with blunt ends demonstrates longitudinal and transverse plasmon resonance peaks at 391 and 506 nm, respectively. The similar optical properties for non-spherical silver NPs were reported before [11, 12].
3.2 Crystal Structure
The crystalline phase and purity of synthesized nanostructures were examined by XRD analysis. The results for spherical, cubic, nanorice, and rod samples are illustrated in Fig. 2. The peaks appeared at 2θ = 38.17, 44.34, 64.55, and 77.45 which are assigned to the (111), (200), (220), and (311) planes, respectively, and correspond to the formation of silver phase according to card no. 01-089-3722 from JCPDS. Crystallographic data obtained from XRD pattern shows the formation of the metallic silver of face center cubic (fcc) lattice structure in all synthesized structures. In order to further identify different shapes of silver nanostructures based on the XRD results, we considered the intensity ratios between the (200) and (111) planes, between the (220) and (111) planes, and between (311) and (111) planes which are shown in Table 1. The intensity ratios of (200)/(111), (220)/(111), and (311)/(111) for four different shapes were all smaller than the standard values (Table 1). This result reveals that the all shapes were abundant in (111) crystalline planes. In other words, the (111) planes are predominant as the basal face in four different shapes of silver NPs [13]. In a similar study, Ashkaran et al. reported that by orienting the Ag structures in some specific directions, especially for rod-like structures, the intensities in some particular directions increase, indicating the preferential growth in those directions [11].
3.3 Size and Morphology of Various Shapes of Silver NPs
In order to determine the size and shapes of synthesized silver NPs, SEM was used. Figure 3 shows SEM images of different shapes of silver NPs including nanorice, nanorod with sharp ends, and nanorod with blunt ends, spherical and cubic. The dimensions of nanoparticles are presented in Table 2. Spherical silver NPs are the smallest, and both nanorods with blunt and sharp ends have the maximum aspect ratio.
There are several studies on green synthesis of silver and gold nanoparticles; however, the dominant shapes of the green synthesized silver NPs are reported as spherical or quasi-spherical [4, 10, 14,15,16]. To the best of our knowledge, there have not been any efforts on the green synthesis of different shapes of silver nanoparticles similar to what is presented in this research.
For the synthesis of silver with various shapes, the temperature, concentration of AgNO3, pH of solution, and time of synthesis are the factors affecting on this process. When nucleus are formed, some of planes are stabilized by interaction with capping or surfactant agents, then other planes will grow. It is possible that atoms of crystallographic planes have different interactions with polymers and surfactants, which lead to anisotropic growth. In this case, asymmetric particles will be formed [17, 18].
In order to synthesis the cubic nanoparticles, concentration of precursor must be high (more than 0.05 M in polysaccharide method and more than 0.1 M in combinatorial method) and the amount of stabilizer should be low. In this case, the nucleation will be fast and time for format of twin defects will be very low. Chitosan like other similar chemical compounds (such as PVP) can reduce surface energy of the planes (100) by interacting and bonding with them, and then surface energy of the (100) is lower than (111) and twin formation will not happen. In this case, cubo octahedron single-crystal seeds will be formed and finally (111) planes will grow. When cube is formed, all the planes have the same surface energy; therefore, growth rate of all planes will be the same [11, 17, 19].
For the synthesis of other shapes of silver nanostructures by green methods, in low concentration of precursor (0.1 M in combinatorial method), chemical potential for crystallization is reduced, and multiple twined decahedra seeds is formed, because they are the most thermodynamically stable seed. In this study, starch and culture supernatant of Fusarium fungus were used, which act in a way similar to CTAB that covered (100) facets and act as both of stabilizer and reducer. Starch and supernatant of F. oxysporum bond more with (100) facets than (111). When pH of the solution is acidic and at short period of time (3 days), nanorice will be synthesized and by increasing the synthesis time (21 days), nanorod with sharp ends will be formed, because with increase the time, silver ions have more time to place on two ends of nanorice and then will grow.
During nanorod formation as aforementioned for nanocube, early nucleus will be single-crystal and stages of growth repeat. When cube formed, growth of nanoparticle will be on (110) axis. In combinatorial method, starch and supernatant of F. oxysporum covered (100) facets will lead to the preferential addition of silver atoms to other facets and early nucleus will have growth on (110) axis [20].
The size of silver nanostructures can be controlled by changing the concentration and ratio of capping agent to AgNO3 [11]. In this study, spherical silver nanoparticle was obtained at low concentration of silver nitrate (0.08 M) relative to concentration of starch as capping agent; in this case, a thick film of starch covers the entire surface of the formed seeds and prevents the growth of nanoparticles.
3.4 Antibacterial Activities
The MICs of different shapes of silver NPs for the test bacterial strains were used to determine and compare their antibacterial potencies. Figure 4 shows the results of this assay. All shapes of silver NPs showed antimicrobial activities against both the Gram-positive and Gram-negative bacteria. When bacterial strains were exposed to different concentration of nanosilver solutions, silver solution of lower than 10 ppm for cubic structure was enough to inhibit all viable cells of bacteria within 24 h. However, this low concentration for other shapes of nanosilver was not enough to inhibit bacteria, and at higher concentration, the growth inhibition happened.
As it is shown in Fig. 4, the obtained antibacterial results suggest that physicochemical properties of nanosilver such as shape have significant influence on the antibacterial activity of silver nanostructures. More specifically, as presented in Fig. 4, cubic and rod with sharp ends exhibited the most significant antibacterial activity among the five shapes of silver nanostructures against bacteria. The results demonstrated that the lowest MIC value was associated with these nanostructures (cubic and sharp-rod) and may be due to the existence of sharp vertex with less angle which causes increase in the strength of electric field intensity. As a result, sharp vertexes in these nanostructures have stronger interaction with bacterial cell membrane which is in agreement of previously reported results [21]. Nanorice and spherical NPs showed the antibacterial activities against bacteria after cubic and rod; however, they have smaller size than cubic and sharp-rod. According to these results, it turns out that the antibacterial activity of nanosilvers is more dependent on the shape than size of nanostructures. In another study, Pal et al. reported the effect of spherical, rod, and triangular silver nanoparticles synthesized by citrate reduction against E. coli at different concentrations. It was found that triangular nanoparticles with a (111) lattice plane as the basal plane displayed the strongest biocidal action, compared with other nanoparticles [22].
3.5 Cell Viability Assay
The cell viability for silver nanostructures with different shapes is shown in the bar charts of Fig. 5. Interestingly, for concentrations lower and equal to 10 μg/ml, silver nanostructures have no significant effect on the cell viability. This indicates that silver nanostructures with different shapes are not inherently toxic to human cells at low concentrations. However, for concentrations larger than 10 μg/ml, the silver treatment limits cell viability in a shape and dose dependent manner. Silver nanocube, nanorice, and sharp-nanorod decreased cell viability at concentrations of 25 and 35 μg/ml as compared with the control (p value <0.05), whereas nanosphere and blunt-nanorod showed no significant cytotoxic effect at these concentrations (p value >0.05). Within the tested range (5–75 μg/ml), MCF-7 cells retained over 60% viability when exposed to silver nanosphere up to 75 μg/ml over 24 h, whereas cell viabilities lower than 30% can be observed with silver nanocubes at this concentration. As it is shown in Fig. 5, the results indicate that there is significant differences between cytotoxicity of different shapes of silver nanostructures. Silver nanosphere has lower toxicity to human cells in comparison with cubic and sharp-rod nanostructures.
3.6 Silver Nanoparticle–Protein Interaction
Silver NPs with various shapes were exposed to human serum albumin for 1 h. The concentration of silver nanostructures was the same (30 μg/ml) for all shapes. The comparison of SDS–PAGE analysis of protein interaction with various shapes and morphologies of silver nanostructures is presented in Fig. 6. The most prominent observation is that cubic nanostructures show significant protein damage and band changes. However, the protein patterns of other nanostructures (i.e., sphere, rods, and nanorice) have not significant change, suggesting that these nanostructures have less binding affinity for proteins. Indeed, cubic structures with sharp ends act as aggressive binders and interact with protein lead to denaturation of proteins. This result is in agreement with earlier study [11]. Ashkaran et al. demonstrated that the competition of proteins for attachments to the sharp edges at the surface of silver NPs is much higher than that of the smooth surfaces [11].
Since silver NPs as antibacterial agent should have high toxic effect on bacteria and no toxic (biocompatible) effect on human cells, the investigation on their corresponding protein interaction is of crucial interest.
It is noteworthy to mention that research on silver NPs to date has been primarily focused on spherical nanoparticles and little has been known regarding the various shapes of silver nanostructures that have broad implications for understanding the mechanism of action and their interaction with biological environments. Here, it was shown how cubic shape of nanosilver can degrade the structure of proteins. This observation is consistent with MTT assay data. According to these results obtained from biological activity of various shapes of silver nanostructures, the toxicity of these nanostructures is shape and dose dependent. It seems that silver NPs can act as a double-edged sword. Silver NPs can be used as antimicrobial agents in medicine to fight against pathogen bacteria. On the other hand, the interaction with biological environment and proteins can cause damage to the host cell. Therefore, in designing of silver NPs-based antibiotic, this issue will require more consideration in the future. It can be proposed that silver nanostructures with smooth surfaces could be considered as antibacterial agents in pharmaceutical fields, and silver nanostructures with sharp edges and more aggressive activity could be a promising candidates to fight against biofilm formation in industrial instruments.
4 Conclusion
In this study, silver nanostructures with different shapes including spherical, cubic, nanorice, sharp-rod, and blunt-rod were synthesized by green methods and their biological activities were investigated. Since silver nanoparticles are finding increasing applications in biological systems, for example, as antimicrobial agents, the study of interaction of these nanoparticles with host cell and biomolecules is of considerable importance. In this study, we conclude that silver nanostructures at concentrations higher than 10 ppm have the potential to cause toxicity on human cells which is in a shape and dose dependent manner. The present study concludes that cubic silver nanostructure has antibacterial activity with MIC of lower than 10 ppm and decreases the cell viability of human cells in higher dose than MIC for bacteria. At concentration that cubic nanostructures causes cell toxicity, the interaction with protein was also investigated and showed that at this dose (30 ppm), cubic nanostructure completely has degraded protein; however, at this dose, other shapes of nanosilver have no denaturation effect on protein structure. On the basis of the aforementioned results, we conclude that due to stronger vertex in sharp ends of nanosilvers, some geometries of nanosilver especially cubic structures have more interaction with bacterial cell membrane and proteins which leads to stronger biological activity. Future application of silver nanostructures as antibacterial agent could be limited by the fact that they are toxic to human cells. However, more investigations should be conducted in this field to achieve the deeper understanding of nanosilver toxicity. We suggest that for the synthesis and application of nanosilver as antibacterial agents, shape and concentration should be considered accordingly to maximize the antibacterial activity and minimize their toxicity on host cells.
References
Rai, M., et al. (2012). Silver nanoparticles: the powerful nanoweapon against multidrug-resistant bacteria. Journal of Applied Microbiology, 112(5), 841–852.
Chen, X., & Schluesener, H. (2008). Nanosilver: a nanoproduct in medical application. Toxicology Letters, 176(1), 1–12.
Cho, K.-H., et al. (2005). The study of antimicrobial activity and preservative effects of nanosilver ingredient. Electrochimica Acta, 51(5), 956–960.
Sharma, V. K., Yngard, R. A., & Lin, Y. (2009). Silver nanoparticles: green synthesis and their antimicrobial activities. Advances in Colloid and Interface Science, 145(1), 83–96.
Haizhen Huang, X. Y. (2004). Synthesis of polysaccharide-stabilized gold and silver nanoparticles: a green method. Carbohydrate Research, 339(15), 2627–2631.
Mohammadian, A., Shojaosadati, S., & Habibi Rezaee, M. (2007). Fusarium oxysporum mediates photogeneration of silver nanoparticles. Scientia Iranica, 14(4), 323–326.
Khosravi, A., & Shojaosadati, S. (2009). Evaluation of silver nanoparticles produced by fungus Fusarium oxysporum. International Journal of Nanotechnology, 6(10–11), 973–983.
Ghaseminezhad, S. M., Hamedi, S., & Shojaosadati, S. A. (2012). Green synthesis of silver nanoparticles by a novel method: comparative study of their properties. Carbohydrate Polymers, 89(2), 467–472.
Hamedi, S., et al. (2014). Extracellular biosynthesis of silver nanoparticles using a novel and non-pathogenic fungus, Neurospora intermedia: controlled synthesis and antibacterial activity. World Journal of Microbiology and Biotechnology, 30(2), 693–704.
Wei, D., & Qian, W. (2008). Facile synthesis of Ag and Au nanoparticles utilizing chitosan as a mediator agent. Colloids and Surfaces B: Biointerfaces, 62(1), 136–142.
Ashkarran, A. A., et al. (2012). Bacterial effects and protein corona evaluations: crucial ignored factors in the prediction of bio-efficacy of various forms of silver nanoparticles. Chemical Research in Toxicology, 25(6), 1231–1242.
Rycenga, M., et al. (2011). Controlling the synthesis and assembly of silver nanostructures for plasmonic applications. Chemical Reviews, 111(6), 3669–3712.
Sun, Y., & Xia, Y. (1991). Large-scale synthesis of uniform silver nanowires through a soft, self-seeding, polyol process. Nature, 353(1991), 737.
Venkatesham, M., et al. (2014). A novel green one-step synthesis of silver nanoparticles using chitosan: catalytic activity and antimicrobial studies. Applied Nanoscience, 4(1), 113–119.
Mohan, S., et al. (2014). Completely green synthesis of dextrose reduced silver nanoparticles, its antimicrobial and sensing properties. Carbohydrate Polymers, 106, 469–474.
Birla, S.S., et al., Rapid synthesis of silver nanoparticles from Fusarium oxysporum by optimizing physicocultural conditions. The Scientific World Journal, 2013. 2013.
Krutyakov, Y. A., Kudrinskiy, A. A., Olenin, A. Y., & Lisichkin, G. V. (2008). Synthesis and properties of AgNPs: advances and prospects. Russian Chemical Reviews, 77, 233–257.
Skrabalak, S. E., et al. (2007). Facile synthesis of Ag nanocubes and Au nanocages. Nature Protocols, 2(9), 2182–2190.
Wiley, B. J., et al. (2006). Maneuvering the surface plasmon resonance of silver nanostructures through shape-controlled synthesis. The Journal of Physical Chemistry B, 110(32), 15666–15675.
Liao, H.-G., et al. (2014). Facet development during platinum nanocube growth. Science, 345(6199), 916–919.
Ojha, A. K., et al. (2013). Synthesis of well–dispersed silver nanorods of different aspect ratios and their antimicrobial properties against gram positive and negative bacterial strains. Journal of Nanobiotechnology, 11(1), 1.
Pal, S., Tak, Y. K., & Song, J. M. (2007). Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram-negative bacterium Escherichia coli. Applied and Environmental Microbiology, 73(6), 1712–1720.
Acknowledgements
The authors wish to acknowledge Iranian nanotechnology initiative council for their financial support towards major research project.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Soleimani, F.F., Saleh, T., Shojaosadati, S.A. et al. Green Synthesis of Different Shapes of Silver Nanostructures and Evaluation of Their Antibacterial and Cytotoxic Activity. BioNanoSci. 8, 72–80 (2018). https://doi.org/10.1007/s12668-017-0423-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12668-017-0423-1