Abstract
Brain tumors are the most common and vigorous cause of death in the modern era. The medical community is working hard to develop effective methods to detect brain tumors in an early stage. Machine learning-based optimized classifiers can provide an efficient, accurate, and timely solution to detect brain tumors. Herein, a three-step least complex optimal linear support vector network-based computer-aided healthcare system for tumor cell detection using magnetic resonance imaging (MRI) is proposed. In the first step, features obtained from the Handcrafted features (HF) and a 14-layered convolutional neural network (CNN) operating in parallel are concatenated. Initially, these combined features are used for tumor classification. In the second step, to reduce the computational complexity, the bag of feature vector (BoFV) technique followed by principal component analysis (PCA) is introduced to select quality features. As this research focuses on the early-stage detection of brain tumors, an optimized linear support vector network (oLSVN) was introduced for classification in the third step. oLSVN sends tumors-classified images for segmentation to detect the exact area of the tumors, whereas the images in which the tumor is not detected due to poor visibility and noise undergo contrast-limited adaptive histogram equalization (CLAHE) process for noise filtration and image enhancement. These enhanced images are classified again for brain tumor detection in an early stage. A comparative analysis shows that the proposed model outperforms some already existing models. The execution time of the proposed model is \(1.32\) seconds with \(98.25\%\) accuracy. As compared to some already existing approaches, the proposed model has an F1-Score of \(98.27\%,\) precision of \(97.28\%\), specificity of \(97.22\%\), and a Mathew's Correlation Coefficient of \(96.52\%.\) These results validate that the proposed state-of-the-art methodology can thus help the medical industry in the timely and efficient detection of brain tumors.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Medical technology is advancing day by day, however, there is still room for improvement in the methodologies and technologies used to detect and classify different diseases. A first step toward improving healthcare systems for real-time application will be to integrate cutting-edge technologies with medical data sets. The second step is to create algorithms for health data management that are accurate, precise, and resourceful. Currently, a benign tumor may turn out to be malignant due to misidentification at an early stage as healthcare biometrics are not in practice everywhere. The best solution for countries that cannot afford expensive equipment to treat patients with brain tumors is to equip a machine integrated with medical data sets that support health data management for precise analysis (Mattila et al. 2021). Healthcare services are now among the best in the world, thanks to the advances in machine learning techniques and the development of the Internet of Medical Things (IoMT). Brain tumors are deadly diseases in humans and there is no way to detect them in their early stages (Garg and Garg 2101). Magnetic resonance imaging (MRI) can easily detect the disease, however, only in the final stages when the tumor is visible. The healthcare sector thus needs advanced machine-learning technology for tumor detection.
Different authors have recently used deep neural networks (DNN), such as CNN and RNN, for tumor classification. In (Liu et al. 2021), the authors used a neutrosophic set–expert maximum fuzzy-sure entropy (NS-EMFSE) for feature enhancement. The authors focus on algorithms that aid in system design to analyze the MRI images for tumor detection. Moreover, the authors used a feedback system in their optimized network for tumor detection in an image even at its early stages. (Ozyurt et al. 2019) used CNN with neutrophils to classify tumors as benign or malignant based on the MRI images. The authors used expert NS-EMFSE of MRI images obtained from the CNN model for segmentation, whereas support vector machine (SVM) and k-nearest neighbors (kNN) classifiers were used for identification.
In the literature, some researchers have proposed the use of imaging sensors to capture higher-quality images. Wireless sensors are ideal for such tasks as they can monitor, record, and transfer physical changes wirelessly. Furthermore, the operation of these imaging sensors is based on real-time applications, which thus aids in brain tumor diagnosis by providing high-quality images. In (Shakeel et al. 2019), the authors proposed wireless infrared imaging sensor-based DNN for brain tumor detection. The authors used a machine learning-based back propagation neural network (ML-BPNN) to acquire clear and precise images. The images were examined using infrared (IR) imaging techniques. The fractal dimension algorithm (FDA) was used in real-time to extract critical features, whereas the remaining exclusively critical features were extracted using dint of multifractal detection. As a result, complexity was reduced and the results obtained were effective and accurate.
Narmatha et al., in Narmatha et al. (2020) suggested a hybrid solution, i.e., a fuzzy brain-storm optimization algorithm (FBSO) for the detection and classification of Glioma brain tumors. The FBSO algorithm obtained using BRATS 2018 was accurate with highly satisfying results. An active deep learning-based model was used for feature selection, recognition, and classification of a brain tumor in Sharif et al. 2020a. In the pre-processing stage, the authors sharpened the contrast, and the images were converted into a binary format which helped in reducing the machine errors. A CNN model was applied and fitted into the dominant rotated local binary patterns (DRLBP) for detailed analysis. BRATS 2017 and 2018 were used for segment validation, whereas BRATS 2013, 2014, 2017, and 2018 were used for classification. (Zhao et al. 2018) used conditional random field with full CNN to identify and classify brain tumors with the help of MRI images. The authors also presented an alternative technique, i.e., a conditional random field with deep medic in Zhao et al. (2018), which further enhanced the results when combined with the first technique. As the manual ways of segmentation are time-consuming and challenging, authors in Pereira et al. (2016) provided an effective and easy way of segmentation using an enhanced CNN. Small kernels were designed for the in-depth details, whereas skull stripping and image enhancement were used in the pre-processing stage.
In (Deepak and Ameer 2019), the authors adopted a pre-trained GoogleNet technique and applied the process of deep transfer learning to extract features from the images. The proposed method applies the concept of classical image restoration, single image super-resolution (SISR), and threshold segregation based on the maximum fuzzy entropy segmentation (MFES) for allocating medical images into different sets. After analyzing the misclassifications, the authors proposed that an advantageous technique would be transfer learning if the medical images are available. (Sert et al. 2019) utilized CNN and SVM to extract valuable features with the help of a pre-trained ResNet structure. The authors showed that SISR presents transparent and accurate information. In (Chaudhary and Bhattacharjee 2020), the researcher utilized a sturdy image processing technique on MRI images. Images were initially segmented using the clustering-based methodology and tumor detection is carried out using SVM.
In (Soltaninejad, et al. 2017), the authors proposed parallel processing of two MRI images using T2 weighted image and fluid-attenuated inversion recovery (FLAIR). Their method is based on the super-pixel technique where the classification of each super-pixel is carried out using FLIAR. An innovative and modified approach for brain tumor detection named BrainMRNet was proposed in Sharif et al. 2020b. BrainMRNet was used in the pre-processing stage and the pre-processed images were transferred to the active attention modules, where the crucial features of the image were highlighted. The image was then moved to the convolutional layers for further processing. The authors in Toğaçar et al. 2020a suggested a classification framework based on data augmentation. A comparative analysis with different ML models proved that CNN is more efficient among them. (Saba et al. 2020) proposed fine-tuned GrabCut method for brain tumor segmentation, whereas a transfer learning approach was used for feature extraction. The author used different segmentation techniques, including k-means clustering, fuzzy entropy, and thresholding. The features obtained were classified and merged into a single vector and were validated on MICCAI using BRATS 2015, 2016, and 2017.
A distinctive collective algorithm, i.e., potential field segmentation (PFS) was proposed in Cabria and Gondra (2017). The results of PFS, potential field clustering (PFC), and force clustering (FC) were merged and tested on BRATS software for evaluation. In (Fazelnia et al. 2020), multi-stage morphological methodologies were designed and presented and several data sets were used to obtain clarity and precision in the images. The images were pre-processed for denoising and converted into the binary form, whereas morphological processing was established for clarity before the final segmentation. Optimized computational programming tools were used to improve the accuracy. The authors in Sharif et al. 2020c proposed a particle swarm optimization (PSO) algorithm for tumor detection. The skull was eliminated in a picture and transferred into the PSO for better segmentation. Asghar et al. proposed high-quality feature selection using deep features obtained from segmented images and local binary patterns (LBP) in Asghar et al. (2020). Features were selected, merged, and classified before segmentation using a genetic algorithm (GA). This data compilation method has shown significant improvement in terms of accuracy and is a gateway for real-time image recognition.
MRI is a common and reliable method used for a detailed view of brain insights and tumor diagnosis. Therefore, it is highly recommended to consider MRI for the identification and classification of a brain tumor as compared to other techniques. This manuscript proposes an optimal support network-based computer-aided healthcare system for the efficient detection of tumor cells. The novel contributions of this manuscript are:
-
A Hybrid features vector is proposed, obtained from the concatenation of Handcrafted features with features extracted from a CNN. The combined feature vector has both high-level and low-level information about the MRI images which allows the model to capture a wider range of information from the images, potentially improving its ability to classify the tumor.
-
The BoFV technique is proposed to select the most important features from the concatenated feature set, which therefore reduces the computational complexity of the proposed model. PCA is then applied to further reduce the feature set while still preserving as much of the important information as possible.
-
An oLSVN algorithm is proposed for the classification of tumor cells in MRI images. The oLSVN is carefully designed to provide accurate results with the least computational complexity. It improves the accuracy of tumor classification and enables the early-stage detection of brain tumors with the help of the CLAHE technique where the uncertainty of the disease exists.
-
After classification, segmentation is applied to the tumor images to identify the exact location of the tumor within the image. This information can be useful for treatment planning and monitoring of the tumor's growth.
-
Simulation results are provided which show the ability of the proposed model to capture important features from the MRI images, reducing computational complexity, improving accuracy, enabling early-stage detection of brain tumors, and improving image quality for more accurate classification.
The rest of this article is structured as: Sect. 2 provides details about the experimental process involving the data set and its pre-processing. Section 3 explains the proposed methodology used to achieve the optimum results. Results and discussion are provided in Sect. 4, whereas the paper is concluded in Sect. 5.
2 Experimental process
2.1 Dataset description
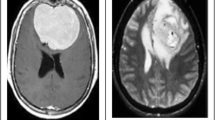
The data set used comprises \(5710\) MRI images that are divided into two sets; a training data set and a testing data set. In the training dataset, there are \(2400\) images for training, which is further divided into two folders, namely, 'benign' and 'malignant.' Each image has dimensions of \(180 \times 218\). The testing dataset consists of \(3310\) MRI images for testing. Images in the testing data set are comprised of a mixture of tumor and healthy images as in Toğaçar et al. 2020b. Figure 1 shows an exemplary illustration of different images with and without brain tumors, Fig. 1a, b are samples taken from the healthy class, whereas Fig. 1c, d are taken from the tumor class.
It is worth mentioning that the classifier might get biased during the classification stage if the number of samples of a specific class is significantly higher than the other class. Moreover, the dimension of all the sample images needs to be standard, i.e., \(227 \times 227\) for the AlexNet. Therefore, pre-processing is necessary to re-scale and balance all the images into the same quality and quantity.
2.2 Pre-processing
In the used data set, the number of MRI images with tumor cells is higher than that of the MRI images without tumors, i.e., an imbalanced data set. Therefore, augmentation is applied to the MRI images without tumors to balance the sample dataset. Data augmentation is a useful technique for enhancing the performance of ML-based models and achieving balanced datasets (Toğaçar et al. 2020a). In this study, augmentation methodologies are employed to equalize the number of samples in the standard class of brain tumor MRI images with the total number of brain tumor MRI images. The augmentation techniques presented by Kera’s library (Toğaçar et al. 2020a) were used to equalize all the samples in MATLAB. The turns and rotations, width and stature evolving, fragmentation and magnifying, horizontal pivoting, lighting up, and filling tasks were performed on the average class MRI images. The image pivot's level or degree was arbitrarily generated in the range of \(0-20\). An adjustment under \(0.15\) was made for estimations and shifts in the height and breadth, horizontal pivoting proportion boundaries, and zooming.
3 Methodology
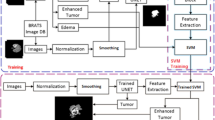
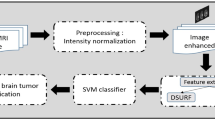
Different procedures and algorithms have been proposed for the diagnosis and classification of brain tumors in the literature. However, most of the proposed procedures and algorithms are computationally demanding, therefore, it is recommended to consider high-quality features that can comprehend precise results in a short interval of time with minimal computational complexity. To address computational complexity, this paper proposes several fundamental steps that guarantee accurate tumor detection while reducing computational overhead, as shown in Fig. 2.
After splitting the feature vector into train and test data, MRI images are processed for resizing and gray scaling, followed by 14-layered CNN and Handcrafted (HF) algorithms working in parallel to extract essential information from the features. These features are concatenated to get a hybrid feature vector. PCA is then used to reduce the computational complexity of the overall process. Table 1 provides the computational complexity of the proposed approach to that of some well-known existing techniques.
3.1 Feature extraction
Feature extraction is a proficient technique for extracting significant visual constituents of the MRI images in brain tumor detection (Narmatha et al. 2020). This is a challenging phase as tumors usually vary in size in every MRI scan and it is relatively hard to extract features of a large-sized tumor.
3.2 Hand crafted features
The proposed approach focuses on forging a texture descriptor robust to variation in scale, rotation, and illumination. A complete joint scale local binary pattern (CJLBP) is considered as the initial point to perform a multi-scale fusion for the generation of a pixel patch of size \(3\times 3\). The pixel patch is obtained by averaging all the defined radii associated with a segment. Using local difference sign magnitude (LDSMT), sign and magnitude components are calculated subject to a multiscale fused patch. The center, sign, and magnitude components of the image are represented with the histogram shown by using operators CJLBP_C, CJLBP_S, and CJLBP_M, respectively. These operators are combined in different ways to devise numerous variants of the former technique, e.g., CJLBP_MC is the consequence of combining CJLBP_M and CJLBP_C, respectively. To achieve accuracy in categorization, CJLBP_S\_MC, CJLBP_SM, and CJLBP_SMC are processed for demonstration.
The suggested approach is classified into three phases. The initial phase of the presented methodology is prevention. It is provided to averse the additive white Gaussian noise and the protection opposed to the framework changes. In the second phase, descriptors are made vigorous to achieve progress with a variable scale. Moreover, the extracted features are also made available in normal conditions in their respective radii. In the final phase, the descriptors become tolerant to the shifts in radiation. The above levels are discussed in detail in the given subsections.
Level 1
In the proposed methodology, two-dimensional discrete wavelet transform (2D-DWT) is first used to decompose the image under analysis. Single-level decomposition of the estimated coefficients is obtained by waveform Dmey (Discrete approximation of Meyer) of wavelet. The values of the feature vector that are close to the image under analysis are extricated. Individual segments of the low-frequency sub-band (LL) are then calculated as
where \({X}_{i,j}\) displays a segment of the approximated image that corresponds to the center pixel. The position of the segment in the approximated image is interpreted by row (\(i\)) and column (\(j\)). The variables \(i\) and \(j\) are restricted to change over a certain range defined by the interval \(\left[{y}_{1}+ {R}_{max}, {y}_{n}- {R}_{max}\right]\) and \(\left[{x}_{1}+ {R}_{max}, {x}_{n}- {R}_{max}\right]\) as it is impossible to pull out a segment \({X}_{i,j}\) near the boundary of the image. \({y}_{n}\) and \({x}_{n}\) depicts the end row and column position of the approximated image, whereas \({R}_{max}\), is the maximum radius of the segment \({X}_{i, j}\). Variables \(u\) and \(v\) in Eq. 1 define the coordinates of the segment in the approximated image. It is worth mentioning that both \(u\) and \(v\) span over a range of \(\left[-{R}_{max} +{R}_{max}\right]\). Dimensions of \({X}_{i, j}\) in the pixels equal \(\left(1+ 2 {R}_{max}\right)\times \left(1+ 2 {R}_{max}\right)\) segment of the approximation.
Figure 3 shows the intersection of the multi-oriented fusion process of the pixels, generating a topological formation of 8 specimens for all the 3 scales. The intersection of the multi-oriented fusion process is carried out on the pixels that are placed in radii 2 and 3 as given in Eqs. 2 and 3, respectively. Specimen points \({P}_{k}^{3}\), \({P}_{k}^{2}\), and \({P}_{k}^{1}\) in Eqs. 2, 3, and 4 show the topological composition of 8 specimens that have radius 3, 2, and 1, respectively.
where \(J\) represents pixels of the segment \({X}_{i, j}\), the subscript of \(P\) is the orientation angle and the superscript is the radius.
Level 2
To make the descriptor resistant to scale fluctuation, a two-step multi-scale fusion operation is performed as shown in Fig. 4.
Samples from radii 1 and 2 are fused in the initial step using the mean operation followed by mixing samples from radii 2 and 3. For each segment \({X}_{i, j}\), the multi-scale fusion depends on the radius \(r\) and the sample point \(P\), defined as
Level 3
The third stage involves the extraction of feature vectors from each topological structure as shown in Fig. 5. From \({X}_{i, j}\), the sign and magnitude component are calculated using local difference sign-magnitude transform (LDSMT), defined as
where \({d}_{i, j}\) is the set of local differences, \({\widetilde{X}}_{i,j}^{k}\) shows the \({k}^{th}\) sample point of the multi-scale fused patch, and \({\widetilde{X}}_{i ,j}\) denotes the intensity value of the center pixel analogous to segment \({C}_{i, j}\). \({d}_{i,j}^{k}\) shows the \({k}^{th}\) sign element with a value 1 when \({d}_{i,j}^{k} \ge 0\), and \(-1\) when \({d}_{i,j}^{k} \le 0\).
The magnitude component \({m}_{i,j}\) contains absolute values of the local difference \({d}_{i,j}\). The rotation invariant operator \(OMTLB{P}_{{M}_{P,r}^{riu2}}\) can be calculated through the sign component \({s}_{i,j}\) of the local difference \({d}_{i,j}\). Its encoding is formulated as \({\mathrm{LBP}}_{\mathrm{P},\mathrm{r}}^{\mathrm{riu}2}\) (Khan et al. 2019). Using magnitude component of the local difference \({d}_{i,j}\), the rotation invariant operator \(OMTLB{P}_{{M}_{P,r}^{riu2}}\) can be calculated as
where \(\psi (y,x)\) is 1 for \(y\ge x\) and 0 for \(y < x\). The \({\mathrm{m}}_{\mathrm{i},\mathrm{j}}^{\mathrm{p}}\) is the \({\mathrm{k}}^{\mathrm{th}}\) unit component, and \({\upmu }_{\mathrm{m}}\) is the average value of the factor component of the segment at location \((i,j)\) for the given image. The center pixel's grey level \({OMTLB{P}_{C}}_{P,r}\) is mainly considered along with the pattern of magnitude and sign, represented as
where \({\upmu }_{\mathrm{I}}\) shows the average value of the entire input image. The combined value of operators \(OMTLB{P}_{{S}_{P,r}^{riu2}}\), \(OMTLB{P}_{{M}_{P,r}^{riu2}}\), and \({OMTLB{P}_{C}}_{P,r}\) is represented by \(OMTLB{P}_{S{MC}_{P,r}^{riu2}}\). The train and test feature vectors are acquired with the help of the operator \(OMTLB{P}_{S{MC}_{P,r}^{riu2}}\). Chi-square distance is utilized as a dissimilarity measure and the classification accuracy is obtained using Eq. 11.
Here \({T}_{infected}\) means true positive, \({T}_{uninfected}\) shows true negative, \({F}_{uninfected}\) is a false negative, and \({F}_{infected}\) means false positive samples of the data set.
3.3 Deep features
Feature extraction technique helps to recognize the shape and location of a tumor in MRI images (Ozyurt et al. 2019). For feature extraction, a 14-layered CNN model is well appreciated in the field of artificial intelligence and medical imaging (Shakeel et al. 2019). Machine learning utilizes CNN to recognize and authenticate things on its own. In the proposed model, the HF and 14-layered CNN models are used in parallel for feature extraction. The CNN layers are designed explicitly and consist of 5 convolutional layers, 2 fully connected layers, 2 dropouts, 3 max-pooling layers, and a single output layer. Dimension parameters of the input layer are \(224\times 224\), whereas the received output dimensions are presented in Table 1. The hierarchy of the proposed 14-layered CNN models is as presented in Fig. 2. The acquired vector is comprised of 1000 characteristics, including a fifty percent exclusion layer for every fully convolutional layer to eliminate the oversizing issues. Therefore, the output feature vector has a dimension of \(2400\times 1000\) for training the network and \(3310\times 1000\) for testing and validation. Moreover, to validate the model with the pre-trained AlexNet and GoogleNet networks, the in-depth features of both networks were calculated and came out to be \(3310\times 4096\) and \(3310\times 1000\), respectively. The dimension of the feature vector at each stage with each model is provided in Table 1.
3.4 High-quality feature selection
High-dimension feature vectors are computationally complex and take a long time in processing. Therefore, it is essential to reduce the size of the feature vectors which will in turn reduce the computational complexity. To reduce the size of the features, it is important to choose high-quality features when a network is trained along with better classification performance. In the proposed model, flexible analytical wavelet transformations are used to attain temporal and spatial representation for achieving the temporal boundaries. In the proposed technique for feature selection, an image with the presence of a tumor is decomposed and expressed as a spatial–temporal image. Then, for recognition of the abnormal behavior, the right channel is selected by using combinations of different functions of four neural networks. Deep feature clustering is used to select the feature vector obtained through the selected channel.
3.5 Hybrid feature vector
All the geometrical models involved in handcrafted feature extraction yield the respective features in the third stage of the HF model. The Hybrid feature vector is obtained by concatenating the features from the HF and 14-layered CNN operating in parallel, for the early detection and proper treatment of brain tumors. The parallel concatenation will save time and effort for the overall process as concatenation helps to address the features as a single vector. These features undergo principal component analysis for dimensionality reduction. PCA is applied as a clustering-based algorithm that helps in reducing the input dimensions of the hybrid feature vector to develop a particular data set. It integrates vital information and provides a high-quality data set while discarding irrelevant features. The simulations prove that the hybrid feature vector approach provides effective and accurate results.
3.6 Bag of feature vector: feature selection
Bag-of-feature (BoF) method is applied for image classification that shows images as a collection of features without any order in medical imaging and reduces them to an appropriate value for proper training of the data set. Processing time is higher for a higher number of significant features, whereas the classification does not remain accurate as well. The BoF comprises two sets where the features are initially extracted using k-mean clustering with the help of the AlexNet model. A single feature vector is chosen by grouping the identical features. The features are reduced in the first stage using the mean clustering approach and are further optimized for computing the features of the histogram in the final stage. This model helps in selecting the desired features by eliminating redundancy in the high-quality feature vector obtained from the PCA.
3.7 Optimized linear support vector network
Optimized Linear Support Vector Network (oLSVN) is a new learning model for classification and segmentation. The overall computational cost of the oLSVN model makes it an immensely cost-effective model as no unnecessary or insignificant techniques are applied to the images.
The MRI images are usually not explicitly obvious and at times the tumor is not visible in the initial stages with certainty. Therefore, they are passed through the enhancement process to verify whether the tumor exists. Moreover, images, where the tumor is not visible, are enhanced before segmentation. However, the images with tumors are classified easily and need not be enhanced which thus reduces the computational complexity. Moreover, applying enhancement on the white portion of the images, which shows the tumor and diseased area, distorts pixels and makes it difficult to find out the exact area of the brain tumor.
Before the oLSVN, a feature vector is introduced as input data and is integrated into the test and training data sets. Both sets of images are tested for tumors. If the results are positive, the image is processed for segmentation as well as classification, otherwise, the image is enhanced for further certainty. After enhancement, if the applied algorithm provides negative results, i.e., the tumor is not detected after enhancement, it is classified as healthy.
3.8 Enhancement of images
In the enhancement section, the CLAHE algorithm is primarily applied to the Green Channel. The green channel is chosen for its ability to enhance image quality, increase contrast, and intensify image features. A salt and pepper noise is added to the images that are classified as healthy. The salt and pepper noise superimposes the original noise of the processed image. A pair of uninterrupted filters, i.e., Median and Morphology filters are utilized to filter out the merged noises. Most of the noise is filtered out using these two consecutive filters. Enhanced images are obtained after applying gray scaling on the filtered images as shown in Fig. 6.
4 Experimental results and discussion
This section presents simulation results of the proposed techniques using MATLAB R2022a. The performance of the proposed technique is evaluated and compared to well-known techniques available in the literature. The performance parameters used for evaluation are precision, specificity, sensitivity, F1-score, and Matthew’s correlation coefficient (MCC), obtained manually from the confusion matrix, Table 2.
Precision of the proposed model can be calculated as:
where \({T}_{infected}\) is true positive, i.e., a tumor is anticipated to be present and it exists indeed. \({F}_{infected}\) represents a false positive, i.e., the region where it is supposed that the tumor will be there but it is not present.
The specificity of the proposed model can be evaluated as:
\({F}_{uninfected}\) represents false negative, i.e., it is anticipated that the tumor is not detected but it was present. Sensitivity and specificity are the basic foundations used to validate the accuracy and precision of the overall framework. Sensitivity helps to accurately test and identify the patients with tumors, whereas specificity aids in correctly identifying the tests without tumors.
The sensitivity of the proposed model is calculated as:
where \({T}_{uninfected}\) represents True negative, i.e., it is estimated that the tumor is not detected and is not present.
F1-Score measures the accuracy of the data set of the model and conducts the fairness between specificity and sensitivity, defined as
MCC helps machine learning techniques to measure the effectiveness of classifiers and averages down the spatial features with other features using various classifiers. MCC for the proposed model is calculated using Eq. 16.
Figure 7 provides a comparative analysis of the proposed model to some already existing algorithms in the literature based on the aforementioned parameters. The pre-trained models used are GoogLeNet and AlexNet, where GoogLeNet has \(3310\times 1000\) feature dimensions with 7 M parameters for the 22 layers model. It has an accuracy of 94.61%, whereas its execution time is 260 s. The feature dimensions of AlexNet is \(3310\times 4096\) and have eight layers with 61 M parameters. Its execution time is 119.7 s and the results are accurate up to 91.43%. On the other hand, the first step of the proposed model, which is a concatenation of features obtained from CNN and HF to get the hybrid feature, has an execution time of 243.6 s with an accuracy of 87.4%. To enhance accuracy while reducing computational complexity, BoF is integrated into the second step of the proposed model, resulting in a combination of CNN, HF, and BoF. The results are improved up to 89.92% with an execution time of 1.2 s, which depicts a considerable reduction in the computational complexity of the proposed models by removing the redundant features. To further improve the accuracy of the model, in the third step oLSVN is utilized with CNN, HF, and BoF. With the addition of the oLSVN, the accuracy is improved up to 98.25% with an execution time of 1.32. Therefore, utilization of oLSVN will make a noticeable contribution to the medical imaging and optimization techniques for improving the system performance in terms of accuracy. A comparative analysis of the proposed algorithm to GoogleNet and AlexNet is provided in Table. 1
Besides accuracy and the execution time, the proposed model was also evaluated manually using the confusion matrix obtained after the testing dataset. It provides noticeable results as shown in Table 2. Utilizing values from the confusion matrix in Eqs. (12), (13), (14), (15), and (16), a precision of 97.28%, specificity of 97.22%, sensitivity of 99.27%, an F1-Score of 98.27%, and a Matthews Correlation Coefficient (MCC) of 96.52% were calculated, as depicted in Fig. 7.
The primary goal of this research paper was to design and implement an efficient model for brain tumor detection in the 215 images falling in the category of false negatives (tumors exist but the model considered them healthy). Figure 8a, b, and c shows exemplary images before enhancement from the false negative category. In the model, these 215 images were enhanced and the tumor became exclusively visible after enhancement in 168 images, as shown in Fig. 8d, e, and f. Enhancement thus improved the visibility of the tumor in these 168 images and a total of 1643 positive cases were classified as unhealthy which were 1536 previously.
Table 3 depicts the practical results obtained using CLAHE technique. CLAHE uses filtration and commotion of noises to produce the final image. Through CLAHE, one can determine the smallest Mean Square Error (MSE) between 0.0539 and 1.739, respectively. Moreover, for culmination, the high to low SNR value is calculated as 49.4533 and 57.7621, respectively. The median average for MSE and Peak Signal to Noise ratio (PSNR) is 0.6813 and 49.99, respectively.
The segmentation process is a critical stage for a neuro-patient. If the tumor tissue is operated ineffectively, the result might be a permanent loss of sensory neurons for the patient. The system is thoroughly scanned, and numerous copies are stored for further identification of the tumor's exact location. In this study, both supervised and unsupervised techniques are employed to detect brain tumors. The supervised methodologies help in identifying the tumor areas, whereas the localization of the tumor in brain MRI images is performed with unsupervised techniques. The dispersed and distorted pixels of the tumor image is labeled by k-mean clustering. The process of locating a tumor is justified by determining the rotation of pixels found in the MRI images. The images are processed with structural operations before the segmentation and clustering to remove the skull's outline. Tumor tissues are determined by k-mean clustering and a unit of three clusters is identified using the same means. The cluster showing an excellent centroid value in the set is chosen for the localization of the tumor in brain MRI images. The brain MRI images after the segmentation (for k = 3) process are shown in Fig. 9.
The color-keying in a grayscale image is comprehended to determine the dimensional parameters of the tumor in proposed algorithm. The colored image inserted initially is transformed into a grayscale image by establishing an average sum of the Red–Green–Blue color constituents. The information related to saturation and shades is discarded while the fluorescent dazzle is retained to convert the image into grayscale. A grayscale map is received due to the process, which is subsequently stored in fluctuating image Ig. The area of tumor classified and segmented is shown in Table 4.
5 Conclusions
A novel least complex approach for brain tumor detection using oLSVN is proposed. Initially, a 14-layered CNN model is used in parallel with the Handcrafted model for feature extraction. The features of both models were concatenated and used for classification which provided an accuracy of \(87.4\%\) with an execution time of 243.6 s. To improve the accuracy as well as reduce the computational complexity, the BoF technique was combined with CNN and HF in parallel to PCA. PCA was used to select quality features from the hybrid feature vector by eliminating redundant features, thus, decreasing the computational complexity (execution time to 1.2 s) and slightly improving the accuracy to \(89.92\%\). The primary goal of this research was to design a model for the early-stage detection of tumors, for which oLSVN with CNN + HF + BoF was introduced. oLSVN is used to classify the brain tumor with the help of high-quality images obtained from the PCA, whereas segmentation is performed to detect the exact area of the tumor after classification. To detect tumors at an early stage, CLAHE technique was used to enhance the quality of the image and reduce noise from the images where the tumor was not found previously due to poor visibility. This additional step slightly increased the execution time to 1.32 s, however, the accuracy of the proposed model was improved to \(98.25\%\). As compared to some already existing approaches, the proposed model has F1-Score = \(98.27\%\), precision = \(97.28\%\) precision, specificity = \(97.22\%\), and \(96.52\%\) Matthew’s Correlation Coefficient which validates that the proposed solution could be a best fit for practical health care systems.
References
Asghar MA, Khan MJ, Rizwan M, Mehmood RM, Kim SH (2020) An innovative multi-model neural network approach for feature selection in emotion recognition using deep feature clustering. Sensors 20(13):3765
Cabria I, Gondra I (2017) MRI segmentation fusion for brain tumor detection. Information Fusion 36:1–9
Chaudhary A, Bhattacharjee V (2020) An efficient method for brain tumor detection and categorization using MRI images by K-means clustering and DWT. Int J Inf Technol 12(1):141–148
Deepak S, Ameer PM (2019) Brain tumor classification using deep CNN features via transfer learning. Comput Biol Med 111:103345
Fazelnia A, Masoumi H, Fatehi MH, Jamali J (2020) Brain tumor detection using segmentation of MRI images. J Adv Pharmacy Educ Res 10(S4)
Garg G, Garg R Brain Tumor Detection and Classification based on Hybrid Ensemble Classifier arXiv preprint arXiv:2101.00216 2021.
Khan MJ et al (2019) Texture representation through overlapped multi-oriented tri-scale local binary pattern. IEEE Access 7:66668–66679. https://doi.org/10.1109/ACCESS.2019.2918004
Khan HA, Jue W, Mushtaq M, Mushtaq MU (2020) Brain tumor classification in MRI image using convolutional neural network. Math Biosci Eng 17(5):6203
Liu T, Yuan Z, Wu L, Badami B (2021) An optimal brain tumor detection by convolutional neural network and Enhanced Sparrow Search Algorithm. Proc Inst Mech Eng [h] 235(4):459–469
Mattila PO, Ahmad R, Hasan SS, ud. Babar Z (2021) Availability, affordability, access, and pricing of anti-cancer medicines in low-and middle-income countries: a systematic review of literature. Front Publ Health 9 https://doi.org/10.3389/fpubh.2021.628744.
Mittal N, Tayal S (2021) Advance computer analysis of magnetic resonance imaging (MRI) for early brain tumor detection. Int J Neurosci 131(6):1–16. https://doi.org/10.1080/00207454.2020.1750390
Narmatha C, Eljack SM, Tuka AARM, Manimurugan S, Mustafa M A hybrid fuzzy brain-storm optimization algorithm for the classification of brain tumor MRI images, J Ambient Intell Human Comput pp. 1–9, 2020, https://doi.org/10.1007/s12652-020-02470-5.
Ojala T, Pietikainen M, Maenpaa T (2002) Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell 7:971–987
Ozyurt F, Sert E, Avci E, Dogantekin E (2019) Brain tumor detection based on convolutional neural network with neutrosophic expert maximum fuzzy sure entropy. Measurement 147:106830. https://doi.org/10.1016/j.measurement.2019.07.058
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35(5):1240–1251
Saba T, Mohamed AS, El-Affendi M, Amin J, Sharif M (2020) Brain tumor detection using fusion of hand crafted and deep learning features. Cogn Syst Res 59:221–230
Sert E, Özyurt F, Doğantekin A (2019) A new approach for brain tumor diagnosis system: Single image super resolution based maximum fuzzy entropy segmentation and convolutional neural network, Med Hypotheses 133.
Shakeel PM, Tobely TEE, Al-Feel H, Manogaran G, Baskar S (2019) Neural network based brain tumor detection using wireless infrared imaging sensor. IEEE Access 7:5577–5588. https://doi.org/10.1109/ACCESS.2018.2883957
Sharif MI, Li JP, Khan MA, Saleem MA (2020a) Active deep neural network features selection for segmentation and recognition of brain tumors using MRI images. Pattern Recogn Lett 129:181–189
Sharif M, Amin J, Nisar MW, Anjum MA, Muhammad N, Shad SA (2020b) A unified patch-based method for brain tumor detection using features fusion. Cogn Syst Res 59:273–286
Sharif M, Amin J, Raza M, Yasmin M, Satapathy SC (2020c) An integrated design of particle swarm optimization (PSO) with fusion of features for detection of brain tumor. Pattern Recogn Lett 129:150–157
Soltaninejad M (2017) et al. Automated brain tumor detection and segmentation using superpixel-based extremely randomized trees in FLAIR MRI. Int J Comput Assisted Radiol Surg 12.
Toğaçar M, Ergen B, Cömert Z (2020a) BrainMRNet: Brain tumor detection using magnetic resonance images with a novel convolutional neural network model. Med Hypotheses 134:109531
Toğaçar M, Cömert Z, Ergen B (2020b) Classification of brain MRI using hyper column technique with convolutional neural network and feature selection method. Expert Syst Appl 149:113274
Zhao X, Yihong Wu, Song G, Li Z, Zhang Y, Fan Y (2018) A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med Image Anal 43:98–111
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Razzaq, S., Asghar, M.A., Wakeel, A. et al. Least complex oLSVN-based computer-aided healthcare system for brain tumor detection using MRI images. J Ambient Intell Human Comput 15, 683–695 (2024). https://doi.org/10.1007/s12652-023-04725-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-023-04725-3