Abstract
Whey proteins are abundant in peptides that possess various biological activities. In order to enhance the biological properties of protein hydrolysates, it is essential to optimize the conditions of the hydrolysis process. In this study, optimal conditions for hydrolysis of whey protein concentrate (WPC) using filtered trypsin-like protease (FTLP) was determined in vitro. Further, the ability of optimized whey protein hydrolysates to inhibit dipeptidyl peptidase-4 (DiPP4) in vitro was examined. An optimum degree of hydrolysis (DH 42.9%) was obtained with an E:S ratio of 5:100 (w/w), 8.6 h, and a temperature of 40 °C. The factual DH under ideal conditions was 42.04%, indicating the efficiency of the selected model (p ≤ 0.05). Hydrolysates of WPC generated by FTLP, including both the unfractionated section and the fractions obtained via ultrafiltration using 10- and 5-kDa cut-off membranes, exhibited anti-diabetic characteristics. However, the fractions exhibited greater inhibitory effects against the DiPP4 enzyme, with IC50 values of 1.98, 1.19, and 0.9 mg/mL for the unfractionated, 10-kDa and 5-kDa fractions, respectively. Moreover, the results indicated that probiotic L. plantarum subsp. plantarum PTCC 1896 or its components may provide opportunities for future management of type-II diabetes by inhibiting DiPP4.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Whey is a protein-rich liquid that is produced as a byproduct of the cheese and casein manufacturing process in the dairy industry. It is commonly sold in the commercial market as either whey protein isolate (WPI) or whey protein concentrate (WPC). Whey proteins probably make up 20% of all cow’s milk proteins (Lacroix and Chan, 2012). In recent years, whey proteins, a product rich in protein and peptides with biological activities, have been recognized as the essential food and medicine sources that have greatly impacted human health. Whey protein is composed of several major components, including α-lactalbumin, β-lactoglobulin, serum albumin, and immunoglobulin. These are all well-known proteins that have been extensively studied for their nutritional and functional properties (Nilsson et al. 2007; Mota et al. 2006). Due to the increasing number of cheese production factories in the world and on the other hand, due to the high nutritional value of whey protein, it is necessary to carry out extensive research on the use of this product in the formulation of various combinations and special diets. Production of hydrolysates from whey protein concentrate can be an exciting way to enhance its value (Mota et al. 2006). These protein hydrolysates can be produced via enzymatic digestion, proteolysis with plant or microbial enzymes, or fermentation by proteolytic microorganisms. The various protein hydrolysates obtained with proteases often show biological activities and functional properties. These hydrolysates are associated with bioactive components that can have several applications, including medical formulas and dietary supplements. Commercial proteases have shown promising results by producing bioactive peptides from both the casein and whey fractions of milk (O’Loughlin et al. 2013; Brandelli et al. 2015; Morais et al. 2015). A bioactive peptide is a protein element that is inactive in the sequences of the original protein and exhibits some physicochemical roles after released through enzymatic hydrolysis. A significant portion of the research conducted on bioactive peptides pertains to their physiological effects in vivo, including but not limited to antioxidant, immunomodulatory, antimicrobial, antihypertensive, and anti-diabetic activities (Brandelli et al. 2015). Moreover, whey protein hydrolysates exhibit a range of practical applications such as acting as emulsifiers, foaming agents, and gelling factors (Foegeding et al. 2002). Regarding the proteolysis of whey proteins by lactic acid bacteria, some LAB organisms have been investigated for their capability of degrading whey proteins in dairy products, such as Lactobacillus delbruekii ssp., bulgaricus, and Streptococcus thermophiles (Morr and Foegeding 1990; Bertrand-Harb et al. 2003; El-Zahar et al. 2003). Prioult et al. (2005) have suggested that Bifidobacterium lactis NCC362 could potentially serve as a probiotic for preventing bovine milk allergy by breaking down the allergenic portion of β-lactoglobulin. Pancreatic enzymes, especially trypsin and other enzymes from microorganism (such as Trichoderma and Bacillus), have been applied to identify numerous known bioactive peptides (Pihlanto-Leppälä et al. 1998). Although non-GRAS protease products have potential uses in pharmaceuticals, they pose significant hazards. Therefore, utilizing protease derived from GRAS probiotic lactic acid bacteria represents a safer and more viable option for developing bioactive peptides sourced from natural proteins (Mustafa et al. 2020). To date, no studies have explored the capacity of microbial proteases isolated from lactic acid bacteria to hydrolyze whey proteins. This study seeks to fill this gap by investigating the extent of whey protein concentrate degradation through the use of bacterial protease (Filtered Trypsin-Like Protease, or FTLP, derived from Lactobacillus plantarum subsp. plantarum PTCC 1896). Additionally, we aim to optimize the hydrolysis conditions to produce bioactive peptides that can serve as natural DiPP4 inhibitors for the treatment of type-II diabetes. The potential for dipeptidyl peptidase-4 (DiPP4) inhibitor activity has also been reported in lactic acid bacteria (Zeng et al. 2016), which makes the new discovery of Lactobacillus with DiPP4 inhibition potential an exciting development. This discovery could significantly enhance research efforts aimed at detecting DiPP4 inhibitors from probiotics. Thus, the second objective of this study was to evaluate the effectiveness of L. plantarum subsp. plantarum PTCC 1896 as a DiPP4 inhibitor and compare its efficacy that of bioactive peptides produced by FTLP under in vitro conditions.
Materials and Methods
Chemicals and Equipment
Coomassie Brilliant Blue G-250, BSA, Amicon Ultra centrifugal filters, protease inhibitors, and dialysis tubing, Ammonium Sulfate ((NH4)2SO4), and Tris (hydroxymethyl) aminomethane hydrochloride (Tris–HCl) were purchased from Sigma-Aldrich Merck. MRS agar (de MAN, ROGOSA, and SHARPE), Yeast Extract, Glucose, DiPP4 (8 U/L, porcine kidney, ≥ 10 units/mg protein), Gly-Pro-p-nitroanilide (GPpNA), Diprotin A tripeptide (TPDA: Ile-Pro-Ile), and Coomassie Brilliant Blue (CBB-G250) were also purchased from Sigma-Aldrich Merck. Skimmed milk powder and whey protein concentrate (WPC-80) were acquired from Zarrin-Shad Sepahan Co., Iran. The equipment used in this study included an autoclave (80-HG, Japan), a spectrophotometer (UV-2100, Unico, Japan), a laboratory freezer (Danesh Pajoohesh Fajr Co, Iran), a centrifuge (Hettich Zentrifugen Universal 320 R, Germany), a pH meter (Aratajhiz Co, Iran), Microplate reader (DANA-3200, Iran), Amicon ultra-5 and 10 (kDa) centrifugal filter device (Millipore), a shaker incubator (SKIR-601, Korea), and a water bath (Memmert, Germany).
Experimental Bacterial Strains
The bacterial strain utilized for this study was L. plantarum subsp. plantarum PTCC 1896, which was initially isolated from a breastfeeding infant and has demonstrated probiotic properties in numerous investigations (Mirlohi et al. 2008). For the working inoculum, a 1:100 dilution of L. plantarum subsp. plantarum PTCC 1896 preculture was transferred to 10 ml of MRS broth and incubated at 37 °C with shaking at 50 rpm for 18 h. The resulting bacterial suspension was centrifuged at 8000 g for 20 min at 4 °C, and the cell pellet was washed twice with sterile saline solution before being utilized as an inoculum for the culture medium.
Culture Medium for TLP Production and Fermentation Conditions
To prepare TLP from L. plantarum subsp. plantarum PTCC 1896, a Skimmed Milk Broth (SMB) medium was used. The SMB medium was prepared by dissolving 6% (w/v) skimmed milk powder in distilled water and adjusting the pH to 8. This mixture was then steam-sterilized at 110 °C for 10 min. Next, 1.2% filtered glucose and 3% autoclaved yeast extract were added to the sterilized SMB medium. Fermentation was carried out in Lab bottles with screw tops at 37 °C for 14 h with an agitation speed of 100 rpm. After fermentation, the mixture was centrifuged (8000 g at 4 °C for 20 min) to eliminate bacterial cells. The resulting supernatant was filtered using a 0.22 μm syringe filter and designated as a crude TLP substance. Next, ammonium sulfate precipitation was carried out on the supernatant, and the obtained precipitate was collected through centrifugation (12,000 rpm at 4 °C for 30 min). The pellet was then dissolved in Tris-HCl buffer (50 mM, pH 8) and dialyzed overnight with the same buffer at 4 °C using a dialysis bag with an 8 kDa molecular weight cut-off. Three changes of dialysis buffer were performed during this process. The dialysate was filtered using an Amicon Ultra centrifugal filter with membranes having a molecular weight cut-off of 30 kDa and designated as filtered TLP or FTLP (Mustafa et al. 2020).
Evaluation of Total Protease Activity in the SMB Medium Supernatant
To determine the total protease activity in the fermentation medium supernatant (SMB), we followed the method described by Vijayaraghavan et al. (2013) using sterilized skimmed milk agar medium (SMA) which was steam-sterilized at 110 °C for 10 min. The SMA medium contained the following components per liter: 5.0 g skimmed milk powder, 5.0 g casein enzymic hydrolysate, 2.5 g yeast extract, 1.0 g dextrose, and 15 g agar, with a final pH of 8. The SMA medium-containing petri dish was solidified and six 6.4 mm diameter holes were then punched into the petri dish. To establish the standard curve, four different concentrations of commercial standard trypsin (namely, 400, 200, 100, and 50 µg/100µl) were utilized, while distilled water served as the negative control. Specifically, 100 µl of crude TLP compound, standard trypsin concentrations, and the negative control were loaded into the perforated holes, after which the petri dish was incubated at 37 °C for 24 h. The clear zone diameters corresponding to the standard trypsin concentrations were used for constructing the standard curve. The concentration of total protease in the supernatant of the examined medium was then estimated by the equation obtained from the standard curve.
The FTLP Inhibition Assay
The study evaluated the impact of different inhibitors on FTLP activity, using the method described by Klomklao et al. (2007). The FTLP was exposed to equivalent amounts of three different protease inhibitor solutions (E-64, SBTI, and TLCK) to achieve targeted final concentrations (0.1 mM E-64, 0.05 mM SBTI, 5 mM TLCK) and incubated at ambient temperature for 15 min. Following incubation, FTLP activity was measured. A control sample was included, where sterilized deionized water was used instead of inhibitors. The percent inhibition was calculated using the following equation (Eq. 1) :
Where I is the percent inhibition, Ani and Ai are the activity of FTLP in the control and inhibitor samples, respectively.
Enzymatic Hydrolysis of WPC Using FTLP
The 10% protein (w/v) Fresh WPC-80 solution in distilled water was pasteurized by heating to 90 °C for 10 min after adjusting the pH to 8.0 using 6 M NaOH. To adhere to the design conditions outlined in Table 1, hydrolysis was performed at their specified time and temperature while maintaining a constant pH of 8.0 through adjustments made every 20 min. To deactivate the FTLP, hydrolysis was halted by immersing the reaction mixture in boiling water for 10 min (Naik et al. 2013). The resulting hydrolysates were then subjected to centrifugation at ambient temperature for 10 min at 12,100 g. The supernatants were collected, stored at − 20 °C, and later analyzed for degree of hydrolysis (DH). The calculation formula of the hydrolysis was as follows (Eq. 2):
Where Pb and Ps were protein content of unhydrolyzed and hydrolyzed whey protein, respectively.
Design of Experiments
To optimize the enzymatic activity for maximum WPC hydrolysis using FTLP produced by L. plantarum subsp. plantarum PTCC 1896, a statistical approach was employed. Central Composite Design (CCD) with response surface methodology (RSM) was utilized, with the independent variables being E:S (X1), hydrolysis time (X2), and temperature (X3). The aim was to optimize the response, which is WPC hydrolysis. A total of 20 fermentation tests were conducted, which included 14 unique combinations and 6 replications of the three variables at five levels (− α, − 1, 0, + 1, +α). Table 1 provides information on the range and levels of the variables examined, as well as the experimental RSM runs. Using a second-order polynomial equation, the relationships and interactions between the variables were determined. The equation takes into account linear, quadratic, and cross-product terms to assess the impact of the variables (Eq. 3):
The estimated response variable, which is the Degree of hydrolysis (DH), is represented by Y. The constant terms are denoted by β°, the coefficient of linear terms by βi, the coefficient of quadratic terms by βii, and the coefficient of interaction terms by βij. The independent variables are symbolized by Xi and Xj. The coefficient of determination, R2, indicates the consistency and fitness of the model equation. All the calculations, including graphical analysis, were performed using the Minitab software package (version 16).
Protein Content Determination
In this experiment, protein content was assessed via the Bradford method (1976), using bovine serum albumin (BSA) as the reference standard. The standard used for calibration was 10, 20, 40, 60, 80, 100, 120, and 140 µg BSA/mL. To quantify the protein content, 20 µL of sample/blank was mixed with 1 mL of diluted Bradford reagent and incubated for 5 min. The resulting absorbance was then measured at a wavelength of 595 nm.
Preparation of WPC Hydrolysates
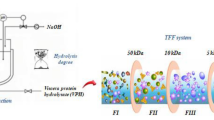
To assess the inhibitory effect of DiPP4, we conducted WPC hydrolysis via FTLP under optimal conditions with constant stirring. Following this, the resulting hydrolyzate was immersed in a boiling water bath for 10 min to inactivate the FTLP. The mixture was then centrifuged (12,100 g at ambient temperature for 10 min), and the supernatant was separated using an ultrafiltration system equipped with 5 and 10 KDa molecular weight cut-off membranes. The resulting permeates were collected and stored at − 20 °C prior to being tested for DiPP4 inhibitory activity.
Determination of DiPP4 Inhibitory Ability
The DiPP4 inhibitory activity assay was conducted using the Liu et al. (2017) method in a 96 well ELISA microplate. In brief, a 25 µL test sample was preincubated with 25 µL of GPpNA substrate (1.6 mM) at 37 °C for 10 min. Following this, 50 µL of DiPP4 was added, and the mixture was incubated at 37 °C for 60 min. The enzymatic reaction was halted by adding 100 µL of NaOAc buffer (1 M, pH 4), and the absorbance of the released p-nitroanilide (pNA) was measured at λ = 405 nm. The percentage of DiPP4 inhibition was calculated using the following equation (Eq. 4):
Where I is the percent of DiPP4 inhibition; Ac and Ai are absorbances of control and inhibitor, respectively.
The DiPP4 inhibitory activity was expressed as IC50, which represents the hydrolysate concentration required to achieve a 50% inhibition of DiPP4 activity. The hydrolysate concentrations used for the assay ranged from 1.25 to 0.0125 mg/mL. To serve as a reference inhibitor, TPDA (Ile-Pro-Ile) was diluted using Tris-HCl buffer (100 mM, pH 8) to varying concentrations (0.2, 0.4, 0.8, 1.6, 3.2, and 6.4 µg/mL).
DiPP4 Inhibitory Activity of the Bacterium
L. plantarum subsp. plantarum PTCC 1896 (1%, v/v) was inoculated into a 200 mL MRSB medium and incubated with a 5% CO2 injection at 37 °C for 12 h. Then 10 mL of the bacterial suspension was collected by centrifugation (12,000 g, 15 min), and the obtained cell pellet was used for the subsequent experiments (Zeng et al. 2016).
Preparation of Cell-Free Culture Supernatant (CFCS)
To obtain the CFCS, the cell pellets (8 × 109 CFU mL−1) were washed thrice with PBS, resuspended in PBS, and further incubated for 6 h. The mixture was then subjected to centrifugation (12,000 g for 15 min) to eliminate bacterial cells. The resulting CFCS was preserved at − 80 °C until testing. It has been demonstrated that the maximum DiPP4 inhibitory activity is obtained when the cells are exposed to PBS for 6 h (Zeng et al. 2016).
Preparation of Cell-Free Intracellular Extract (CFIE)
After PBS washing, the cell pellets (8 × 109 CFU mL−1) were resuspended in fresh PBS and subjected to ultrasonic disruption. Sonication was carried out for 20 min in an ice bath. Following this, cell debris was removed through centrifugation at 12,000 g for 15 min, and the resulting CFIE was stored at − 80 °C until required for further assays.
Statistical Analysis of Data
Data related to protease activity, enzyme inhibition, and DiPP4 inhibitory activity was expressed as a percentage (mean + SE, n = 3) in all assays. A probability value of p < 0.05 was considered statistically significant. The IC50 values were determined by fitting them to a sigmoidal dose-response equation with variable slope, using nonlinear regression analysis.
Results and Discussion
Evaluation of Total Protease Activity
The total hydrolysis activity of L. plantarum secreted protease (Crude TLP) was qualitatively and quantitatively determined using a zone diffusion assay. The basis of this method is based on the hydrolysis of the protein contained in the SMA medium with the secreted protease (Crude TLP) in the SMB medium. When milk protein is hydrolyzed, an obvious protein hydrolysis circle appears around the hole punched into the SMA, indicating that the secreted protease has promising proteolytic activity. As is evident in Fig. 1; Table 2, the proteolytic activity was observed with protease produced in the SMB medium. The protease production in SMB medium may be attributed to the fact that milk is poor regarding free amino acids and peptides, thus lactobacilli have to utilize a complex system of proteases and peptidase to meet growth needs which results in more protease production (Courtin and Rul 2004).
Degradation of SMA substrate using different commercial trypsin concentrations and the protease secreted by L. plantarum subsp. plantarum PTCC1896. (1). Commercial trypsin (400 µg/100µl), (2). Commercial trypsin (200 µg/100µl), (3). Commercial trypsin (100 µg/100µl), (4). Commercial trypsin (50 µg/100µl), (5). L. plantarum protease (Crude TLP) in SMB medium, (6). Sterilized distillated water
Effect of Different Protease Inhibitors on the FTLP Activity
The effects of different protease inhibitors on FTLP activity is shown in Table 3. Protease inhibitors are valuable tools for classifying the types of proteases (Kishimura, al., 2006). The results showed that the FTLP was remarkably inhibited by SBTI (a serine protease inhibitor) and a considerable inhibition was also observed with TLCK (a trypsin-specific inhibitor). On the contrary, E-64 (a cysteine protease inhibitor) showed a small inhibitory effect. Therefore, it was concluded that the FTLP secreted by L. plantarum subsp. plantarum PTCC 1896 most probably belonged to the serine-type proteases.
Analysis of the Model
Table 1 displays the diverse range of DH values achieved through the use of the protein content method to assess the action of FTLP. As demonstrated in Table 1, the DH of WPC varied from 6.07 to 42.78%, depending on factors such as the E:S ratio, hydrolysis time, and temperature.The maximum DH (42.78%) and the lowest absorption obtained at 595 nm using the method of measuring the insoluble protein concentration was in the fourth run. This result indicates that run # 4 has the highest amount of small peptides because it is known that, with Coomassie dye reagents, free amino acids, peptides, and proteins of low molecular weight do not emit color. It should be noted that although measuring the intensity of proteolysis with Ortho-phthalaldehyde (OPA) is a fast and widely used method, usually the presence of a large amount of free amino acid in the sample can increase the concentration of the measured peptide (Pripp et al. 2006). Often, as fermentation progresses, the concentration of peptides gradually decreases, which can be due to the breakdown of peptides into amino acids. Therefore, the results obtained from the OPA reagent do not mean the exact concentration of peptides but can be a set of peptides and amino acids. Run #19 was found to have the lowest hydrolysis rate. In a study conducted by da Silva et al. (2009) utilizing proteases from B. subtilis and B. amyloquefaciens to hydrolyze WPC under different conditions than those employed in this study, DH values of 20 and 14%, respectively, were obtained, which is less than the maximum value achieved in our study (42.78%). Another study by Silva et al. (2013) investigated the hydrolysis of WPC using proteases from B. licheniformis and Aspergillus sojae. According to their results, the highest values obtained were 44.7 and 20.17%, respectively, which is higher than the maximum values obtained in our study. Differences in DH values obtained by the same research group may be attributed to variations in parameters used during hydrolyzate preparation (pH, temperature, and E:S ratio) as well as the use of diverse measurement methods. Table 4 shows that there was no significant difference (p > 0.05) between the maximum DH obtained (42.78%) and the predicted value (41.72%). For confirming the accuracy of the results obtained and optimization of hydrolysis conditions, first, the model should be determined to describe the amount of hydrolysis. Then the accuracy of the determined model should be verified using the obtained data.
Model Fitting and Statistical Analysis
In the present study, three important parameters, E:S ratio (X1), hydrolysis time (X2), and temperature (X3), were considered.The DH obtained from all experiments is reported in Table 1 based on the RSM design. Multiple regression coefficients listed in Table 4 were used to predict a second-order polynomial model.The statistical significance of these coefficients was evaluated through Fisher’s F test and p-value. The F-value is a test that compares the variance of curvature with residual variance, while the probability (p-value) is the likelihood of observing the F-value if the null hypothesis is valid (Bayraktar 2001). Among the tested variables, X1 (E:S), X12 (E:S × E:S), X2 (hydrolysis time), X22 (hydrolysis time × hydrolysis time), X3 (temperature), X32 (temperature × temperature), X1 × 2 (E:S × hydrolysis time), and X1 × 3 (E:S × temperature) are significant model terms with p ≤ 0.05, and they have a substantial effect on whey protein hydrolysis. On the other hand, the X2 × 3 (hydrolysis time × temperature) term is insignificant and has no notable effect on the hydrolysis process (p > 0.05). Based on these results, we propose a polynomial model for whey protein hydrolysis as a function of the most significant variables, which is shown in the following equation:
Y = 20.509 + 9.665 × 1 + 13.668 × 2 + 6.123 × 3 + 4.844 × 12 + 7.544 × 22 – 6.596 × 32 + 3.987 × 1 × 2–6.873 × 1 × 3.
The study utilized ANOVA to examine the significance ratio between mean square deviation and mean square residual error, as shown in Table 5. ANOVA is a statistical technique that subdivides the total variation in a dataset into component parts associated with specific sources of variance, allowing for the analysis of hypotheses on model parameters (Torrades and García-Montaño 2014). The results indicate that the linear, square, and interaction terms are statistically significant at p ≤ 0.05, implying that these variables may have a considerable effect on the model. Additionally, the high F-values obtained for all regressions suggest that the regression equation can explain most variations in the response variable.
Evaluation of the Validity of the Selected Model
One of the methods to check the appropriateness of the method is the Lack of Fit testing coefficient. The Lack of Fit test involves comparing the residual error to the pure error from repeated design points, as described by Arslan-Alaton et al. (2010). In this case, the lack of fit F-value is 1.11 and the p-value is greater than 0.05, indicating that the lack of fit is not statistically significant. This suggests that the selected model is competent. The correlations between the actual data and the predicted data for DH have also been investigated. R2 suggests a positive correlation between the expected values and the experimental data points, the top value of which demonstrates the validity of the recommended model. R2, with a value of 0.98 showed the validity of the proposed model.
Optimization of WPC Hydrolysis
The technique of pre-hydrolysis thermal processing can lead to structural reforms in the protein substrate, which can have an impact on the rate of the hydrolysis reaction. Kim stated that pre-hydrolysis heat treatment improves the enzymatic (peptic and tryptic) hydrolysis of WPC. Therefore, in the study, WPC also was exposed to heat treatment before hydrolysis (90 °C) (Kim et al. 2007). The present study was performed to investigate whether the degradation of whey proteins was influenced by environmental conditions such as temperature and hydrolysis time, or if it solely depended on the E:S ratio. The optimum E:S ratio, hydrolysis time, and incubation temperature for hydrolysis of WPC were determined to be 5:100, 8.6 h, and 40 °C, respectively, with an expected optimal DH level (42.9%). A test was conducted based on RSM to validate the measured values. The optimized level of the parameters was used to replicate the run. The factual DH under ideal conditions was 42.04%, which was following the predicted value. The optimum temperature for hydrolysis of WPC substrate is almost in line with our observations shown in a previous study (Mustafa et al. 2020) that the optimum temperature for hydrolysis of BAPNA substrate by the same enzyme (FTLP) was 39 °C. Therefore, it can be concluded that a temperature above 40 °C is probably unsuitable for WPC hydrolysis like BAPNA. The temperature does not affect the release of peptides, but rather the enzyme activity. Indeed, at very high temperatures from the optimal point of enzyme activity, much fewer small peptide fragments are produced; this is because very high temperatures lead to rapid denaturation of the enzyme (Cheison et al. 2011). Several reports in the literature deal with the optimization of WPC hydrolysis. However, the optimal hydrolysis conditions vary depending on the type of whey and enzyme used, in addition, the hydrolysis conditions (temperature, pH, E:S ratio, and hydrerolysis time). In a study performed by da Silva et al. (2009), the best conditions obtained for WPC hydrolysis by Aspergillus oryza protease were E:S 1:100, pH 7, 50 °C, 5 h. In another study for Silva et al. (2010) outstanding hydrolysis conditions for hydrolyzing WPC by using papain and pancreatin were (E:S 2:100, pH 7, 55 °C, 5 h) and (3:100; pH 7, 50 °C, 5 h) respectively. Also, in another study carried out by Mota et al. (2006), the best conditions for β-lg hydrolysis for five h by pancreatic trypsin with an activity of 1020 BAEE units/mg protein were pH 8, 50 °C.
Effects of the Enzyme: Substrate Ratio
The study of the effect of the E:S ratio is one of the most critical factors affecting the hydrolysis of the WPC process. In this study, the effect of FTLP on the hydrolysis of WPC with different E:S ratios was considered. The surface plots presented in Fig. 2A–B illustrate the correlation between the E:S ratio, hydrolysis time, and incubation temperature, and how they collectively affect the hydrolysis of WPC. When the E:S ratio is increased from 4:100 to 5:100, the rate of hydrolysis also increases. However, as the E:S ratio continues to increase beyond 5:100, the hydrolysis process approaches a stationary phase. In previous studies on producing bioactive peptides DPP-IV inhibitors using whey protein substrate, the enzyme ratio used was around 4:100 to 5:100 (w/w) for either pepsin or trypsin (Lacroix and Li-Chan 2012; Silveira et al. 2013). Therefore, in this study, the observed rate of FTLP did not exceed 5:100. Nonetheless, it has been suggested that using a lower E:S ratio can be beneficial for generating protein hydrolysates with higher levels of small peptides and free amino acids and fewer large peptides (Silvestre et al. 2012). It is important to note, however, that our findings conflict with this conclusion. The beneficial use of a lower E:S ratio was noticed in some cases for the three enzymes (Aspergillus oryzae, Aspergillus sojae, and pancreatin), while the same state did not show for the protease from Bacillus licheniformis, which produced the lowest amount of large peptides using the highest of E: S ratio (E:S 8:100) (Morais et al. 2015).
Effect of the Temperature
Besides E:S ratio, incubation temperature is one of the most important factors affecting the hydrolysis process. As shown in the curve of the simultaneous effect of E:S ratio and temperature (Fig. 2A), with increasing temperature from 34 to 37 °C, the rate of hydrolysis increases due to the breakdown of the peptide chain. With the advancement of the reaction up to 39 °C, the hydrolysis rate curve reaches the phase of stationary, until, at a temperature above 39 °C, the hydrolysis process is significantly reduced. One of the reasons for the decrease in the DH at higher temperatures can be explained by the reduction of intact peptide bands and decreasing enzyme activity at higher temperatures (Kristinsson and Rasco, 2000). The highest DH rate was observed at 37 °C. This result is relevant to the general knowledge that the optimum temperature for trypsin activity is 37 °C (Koutsopoulos et al. 2007).
Effect of the Hydrolysis Time
The simultaneous effect of concentration of E:S and time (Fig. 2B), clearly shows that there is no significant change in the DH during initial reaction times of up to 4 h and 30 min. This result is consistent with a study performed by Custódio et al. (2009), whose electrophoretic analysis showed that neither trypsin nor chymotrypsin alone could hydrolyze whey proteins in less than 3 h. The change in the DH intensified after about 5 h and reached its maximum after about 9 h, so the highest rate observed in long-term hydrolysis may be due to the increase in the content of small peptides (da Costa et al. 2007). The rate of hydrolysis in whey proteins, whether heated or native, increases with increasing hydrolysis time. Kim et al. (2007) reported that with increasing hydrolysis time and increasing the level of hydrolysis enzyme, the rate of hydrolysis in heated whey proteins increases.
DiPP4 Inhibitory Activity of WPC Hydrolysates
According to the results, WPC did not show any inhibitory activity of DiPP4 without hydrolysis. In contrast, hydrolyzed WPC and 10 and 5 kDa permeates had IC50 values equal to 1.98, 1.19, and 0.9 mg/mL, respectively (Fig. 3; Table 6). This result confirms that some of the peptides liberated by FTLP may act as inhibitors of DiPP4. Silvera et al. (2013), investigated WPC hydrolysis with commercial trypsin for 3 h at 37 °C. According to the results of their study, after hydrolysis, the inhibitory activity of the DiPP4 enzyme was also observed and the IC50 value was equal to 1.51 mg/mL, which corresponds with the results obtained in the present study. In another study performed by Lacroix et al. (2012), it was reported that a peptide fraction obtained from pepsin WPI hydrolysate (1 h hydrolysis) could be an effectual DiPP4 inhibitor (IC50: 75 µg/mL).
A lower IC50 value obtained for the 5 and 10 kDa fractions compared to hydrolyzed WPC indicates that the fractions have a relatively better ability to inhibit the DiPP4 enzyme. This is consistent with the results obtained by Konrad et al. (2014), who offered similar results for peptide fractions derived from enzymatic hydrolysis of whey proteins by plant serine proteases (Cucurbita ficifolia) The inhibition of DiPP4 enzyme for peptides obtained by WPC hydrolysis with FTLP was measured in the mg/ml range, while most studies were reported in the µg/mL range. Since whey protein hydrolysis consists of a combination of peptides, only some of them exhibit DiPP4 inhibitory activity, so it is not surprising that they are less effective than those reported values for derived peptides or synthetic drugs. (Lacroix et al., 2012).
The purpose of the present study was to investigate the ability of WPC to produce DiPP4 inhibitory peptides regardless of the domain of inhibitory activity. Preparation of small peptide fractions using RP-HPLC and RP-FPLC may increase activity up to µg/ml. It is noteworthy that the preparation of peptide fractions based on molecular weight can improve inhibitory effect (Silveira et al. 2013). This rule may not always be true because in some studies smaller peptides were not the most important DiPP4 inhibitors. Since many of the DiPP4 inhibitory peptides introduced in the studies are di and tripeptides (Yan et al. 1992; Cohen et al. 2004), it is strange that some low molecular weight peptides have lower DiPP4 inhibitory activity. In research conducted by Lacroix et al. (2012), the lower inhibitory activity of a peptide fraction < 1 kDa induced by WPI hydrolysis by pepsin was reported compared with higher molecular weight fractions.
In some studies, several bioactive peptides from different whey proteins hydrolyzed by various proteolytic enzymes have been introduced as suitable inhibitors of DiPP4. Silvira et al. (2013) identified several short-chain peptide fragments from WPC hydrolysis with commercial trypsin as highly potent peptides (IC50 86, 367 µg/mL) to inhibit DiPP4. In research done by Lacroix et al. (2012) several peptide fractions of whey proteins hydrolyzed by pepsin as a DiPP4 inhibitor were also introduced. Although the inhibitory activity of the DiPP4 enzyme was noted in all whey proteins, the hydrolysis of alpha -lactalbumin with an IC50 value equal to 36 µg/mL showed greater potency. In another study performed by Uchida et al. (2011), a six-amino acid peptide (Val-Ala-Gly-Thr-Trp-Tyr) as a DiPP4 inhibitor was obtained from the hydrolysis of beta -lactoglobulin by the enzyme trypsin. This variation in activity observed between various researches may be due to variations in experimental conditions, such as the nature of the substrate and the origin of the protease enzyme used to evaluate the inhibitory function.
DiPP4 Inhibitory Activity of TPDA
TPDA, one of the first known DiPP4 inhibitors, has been derived from the Bacillus cereus bacterium (Umezawa 1984). TPDA acts as an inhibitor of DiPP4 stimulatory activity in the human central nervous system (HCNS), the immune system, and DiPP4 in the endocrine system (Maes et al. 2007). In this study, the IC50 value for TPDA, the DiPP4 reference inhibitor, was also measured and was equal to be 1.73 µg/mL (Fig. 4; Table 7). The reason for the high inhibitory activity of TPDA may be due to its unique purity and specificity (Maes et al. 2007). This result is almost consistent with the results reported in other studies, which indicates the correctness of the method of measuring DiPP4 inhibitory activity during the experiment. The IC50 for TPDA was stated by Bharti et al. (2012) and Chakrabarti et al. (2016) as equal to 1.62 and 1.54 µg/mL, respectively.
DiPP4 Inhibitory Activity of L. Plantarum subsp. Plantarum PTCC 1896
DiPP4 inhibitory activity is found in bacteria due to the presence of an enzyme called x-prolyl dipeptidyl aminopeptidase (X-PDAP), which has approximately the same enzyme activity as DiPP4. This activity can be used to adjust enzymatic activity or microbial metabolism or to support the competition of bacterium with other microbial species (Panwar et al. 2016). Recently, the potential for DiPP4 inhibitory activity in lactic acid bacteria has also been reported. Panwar et al. (2016) reported that some Lactobacillus species such as L. plantarum (12–25%) and L. fermentum (14%) are potential sources of DiPP4 inhibitory activity. Therefore, in this study also, the DiPP4 inhibitory activity in vitro L. plantarum subsp. plantarum PTCC 1896 was investigated. Both CFCS and CFIE (from 8 × 109 CFU/mL, 6 h) showed different degrees of DiPP4 inhibitory activity (10.9 and 8.4%, respectively, p < 0.05). The result obtained was consistent with the result of Zeng et al. (2016). Based on the results of their study, the DiPP4 inhibitory activity in both CFCS and CFIE samples of L. plantarum IF2-14 (from 5 × 1010 CFU/mL, 6 h) was 20.7, and 14.8%, respectively. Therefore, it is concluded that L. plantarum subsp. plantarum PTCC 1896 had a reasonable amount of DiPP4 inhibitory activity. The main components of the DiPP4 inhibitors secreted by Lactobacillus are peptide compounds and are not produced by bacterial metabolism/fermentation. Zeng et al. (2016) showed that the treatment of CFCS with trypsin considerably improved the inhibitory activity of DiPP4, which could be imputed to the hydrolysis-induced peptides. Thus, it may be assumed that the structural components responsible for inhibiting DiPP4 in Lactobacillus bacteria are peptides (Yamamoto et al. 1994).
Conclusion
In this study, the validity of a simple and rapid method for determining proteolytic activity has been investigated. TLP secreted by L. plantarum subsp. plantarum PTCC 1896 has shown both qualitatively and quantitatively proteolytic activity on the SMA medium, therefore, this bacterium was identified as a protease-producing strain. In addition, in this study, the ability of hydrolysis of whey protein concentrate (WPC) by filtered trypsin-like protease (FTLP) under in vitro conditions for its use in the production of safe bioactive peptide components was investigated. In order to predict the best hydrolysis conditions, optimization was performed using response surface design. The results showed that the RSM statistical method was a good and reliable method for selecting the optimum levels of the investigated factors because the DH rate actually measured in vitro was in line with the predicted DH rate in the RSM. Based on the obtained results, the data revealed that the WPC hydrolysis was significantly affected by the E∶S ratio and temperature. Indeed, the temperature does not affect the release of peptides, but rather the enzyme activity. DiPP4 inhibitory activity of bioactive peptides induced by hydrolysis of whey protein by proteases such as trypsin-like protease from lactic acid bacteria has not been previously reported. In this study, it was found for the first time that WPC hydrolyzed by FTLP had acceptable IC50 values. In addition, the results showed that L. plantarum subsp. plantarum PTCC 1896 had a reasonable amount of DiPP4 inhibitory activity. Therefore, it was concluded that in the future there may be a possibility to use the probiotic bacterium L. plantarum subsp. plantarum PTCC 1896 in the management of T-IID. However, based on findings from previous studies, it is important to note that the in vitro IC50 value may not necessarily correlate directly with the effect of diabetes in vivo, so it is recommended that the results of this study be evaluated in laboratory mice. It is also encouraged that the ability of FTLP to hydrolyze other proteins to gain a more comprehensive understanding of its enzymatic activity.
5. References
Arslan-Alaton, I., Akin, A., Olmez‐Hanci, T.: An optimization and modeling approach for H2O2/UV‐C oxidation of a commercial non‐ionic textile surfactant using central composite design. J. Chem. Technol. Biotechnol. 85, 493–501 (2010)
Bayraktar, E.: Response surface optimization of the separation of DL-tryptophan using an emulsion liquid membrane. Process Biochem. 37(2), 169–175 (2001)
Bertrand-Harb, C., Ivanova, I.V., Dalgalarrondo, M., Haertllé, T.: Evolution of β-lactoglobulin and α-lactalbumin content during yoghurt fermentation. Int. Dairy. J. 13, 39–45 (2003)
Bharti, S.K., Krishnan, S., Kumar, A., Rajak, K.K., Murari, K., Bharti, B.K., Gupta, A.K.: Antihyperglycemic activity with DPP-IV inhibition of alkaloids from seed extract of Castanospermum australe: Investigation by experimental validation and molecular docking. Phytomedicine. 20, 24–31 (2012)
Bradford, M.M.: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976)
Brandelli, A., Daroit, D.J., Corrêa, A.P.F.: Whey as a source of peptides with remarkable biological activities. Food Res. Int. 73, 149–161 (2015)
Chakrabarti, S., Wu, J.: Bioactive peptides on endothelial function. Food Sci. Human Well. 5, 1–7 (2016)
Cheison, S.C., Lai, M.Y., Leeb, E., Kulozik, U.: Hydrolysis of β-lactoglobulin by trypsin under acidic pH and analysis of the hydrolysates with MALDI–TOF–MS/MS. Food Chem. 125, 1241–1248 (2011)
Cohen, M., Fruitier-Arnaudin, I., Piot, J.: Hemorphins: Substrates and / or inhibitors of dipeptidyl peptidase IV: Hemorphins N-terminus sequence influence on the interaction between hemorphins and DPPIV. Biochimie. 86, 31–37 (2004)
Courtin, P., Rul, F.: Interactions between microorganisms in a simple ecosystem: Yogurt bacteria as a study model. Le Lait. 84, 125–134 (2004)
Custódio, M.F., Goulart, A.J., Marques, D.P., Giordano, R.C., Giordano, R.D.L.C., Monti, R.: Hydrolysis of cheesewhey proteinswith trypsin, chymotrypsinand carboxypeptidase A. Alim Nutr. Araraquara. 16, 105–109 (2009)
da Costa, E.L., da Rocha Gontijo, J.A., Netto, F.M.: Effect of heat and enzymatic treatment on the antihypertensive activity of whey protein hydrolysates. Int. Dairy J. 17(6), 632–640 (2007)
da Silva, M.C., Silva, V.D.M., Lana, A.M.Q., Silvestre, M.P.C.: Degree of hydrolysis and peptide profile of enzymatic hydrolysates obtained from whey protein concentrate. Braz J. Food Technol. 20, 395–403 (2009)
El-Zahar, K., Chobert, J.M., Sitohy, M., Dalgalarrondo, M., Haertlé, T.: Proteolytic degradation of ewe milk proteins during fermentation of yoghurts and storage. Food/Nahrung. 47, 199–206 (2003)
Foegeding, E.A., Davis, J.P., Doucet, D., McGuffey, M.K.: Advances in modifying and understanding whey protein functionality. Trends Food Sci Technol. 13, 151–159 (2002)
Kim, S.B., Ki, K.S., Khan, M.A., Lee, W.S., Lee, H.J., Ahn, B.S., Kim, H.S.: Peptic and tryptic hydrolysis of native and heated whey protein to reduce its antigenicity. Int. J. Dairy. Sci. 90, 4043–4050 (2007)
Kishimura, H., Tokuda, Y., Klomklao, S., Benjakul, S., Ando, S.: Comparative study of enzymatic characteristics of trypsins from the pyloric ceca of yellow tail (Seriola quinqueradiata) and brown hakeling (Physiculus japonicus). J. Food Biochem. 30, 521 (2006)
Klomklao, S., Benjakul, S., Visessanguan, W., Kishimura, H., Simpson, B.K.: Purification and characterisation of trypsins from the spleen of skipjack tuna (Katsuwonus pelamis). Food Chem. 100, 1580–1589 (2007)
Konrad, B., Anna, D., Marek, S., Marta, P., Aleksandra, Z., Józefa, C.: The evaluation of dipeptidyl peptidase (DPP)-IV, α-glucosidase and angiotensin converting enzyme (ACE) inhibitory activities of whey proteins hydrolyzed with serine protease isolated from Asian pumpkin (Cucurbita ficifolia). Int. J. Pept. Res. Ther. 20, 483–491 (2014)
Koutsopoulos, S., Patzsch, K., Bosker, W.T., Norde, W.: Adsorption of trypsin on hydrophilic and hydrophobic surfaces. Langmuir. 23, 2000–2006 (2007)
Kristinsson, H.G., Rasco BA.: Fish protein hydrolysates: production, biochemical, and functional properties. Crit. Rev. Food Sci. Nutr. 40(1), 43–81 (2000)
Lacroix, I.M., Li-Chan, E.C.: Dipeptidyl peptidase-IV inhibitory activity of dairy protein hydrolysates. Int. Dairy. J. 25, 97–102 (2012)
Liu, R., Zhou, L., Zhang, Y., Sheng, N.-J., Wang, Z.-K., Wu, T.-Z., Wang, X.-Z., Wu, H.: Rapid identification of dipeptidyl peptidase-IV (DPP-IV) inhibitory peptides from Ruditapes philippinarum hydrolysate. Molecules. 22, 1714 (2017)
Maes, M.B., Dubois, V., Brandt, I., Lambeir, A.M., Van der Veken, P., Augustyns, K., …, De Meester, I.: Dipeptidyl peptidase 8/9-like activity in human leukocytes. J. Leukoc. Biol. 81(5), 1252–1257 (2007)
Mirlohi, M., Soleimanian-Zad, S., Sheikh-Zeinodin, M.: Identification of Lactobacilli from fecal flora of some Iranian infants. Iran. J Pediatr. 18(4), 357–363 (2008)
Morais, H.A., Silvestre, M.P.C., Silva, M.R., Silva, V.D.M., Batista, M.A., Silva, A.C.S., Silveira, J.N.: Enzymatic hydrolysis of whey protein concentrate: Effect of enzyme type and enzyme: Substrate ratio on peptide profile. J. Food Sci. Technol. 52, 201–210 (2015)
Morr, C.V., Foegeding, E.A.: Composition and functionality of commercial whey and milk protein concentrates and isolates: A status report. Food Technol. 44(4), 100–112 (1990)
Mota, M.V.T., Ferreira, I.M.P.L.V.O., Oliveira, M.B.P., Rocha, C., Teixeira, J.A., Torres, D., Gonçalves, M.P.: Trypsin hydrolysis of whey protein concentrates: Characterization using multivariate data analysis. Food Chem. 94, 278–286 (2006)
Mustafa, M.H., Soleimanian-Zad, S., Sheikh-Zeinoddin, M.: Characterization of a trypsin-like protease 1 produced by a probiotic Lactobacillus plantarum subsp. plantarum PTCC 1896 from skimmed milk based medium. LWT. 119, 108818. (2020)
Naik, L., Mann, B., Bajaj, R., Sangwan, R.B., Sharma, R.: Process optimization for the production of bio-functional whey protein hydrolysates: Adopting response surface methodology. Int. J. Pept. Res. Ther. 19, 231–237 (2013)
Nilsson, M., Holst, J.J., Björck, I.M.E.: Metabolic effects of amino acid mixtures and whey protein in healthy subjects: Studies using glucoseequivalent drinks. Am. J. Clin. Nutr. 85, 996–1004 (2007)
O’Loughlin, I.B., Murray, B.A., Brodkorb, A., Fitzgerald, R.J., Robinson, A.A., Holton, T.A., Kelly, P.M.: Whey protein isolate polydispersity affects enzymatic hydrolysis outcomes. Food Chem. 141, 2334–2342 (2013)
Panwar, H., Calderwood, D., Grant, I.R., Grover, S., Green, B.D.: Lactobacilli possess inhibitory activity against dipeptidyl peptidase-4 (DPP-4). Ann. Microbiol. 66(1), 505–509 (2016)
Pihlanto-Leppälä, A., Rokka, T., Korhonen, H.: Angiotensin I converting enzyme inhibitory peptides derived from bovine milk proteins. Int. Dairy. J. 8, 325–331 (1998)
Prioult, G., Pecquet, S., Fliss, I.: Allergenicity of acidic peptides from bovine β-lactoglobulin is reduced by hydrolysis with Bifidobacterium lactis NCC362 enzymes. Int. Dairy. J. 15, 439–448 (2005)
Pripp, A.H., Sørensen, R., Stepaniak, L., Sørhaug, T.: Relationship between proteolysis and angiotensin-I-converting enzyme inhibition in different cheeses. LWT. 39, 677–683 (2006)
Silva, M.R.: Obtenção De Hidrolisados enzimáticos do concentrado protéico do soro de leite com alto teor de oligopeptídeos e elevada atividade inibitória sobre a enzima conservadora de angiotensina, utilizando a pancreatina e a papaína Tese (Mestrado), Universidade Federal de Minas Gerais, Belo Horizonte, 89 p. (2010)
Silva, A.C.S., Silveira, J.N.: Correlation between the degree of hydrolysis and the peptide profile of whey protein concentrate hydrolysates: Effect of the enzyme type and reaction time. Am. J. Food Technol. 8, 1–16 (2013)
Silveira, S.T., Martínez-Maqueda, D., Recio, I., Hernández-Ledesma, B.: Dipeptidyl peptidase-IV inhibitory peptides generated by tryptic hydrolysis of a whey protein concentrate rich in β-lactoglobulin. Food Chem. 141, 1072–1077 (2013)
Silvestre, M.P.C., Silva, M.R., Silva, V.D.M., Souza, M.W.S., Junior, L., de Oliveira, C., Afonso, W.O.: Analysis of whey protein hydrolysates: Peptide profile and ACE inhibitory activity. Braz J. Pharm. Sci. 48, 747–757 (2012)
Torrades, F., García-Montaño, J.: Using central composite experimental design to optimize the degradation of real dye wastewater by Fenton and photo-Fenton reactions. Dyes Pigm. 100, 184–189 (2014)
Uchida, M., Ohshiba, Y., Mogami, O.: Novel dipeptidyl peptidase-4–inhibiting peptide derived from β-lactoglobulin. J. Pharmacol. Sci. 117(1), 63–66 (2011)
Umezawa, H.: Studies on low molecular weight immunomodifiers produced by microorganisms: Results of ten years effort. Clin. Infect. Dis. 6, 412–420 (1984)
Vijayaraghavan, P., Vincent, S.G.P.: A simple method for the detection of protease activity on agar plates using bromocresol green dye. J. Biochem. Technol. 4, 628–630 (2013)
Yamamoto, N., Akino, A., Takano, T.: Antihypertensive effect of the peptides derived from casein by an extracellular proteinase from Lactobacillus helveticus CP790. J. Dairy. Sci. 77, 917–922 (1994)
Yan, T.-R., Ho, S.C., Hou, C.L.: Catalytic properties of X-prolyl dipeptidyl aminopeptidase from Lactococcus lactis subsp. cremoris nTR. Biosci. Biotechnol. Biochem. 56, 704–707 (1992)
Zeng, Z., Luo, J., Zuo, F., Zhang, Y., Ma, H., Chen, S.: Screening for potential novel probiotic Lactobacillus strains based on high dipeptidyl peptidase IV and α-glucosidase inhibitory activity. J. Funct. Foods. 20, 486–495 (2016)
Acknowledgements
The authors thank all the researchers and students whose articles have been used as sources.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Haj Mustafa, M., Soleimanian-Zad, S. & Albukhaty, S. Whey Protein Concentrate Hydrolyzed by Microbial Protease: Process Optimization and Evaluation of Its Dipeptidyl Peptidase Inhibitory Activity. Waste Biomass Valor 15, 2259–2271 (2024). https://doi.org/10.1007/s12649-023-02306-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12649-023-02306-1