Abstract
The present study investigates the biochemical characterization of an extracellular (phospho)lipase from a wood fungus Peziza sp. (medium optimization, inducer concentration and substrate specificity measurements). The strain was identified on the basis of ITS1/ITS4 primers. A 604 bp fragment was amplified by PCR and the obtained nucleotide sequence, showed 99% identity with the ITS region of isolates named as Peziza sp. Interestingly, Peziza sp. has both lipase and phospholipase activities with the same level which require both the presence of Ca2+ and bile salts. Our result shows that the lipase hydrolyzes preferably the olive oil at 45 °C, pH 8. Whereas, the phospholipase activity was detected on pure PC at 45 °C, pH 9. Lipid extraction from dry biomass using chloroform/methanol (2/1) and quantitative measurement using electron microscope showed that intracellular triglycerides content was significantly high and reaches 20.88%. Gas chromatography analysis shows a majority of C18:1 (76, 98%) and C18:2 (9, 33%). Whereas, saturated fatty acids ranging C16–C20 represent only 11.5% of total lipids composition.
Graphic Abstract
Lipid accumulation test in Pezizales sp under fluorescence microscope

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Statement of Novelty
A new lipolytic and oleaginous fungal strain was isolated from wood.
They are widely distributed in nature including animals, plants and microorganisms. Lipases from microorganisms have been found to be the most important and interesting for industrial applications [3] because they do not requires cofactors and can be produced at high yields.
Therefore, most lipases used for biotechnological purposes have been isolated from yeast and fungi [4]. Actually they are incorporated in many fields such as food (Fat, lipids, bakery and diary), pharmaceutical industry and cosmetic product. Lipases are evenly involved in detergent formulations, effluent treatment, production of diesel [5], perfumery and treatment of malignant tumors [6]. A wide number of filamentous fungi that produces lipases have been characterized such as Rhizopus oryzae [7], Rhizopus homothallicus [8], Penicillium aurantiogriseum [9], Rhizomucor miehei [10], Thermomyces lanuginosus [11], and Fusarium solani [12, 13].
Phospholipases (EC 3.1.1) are soluble enzymes that cleave esters bonds from phospholipids [14]. Depending on the site of attack, these enzymes were classified as phospholipase A, B, C or D. Phospholipase A; enzymes hydrolyze the 1-acyl ester (PLA1) or the 2-acyl ester (PLA2) of phospholipids. In fact, the use of phospholipases in the industry is growing to invade several areas. They are used in the degumming of oils, bread making and emulsifier producing as molecules (lysolecithin and monoglycerides…) by acting on the phospholipids already present in the ingredients. So, the use of chemical emulsifiers may be substituted by the use of phospholipases, which generate natural emulsifiers by hydrolyzing the lipids in the dough. [15].
Oleaginous microorganisms can accumulate until 70% lipid mass fraction on dry cell extraction and lipids are disposed in intracellular compartment called lipid bodies [16].
The lipids from microorganisms can be incorporated in many fields such: Cocoa butter [17], biodiesel production [18], treatment of many diseases such as inflammation, diabetes, cardiovascular disorders, and cancers… [19].
To study lipolytic enzymes from this fungus and to explore their catalytic properties, we describe in the present study the optimization of the production of both lipase and phospholipase from a new strain Peziza sp. isolated from forest wood in north of Tunisia. Qualitative and quantitative lipid accumulation is also investigated.
Introduction
Lipases (triacylglycerol acylhydrolases, E.C. 3.1.1.3) are soluble enzymes characterized with a considerable physiological role and industrial potential [1]. The interest given to lipases is due to their ability to catalyze the hydrolysis of long-chain acylglycerols at oil, water interfaces and the synthesis of esters from glycerol and long-chain fatty acids [2].
Materials and Methods
Fungal Strains
Decaying wood pieces were collected from a forest in the vicinity of Bousalem, Northwest of Tunisia during the winter of 2012 to isolate lipase-producing microorganisms. Wood pieces were inoculated into Petri Plates containing malt extract agar media, were supplemented with ampicillin and streptomycin (0.01%) to avoid bacterial growth [20] and then incubated at 30 °C. The purified fungal strains were cultivated in the same medium and stored at 4 °C.
Isolation and Screening of Lipolytic Fungus
Screening of lipolytic microorganisms: lipase and phospholipase producing was carried out using a plate assay on a Potato Dextrose Agar (PDA), medium containing 1% olive oil and 10% phosphatidylcholine for lipase and phospholipase activity, respectively and 1‰ rhodamine B as colored indicator under UV. The culture plates were incubated at 30 °C, and colonies showing widespread clearing around them were regarded as putative lipase/phospholipase producers. Among the strains retained, only one fungal colony, isolated from forest wood, exhibited high lipase productivity. The identification of this strain was performed using internal transcribed spacer markers (the ITS region) sequence analysis. The biochemical properties and the morphological aspect of this microorganism correspond to Peziza sp.
Molecular Identification of the Isolated Strain BS24 by Sequencing the ITS Region
For DNA extraction, the fungus was grown in a 250 ml flask containing 30 ml of glucose–peptone-yeast extract medium for 5 days. Mycelium was harvested by vacuum filtration, was washed with sterile Milli-Q water, and was lyophilized. Then 2.5 ml of HSE solution (10 mM HEPES, pH 6.9, 0.5 M sucrose and 20 mM EDTA, pH 8.0) and 250 ml of 10% sodium dodecyl sulphate were added to 0.1 g of the powdered mycelium. After incubation at 65 °C for 15 min, 2.5 ml of TEA solution (50 mM Tris–HCl and 20 mM EDTA, pH 8.0) were added, followed by three extractions with 5 ml of phenol/chloroform/isoamyl alcohol (25:24:1). Then a final chloroform/isoamyl alcohol (24:1) extraction was made. 0.5 ml of 3 M sodium acetate and 3 ml isopropanol were added to the remaining aqueous phase and the mixture was stored at room temperature for 30 min. The precipitate was collected by centrifugation (12,000×g, 10 min, 4 °C), was washed with ethanol (70%), dried and was dissolved in TEB solution (1 mM Tris–HCl and 1 mM EDTA, pH 8.0). RNase A (0.1 lg lL-1) was added to the suspension and incubated at 37 °C for 30 min. Then, 0.2 ml of 3 M sodium acetate and 1.2 ml isopropanol were added and the mixture centrifuged (5000 rpm, 5 min, and 4 °C). The pellet was washed with ethanol (70%), was desiccated and was dissolved in sterile Milli-Q water. The extracted DNA was used as the template in a PCR to amplify the ITS1 and ITS4 regions and the 5.8S rRNA gene. The primers used for the amplification were ITS1 (50-TCCGTAGGTGAACCTGCGG-30) and ITS4 (50-TCCT CCGCTTATTGATATGC-30) [20]. PCR was performed with 1 mM MgCl2 (Applied Biosystems), 1 mM of each primer, 50 mM of each deoxynucleoside triphosphate (Promega), 0.2 mg of DNA template and 1.2 U of Taq DNA polymerase (Applied Biosystems), in a final volume of 50 µL, using a GeneAmp PCR System 2400 (Perkin Elmer). Cycling parameters were 95 °C for 3 min followed by 35 cycles of 94 °C for 1 min, 52 °C for 40 s, and 72 °C for 1 min, with a final extension at 72 °C for 10 min. Control reactions lacking template DNA were performed in parallel. Amplified fragments were visualized on 1% agarose gels stained with ethidium bromide and subsequently purified using a GeneClean kit (Q-BIOgene). Products were then sequenced using the two PCR primers, the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems), and the automated ABI Prism 3730 World J Microbiol Biotechnol 123 DNA sequencer (Applied Biosystems). The sequence was compared by BLAST search [21] in GenBank.
Production of Lipase and Phospholipase by Peziza sp. in Shake Flask Culture
Lipase production was studied in 250 ml Erlenmeyer flasks containing 50 ml of the basal medium. Three medium with the following composition (g/l) were tested: 1.YPD, 2. a growth medium (6 g KH2PO4, 1.2 g MgSO4, 1 g NH4Cl, 0.3 g CaCl2 and 1 g yeast extract per liter) and 3.PKME (15 g peptone, 5 g yeast extract, 1.75 g KH2PO4 and 0.5 g MgSO4 per liter. The production medium is composed of the basal medium supplemented with different concentrations of glucose and olive oil whereas the pH of the medium was adjusted to different values (i.e. 5.5, 6.5, and 7.5). According to the final concentration, olive oil, Tween 80 and glycerol were added to the production medium in a concentration of 1% (w/w) as an inducer of lipolytic enzymes expression and carbon source.
The medium was sterilized at 121 °C for 15 min. The flasks were inoculated with 1% of freshly prepared culture of Peziza sp grown in YPD broth at 30 °C for 24 h. The flasks were incubated aerobically for 5 days on a rotary shaker set at 150 rpm and a temperature at 30 °C. Data are the mean of three independent experiments. Before each assay, the cell debris was removed by centrifugation at 10,000×g for 30 min. Next, the obtained clear supernatant was used as a crude enzyme preparation.
Assay of Lipolytic Activity and Biomass Production
After 5 days of incubation, the content of each flask was analyzed. All data were assumed as the average values of three independent experiments.
Lipase Activity
Lipase activity was assayed potentiometrically by automatically titrating the free fatty acids released from mechanically stirred triacylglycerol emulsions using 0.1 N NaOH and a pHstat. Each assay with short-chain (TC4) was performed in a thermostated (37 °C) vessel containing 0.25 ml triglyceride in 25 ml of a 2.5 mM Tris–HCl, pH 8.5, 3 mM CaCl2. Long-chain triglycerides (olive oil) had first pre-emulsified with gum arabic (GA) by mixing 10 ml of olive oil with 90 ml of a 10% (w/v) GA solution prepared as previously described [22]. Ten milliliters of this emulsion were then mixed in the pH-stat vessel with 20 ml of 2.5 mM Tris–HCl (pH 8.5) containing 3 mM CaCl2. When measuring lipase activity in the absence of CaCl2, EDTA was added to the assay system. One unit of lipase activity corresponds to 1 μmol of fatty acid released per minute under the assay conditions used.
Phospholipase Activity
The phospholipase activity was measured titrimetrically at pH 8.5 and at 37 °C with a pH-stat, using egg phosphatidylcholine (PC) (0.1 g) as substrate mixed in 30 ml distilled water in the presence of different concentrations of NaTDC and CaCl2. One unit of phospholipase activity was defined as 1 μmol of fatty acid liberated per minute.
Effect of pH and Temperature on Peziza sp Activity and Stability
The thermal profile of enzyme activity was measured at temperatures between 25 and 55 °C. The incubation was performed in a temperature-controlled cuvette holder. In order to determine the thermal stability of Peziza sp., aliquots of the supernatant solution were incubated successively for 5, 10 and 15 min at 30 °C, 37 °C, 40 °C and 45 °C. Immediately after incubation, both lipase and phospholipase residual activities were determined, after centrifugation, under standard assay method. The pH optimum for were determined at different pHs ranging from 6 to 11. In order to determine the pH stability, pre-incubation was performed at room temperature for 1 h in the following buffers—50 mM sodium acetate buffer (pH 4.0–6.0), 50 mM potassium phosphate buffer (pH 6.0–8.0), 50 mM Tris–HCl buffer (pH 7.0–9.0) and 50 mM glycine–NaOH buffer (pH 8.0–12.0). The residual activities, lipase and phospholipase one, were determined after centrifugation, under standard assay method, and the pH of each buffer was adjusted at 37 °C, the enzyme reaction temperature.
Lipase, Phospholipase and Biomass Production by Peziza sp. Isolates Under Optimized Conditions
The Peziza sp. isolated fungus was grown in 250 ml Erlenmeyer flasks containing 50 ml of optimized medium with the following composition (g/l): Yeast extract 5.0, KH2PO4 1.75 peptone 15, MgSO4 0.5 olive oil 1% and the inoculums size was adjusted to 1% for lipase activity. Whereas, for the optimized medium of the phospholipase production, its composition contains: Yeast extract 5, K2HPO4 1.75, peptone 15, olive oil 1% and the inoculums size was adjusted to 3%. Lipase and phospholipase activities were tested on the appropriate substrate at several bile salt and CaCl2 concentrations. When measuring lipolytic activities in the absence of CaCl2, EDTA was added to the assay.
Lipids Extraction and Analysis
Mycelium Biomass
Peziza sp. was grown in a 250 ml flask. The medium was consisted of (g/l): yeast extract 5.0, K2HPO4 1.75, peptone 15, MgSO4 0.5 and the inoculums size was adjusted to 1%. The pH of the medium was adjusted to 6.0 before autoclaving. The fungal cells were harvested by suction filtration, washed with water twice.
The wet biomass was used for the qualitative analysis (microscope observation), or dried at 80 °C overnight for lipids extraction (quantitative analysis with CPG).
Microscope Observation
Firstly, we added 1 µg/ml cells into the Oil Red O solution, and then it was incubated for 30 min at room temperature in dark. Two water washes were made. Cells suspensions were collected in PBS buffer (10 mM phosphate buffer, pH 7, 0.15 M KCl), before a microscopic observation (Olympus BX61) fluorescence emission a 460–490 nm excitation filter, 500 nm diachronic mirror, 520 nm barrier filter (U-MWB2).
Lipids Extraction
Extraction of lipids from dry biomass was performed according to the procedure of folch [23]. The dry biomass was ground into a fine powder: 1 g powder was blended with 3 ml chloroform/methanol (2:1) and the mixture was agitated during 20 min in an orbital shaker at room temperature. Solvent phase was recovered by centrifugation. The same process was used three times. The whole solvent was evaporated and dried under vacuum.
Assays
Fatty acids were methylated by boron trifluoride in methanol (Sigma) according to the Metchalfe and Schmitz (1961) method. The Rapid Preparation of Fatty Acid Esters for Gas Chromatographic Analysis [24] and fatty acid methyl esters (FAMEs) were analyzed by gas chromatography (GC) on a Shimadzu (GC-2014) equipped with non polar DANI-SPA column (50 m length, 0.32 mm i.d.) and a flame ionization detector. Nitrogen was used as a carrier gas. The temperatures of the column oven, the injection ports, and the detector were maintained at 165, 250, and 250 °C, respectively. Two microliters of the sample was injected into the column. Chromatographic peaks were identified by comparing the retention times with those of known standards, maintained at 165, 250, and 250 °C, respectively. Two microliters of the sample was injected into the column. Chromatographic peaks were identified by comparing the retention times with those of known standards.
Results
Isolation, Identification and Selection of Lipase and Phospholipase Producing Fungi
In a preliminary screening of lipase-producing microorganisms, out of seven isolates the most efficient lipase producer strains was obtained from North Tunisia based on enzyme assay method. Activity was identified as an orange halo zone around colonies under UV light at 350 nm.
Peziza sp., forming the most intense orange fluorescent halo around the colonies on rhodamine olive-oil agar was selected for molecular identification and optimization experiments of lipase production (Fig. 1).
a Time-course of lipase production using olive oil emulsion as substrate. The culture was carried out at 30 °C in 1-l shaking with 200 ml of culture of medium containing 15 g/l casein peptone, 5 g/l yeast extract, 1.75 g/l KH2PO4, 0.5 g/l MgSO4 and 1% olive oil (pH 6). The values represent the average of three cultures. b Time-course of phospholipase production using egg yolk emulsion as substrate. The culture was carried out at 30 °C in 1-l shaking with 200 ml of culture medium containing 15 g/l casein peptone, 5 g/l yeast extract, 1.75 g/l KH2PO4, and 3% olive oil (pH 6). The values represent the average of three cultures
Optimization of Lipase and Phospholipase Production
The aim of this study was the optimization of extracellular lipase/phospholipase production by Peziza sp. Statistical design of experiments is very effective for screening the best candidate factors. Different carbon and nitrogen sources as well as inoculation size, olive oil induction and glucose concentration were evaluated. Maximum and minimum levels of each parameter (glucose concentration, olive oil concentration and inoculums size) were determined after preliminary experiments. In the appropriate medium, the minimum or maximum concentration of glucose and olive oil were added, the inoculums size of the medium adjusted to values 1%, 2% and 3% and the lipase activity was determined after incubation at 30 °C for 5 days. The medium 1 components, namely, (yeast extract 5 g/l, KH2PO4 1.75 g/l, peptone 15 g/l, MgSO4 0.5 g/l, olive oil 1%, and the inoculums size was adjusted to 1%) whereas the medium 2 components, namely, (yeast extract 5 g/l, KH2PO4 1.75 g/l, peptone 15 g/l, PC 1%, and the inoculums size was adjusted to 3%), were selected to investigate the effective variables of the high lipase and phospholipase activity production, respectively (Table 1).
Inducer Selection for Lipase and Phospholipase Production
The effect of three factors; induction sources (olive oil, glycerol and Tween 80), induction concentration (1%, 2% and 3%) and bile salt level (ranging from 0 to 3 mM) were studied on lipase and phospholipase production. These components were varied in the basal medium containing peptone 15 g, yeast extract 5 g, KH2PO4 0.75 g and MgSO4 0.5 g/l. Lipase production was induced with 1% of olive oil (5 U/ml) (Fig. 2a). Little increase of the activity was observed with glycerol and Tween 80 at values of 0, 5%, 1% or 1, 5%. The highest lipase activity was measured at 2 mM bile salt for all the inducers tested. The same behavior was observed for measurement of phospholipase activity, when we tested the same inducers and at the same percentage. However, the optimal activity levels were observed at 3 mM bile salt on egg PC, and in the presence of 10 mM calcium (Fig. 2b).Thus, 1% olive oil seems to be the best inducer for both lipase and phospholipase activity suggesting that these two activities are supported by the same enzyme. These results are similar to those observed with Fusarium solani (phospho)lipase [25].
a Evaluation of different inducers for lipase activity production by Peziza sp. Three different inducers were used: glycerol, Tween 80 and olive oil 1%. Lipase activity was measured using a pH-stat under standard conditions without inducer as control. b Evaluation of the different inducers for phospholipase activity production by Peziza sp. Three different inducers were used: glycerol, Tween 80 and olive oil 1%. Phospholipase activity was measured using a pH-Stat under standard conditions without inducer as control
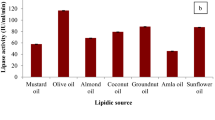
Length Chain Selectivity and Bile Salt Dependence
Lipase and phospholipase substrate specificity was analyzed using different substrates: short chain triacylglycerol (TC4), long chain triacylglycerol (emulsion of olive oil), pure phosphatidylcholine (PC) and egg yolk emulsion. Figure 3a shows that the substrate preferably hydrolyzed by the enzyme was the olive oil (5 U/ml) with a strict dependence to detergent (maximal activity at 2 mM Bile salt). Using TC4 as substrate, the activity reaches 2 U/ml only, with the same amount of bile salt. Whereas, Fig. 3b shows that phospholipase activity was detected nearly with the same amount: 4 U/ml and 5 U/ml on pure PC and egg yolk emulsion, respectively.
Evaluation of Bile Salts concentration on lipase activities (a) and phospholipase activities (b) of Peziza sp. Effect of increasing concentration of bile salt (NaTDC) on lipase activity in presence of 3 mM CaCl2 using olive oil emulsion and (tributyrin)TC4 as substrates (a) and phospholipase activity in presence of 10 mM CaCl2 and phosphatidylcholine and egg yolk PC as substrates (b)
Effects of pH on Peziza sp. Activity and Stability
Enzymatic activities of Peziza sp. were investigated at different pHs using emulsified olive oil or egg yolk PC as substrates. Figure 4a shows that the maximal lipase activity was obtained at pH 8. This activity was reduced drastically at pH 6, 10 and 11. Moreover, maximal phospholipase activity was observed at pH 9 (Fig. 4a).
pH effect on lipase and phospholipase activity (a) and stability (b). Optimal pH was determined using Olive oil emulsion and phosphatidylcholine for lipase respectively. pH stability was analysed after preincubating the pure enzyme for 1 h different buffer solutions at various pH ranging from 4 to 12 using the following buffers: 0.5 M glycine-HCl (pH 3–4); 0.5 M sodium acetate (pH 5–6); 0.5 M Tris–HCl (pH 8–9), and 0.5 M sodium borate (pH 10–11) and the remaining lipase and phospholipase activities were measured. a pH effect on enzyme activity of Peziza sp. Optimal pH was determined using olive oil emulsion and phosphatidylcholine for lipase and phospholipase activities, respectively. b pH stability of lipase and phospholipase activity for Peziza sp. after incubating the supernatant for 15 min in different buffer solutions at various pH from 4 to 12 using the following buffers: 0.5 M glycine–HCl (pH 3–4); 0.5 M sodium acetate (pH 5–6); 0.5 M Tris–HCl (pH 8–9), and 0.5 M sodium borate (pH 10–11). Residual activity was measured at 45 °C. pH 8 and 9 for lipase and phospholipase, respectively
Interestingly, the lipolytic activity of FSL remained stable in the pH range of 8.0–10 after pre-incubation at room temperature for 15 min (Fig. 5b). The remaining residual activity was above 60% at pH 10. However, the enzyme was most stable at pH 9 which shows a residual activity of 95% after incubation in Tris–HCl buffer at room temperature for 15 min.
a Temperature effect on enzyme activity of Peziza sp. Olive oil emulsion and phosphatidylcholine were used for lipase and phospholipase activities, respectively. b.1 Temperature stability of lipase for Peziza sp. Activities were measured under standard conditions. b.2 Temperature stability of phospholipase for Peziza sp. Activities were measured under standard conditions. c Effect of the concentration of Ca2+ on lipase and phospholipase activity. Enzyme activity was measured at increasing concentrations of Ca2+ using olive oil emulsions as substrate for lipase activity in the presence of 2 mM NaTDC and PC as substrate for phospholipase activity in the presence of 3 mM NaTDC. The star indicates the lipase/phospholipase activity measured in the absence of CaCl2 and in the presence of 10 mM EDTA
Effects of Temperature on Peziza sp. Activity and Stability
The lipase and phospholipase activities of Peziza sp. were measured as a function of temperature from 25 to 55 °C (Fig. 5a). Our results showed that the maximum of activity was obtained at 45 °C. The residual lipase activity was maintained at 100% and 70% after treatment at 30 °C, 37 °C for 30 min and 20 min for 45 °C, respectively, indicating that the enzyme was stable up to 37 °C (Fig. 5b). The enzyme activity decreased rapidly at temperature higher than 45 °C, and it was inactivated totally at 50 °C.
Effect of Calcium on Lipase and Phospholipase Activity
The effect of various Ca2+ concentrations on the rate of hydrolysis of olive oil and egg yolk PC as substrates by lipase and phospholipase activity respectively were studied (Fig. 5c). Lipase activity of 2 U/ml was detected in the absence of Ca2+, and in the presence of 10 mM of EDTA (metal chelators). The addition of Ca2+ increases the lipase activity to reach a maximum of 5 U/ml in the presence of 3 mM CaCl2 using olive oil as substrate. The positive effect of Ca2+ is due to the formation of long chain fatty acids during the hydrolysis process and there complexes with the bivalent ions, avoiding the inhibition of the enzyme [26].
Interestingly, using egg yolk PC as substrate, no phospholipase activity was observed in the absence of Ca2+, and in the presence of EDTA. In the absence of chelators, the phospholipase activity of Peziza sp. increases by increasing Ca2+ concentration to reach 5 U/ml at 10 mM CaCl2 at 45 °C and at pH 9.
In fact, cations by binding to the enzyme, modify its conformation by offering a better stability and a greater activity [27].
Lipids Accumulation
Microscope Observation
Nile Red staining (Fig. 6) showed more lipid droplets in Peziza sp. cells compared to the control group (Fusarium solani, which is non oleaginous fungi) which is non oleaginous fungi. Quantitative measurement also clarified that intracellular triglyceride content was significantly high and reaches 20.88%. And thus, Peziza sp. is considerate as an oleaginous strain.
Lipid accumulation test in Peziza sp. under fluorescence microscope (Nile red staining). a Control, shows normal Peziza sp. b Apparent lipid accumulation (bright yellow fluorescence) was observed by using 1 µg/ml cells into the Oil Red O solution in a microscopic observation (Olympus BX61) fluorescence emission a 460–490 nm excitation filter, 500 nm diachronic mirror, 520 nm barrier filter (U-MWB2)
Extraction of Lipids from Fungal Cells: CPG Analyses
Lipids in the fungal cells were obtained by wet extraction and dry extraction. When lipids were extracted from dry biomass, the solvents were chloroform/methanol (2/1) and lipids yield was 20.88%.
Discussion
The obtained nucleotide sequence, showed 99% identity with the ITS region of isolates deposited as Peziza sp. For the molecular identification of the selected fungus, the ribosomal DNA region including ITS1, 5.8S, and ITS2 was sequenced; since it has been described as the suitable target for analysis of fungal phylogeny [21], 604 bp fragments was amplified by PCR using the ITS1/ ITS4 primers.
Culture medium optimization generally, improved the lipase activity and productivity.
So, in order to improve lipolytic activities, a cultivation medium 1 and medium 2 was designed for lipase and phospholipase activities respectively, as shown on Table 1. A maximal lipase and phospholipase activities of 5 U/ml was obtained by growing the strain for 5 days on a rotary shaker set at 150 rpm and a temperature at 30 °C.
The incorporation of oils as inducers or as sole carbon source in medium increased the enzyme production as mentioned in different studies which indicate that using plants oils, triglycerides…increase the total lipase activity [29,30,31,32,33].
In our study, adding 1% olive oil as inducer to both medium 1 and medium 2, increases the lipase and phospholipase activities respectively to reach the highest level of 5 U/ml. In fact, lipase activity was detected at the olive oil at 45 °C, and pH 8. Whereas, the phospholipase activity was detected on pure PC at 45 °C, pH 9.
The lipase and phospholipase activities were negligible before the addition of bile salts which confirms the strict dependence to bile salt, with optimum at 2 mM and 3 mM for lipase and phospholipase activity, respectively.
Like most fungal and yeast lipases that generally have optimal activity in neutral or alkaline pH [33], the lipase of Peziza sp. expresses its maximum activity at pH 8.5–9 on both lipid and phospholipid substrates. However, the pH stability of this lipase differs from those reported in the literature for related lipases. In fact, most fungal lipases were quite stable at pH ranging between 4.0 and 7.0 and showed lower tolerance to alkaline pH [32], as that observed for lipases from Mucor sp. [10]. This property allowed its use in detergent industry.
Concerning the effect of temperature on lipase and phospholipase activities which were confined to the range of 25–55 °C, the optimal temperature was obtained at 45 °C, and the lowest activity was detected at 50 °C. These results are in the range of previous studies which have shown that the optimal temperature was 50 °C for lipase from Talaromyces thermophilus was [34] and R. miehei [35] Meanwhile, lipase from Aspergillus niger had an optimum temperature at 30 °C [36] and Syncephalastrum racemosum at 37 °C [37].
In the absence of calcium and the presence of 10 mM EDTA, lipase and phospholipase activities of Peziza sp. were at low levels but with 2 mM CaCl2 and 10 mM CaCl2 for lipase and phospholipase activities respectively reached their maximum level 5 U/ml.
Similar results were obtained with a (phospho)lipase from S. hyicus [38] which require calcium ions to stabilize its active conformation. So, it seems that Ca2+ has enzyme-activating effect that exert by concentrating the enzyme at the lipid–water interface and stabilizing effect of the enzyme [39]. This behavior towards calcium ions seems to be similar to the known phospholipases A2 (PLA2) which are calcium-dependent. Structural studies of PLA2 in a previous study have reported that calcium was not essential to express lipase activity but essential for phospholipase A1 activity [40].
These two different activities are generated by the same catalytic site. It showed an alkaline stability in which is uncommon in lipases from fungi.
As it was described, fatty Acid Profile Ideal fungal was a good candidate for biodiesel production, which should have suitable fatty acid composition in addition to high lipid content. It is well established that fatty acid profile affects the properties of biodiesel, such as heat of combustion, lubricity, viscosity, low temperature properties and oxidative stability; Most of the vegetable oils have fatty acid profiles in the typical C16–18 range and large proportions of saturated fatty acids, leading to a biodiesel fuel that likely possesses a cloud point >0 °C [41]. As shown in Table 2, the fatty acid distribution, with C18:1 and C18:2 being the majority, 76, 98% and 9, 33%, respectively. Whereas, saturated fatty acids ranging C16–C20 represent only 11.5% of total lipids composition. In general, saturated fatty esters are high in cetane number and thus improve the oxidative stability of the biodiesel, whereas unsaturated fatty esters have better low-temperature properties. It has been suggested that monounsaturated fatty acid ester may act as a balance between oxidative stability and low-temperature properties, and its proportion is considered as an important index to assess the biodiesel quality of microorganism oils [42].
Abbreviations
- CPG:
-
Gas chromatography analysis
- PDA:
-
Potato Dextrose Agar
- PCR:
-
Polymerisation Chain Reaction
- PC:
-
Phosphatidyl choline
References
Sharma, R., Chisti, Y., Banerjee, U.C.: Production, purification, characterization, and applications of lipases. Biotechnol. Adv. 19, 627–662 (2001)
Jaeger, K.E., Reetz, M.T.: Microbial lipases from versatile tools for biotechnology. Trends Biotechnol. 16, 396–403 (1998)
Rudd, E.A., Brockman, H.L.: Pancreatic carboxyl ester lipase (cholesterol esterase). In: Borgstrom, B., Brockman, H.L. (eds.) Lipases, pp. 185–204. Elsevier, Amsterdam (1984)
Verger, R.: ‘Interfacial activation’ of lipases: facts and artifacts. Trends Biotechnol. 15, 32–38 (1997)
Yong, S.K., Lim, B.H., Saleh, S., Tey, L.H.: Optimisation, purification and characterisation of extracellular lipase from Botryococcus sudeticus (UTEX 2629). J. Mol. Catal. B 126, 99–105 (2016)
Sharma, R., Chisti, Y., Banerjee, U.C.: Production, purification, characterization, and applications of lipases. Biotechnol. Adv. 19(8), 627–662 (2001)
Sharma, S., Kanwar, S.S.: Organic solvent tolerant lipases and applications. Sci. World J. 1155, 625258 (2014)
Ben Salah, R., Mosbah, H., Fendri, A., Gargouri, A., Gargouri, Y., Mejdoub, H.: Production of wild-type and peptide fusion cutinases by recombinantSaccharomyces cerevisiae MM01 strains. FEMS Microbiol. Lett. 260, 241–248 (2006)
Diaz, M.J.C., Rodríguez, J.A., Roussos, S., Cordova, J., Abousalham, A., Carrière, F., Bratti, J.: Lipase from the thermotolerant fungus Rhizopus homothallicus is more thermostable when produced using solid state fermentation than liquid fermentation procedures. Enzyme Microb. Technol. 39, 1042–1050 (2006)
Lima, V.M.G., Krieger, N., Mitchell, D.A., Fontana, J.D.: Activity and stability of acrude lipase from Penicillium aurantiogriseum in aqueous media and organic solvents. Biochem. Eng. J. 18, 65–71 (2004)
Derewenda, Z.S., Derewenda, U., Dodson, G.G.: The crystal and molecular structure of the Rhizomucor miehei triacylglyceride lipase at 1.9 A resolution. J. Mol. Biol. 227, 818–839 (1992)
Lafuente, F.R.: Lipase from Thermomyces lanuginosus: uses and prospects as an industrial biocatalyst. J. Mol. Catal. B 63, 17–22 (2010)
Kwon, M.A., Kim, H.S., Yang, T.H., Song, B.K., Song, J.K.: High-level expression andcharacterization of Fusarium solani cutinase in Pichia pastoris. Protein Expr. Purif. 68, 104–109 (2009)
Maia, M.M.D., Heasley, A., Camargo de Morais, M.M., Melo, E.H., Morais Jr., M.A., Ledingham, W.M., Lima Filho, J.L.: Effect of culture conditions on lipaseproduction by Fusarium solani in batch fermentation. Bioresour. Technol. 76, 23–27 (2001)
Jacob, J.S., Miller, K.R.: The effects of galactolipid depletion on the structure of a photosynthetic membrane. J. Cell Biol. 103, 1337–1347 (1986)
De Maria, L., Vind, J., Oxenbøll, K.M., Svendsen, A., Patkar, S.: Phospholipases and their industrial applications. Appl. Microbiol. Biotechnol. 74, 290–300 (2007)
Czabany, T., Athenstaedt, K., Daum, G.: Synthesis, storage and degradation of neutral lipids in yeast. Biochim. Biophys. Acta 1771(3), 299–309 (2007)
Hassan, M., Blanc, P.J., Pareilleux, A., Goma, G.: Selection of fatty acid auxotrophs from the oleaginous yeast Cryptococcus curvatus and production of coca butter equivalents in batch culture. Biotechnol Lett. 16(8), 819–824 (1994)
Zhu, L.Y., Zong, M.H., Wu, H.: Efficient lipid production with Trichosporon fermentans and its use for biodiesel preparation. Bioresour. Technol. 99(16), 7881–7885 (2008)
Demaison, L., Moreau, D.: Dietary n-3 polyunsaturated fatty acids and coronary heart disease-related mortality: a possible mechanism of action. Cell. Mol. Life Sci. 59, 463–477 (2002)
White, T.J., Bruns, T., Lee, S., Taylor, J.: Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J. (eds.) PCR Protocols: A Guide to Methods and Application, pp. 315–322. Academic Press, San Diego (1990)
Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J.: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17), 3389–3402 (1997)
Tiss, A., Carrière, F., Verger, R.: Effects of gum arabic on lipase interfacial binding and activity. Anal. Biochem. 294, 6–43 (2001)
Folch Br, J., Lees, M., Stanley, G.H.S.: A simple method for the isolation and purification of total lipidesfrom animal tissues. J. Biol. Chem. 226, 497 (1957)
Metcalfe, L.D., Schmitz, A.A.: The rapid preparation of fatty acid esters for gas chromatographic analysis. Anal. Chem. 33(3), 363–364 (1961)
Kambourova, M., Kirilova, N., Mandeva, R., Derekova, A.: Purification and properties of thermostable lipase from a thermophilic Bacillus stearothermophilus MC7. J. Mol. Catal. B 22, 307–313 (2003)
Zaliha, R.N., Abd-Rahman, R., Leow, T.C., Salleh, A.B., Basri, M.: Geobacillus zalihae sp. nov., a thermophilic lipolytic bacterium isolated from palm oil mill effluent in Malaysia. BMC Microbiol. 7, 77 (2007)
Kumari, A., Mahapatra, P., Banerjee, R.: Statistical optimization of culture conditions by response surface methodology for synthesis of lipase with Enterobacter aerogenes. Braz. Arch. Biol. Technol. 52, 1349–1356 (2009)
Lopez, E., Deive, F.J., Longo, M.A., Sanroman, M.A.: Culture conditions and investigation of bioreactor configurations for lipase production by Rhizopus oryzae. Chem. Eng. Technol. 33, 1023–1028 (2010)
Lima, V.M.G., Krieger, N., Sarquis, M.I.M., Mitchell, D.A., Ramos, L.P., Fontana, J.D.: Effect of nitrogen and carbon sources on lipase production by Penicillium aurantiogriseum. Food Technol. Biotechnol. 41, 105–110 (2003)
Treichel, H., de Oliveira, D., Mazutti, M.A., Di Luccio, M., Oliveira, J.V.: A review on microbial lipases production. Food Bioprocess Technol. 3, 182–196 (2010)
Wang, D., Xu, Y., Shan, T.: Effects of oils and oil-related substrates on the synthetic activity of membrane-bound lipase from Rhizopus chinensis and optimization of the lipase fermentation media. Biochem. Eng. J. 41, 30–37 (2008)
Abbas, H., Abel, H., Deyris, V., Comeau, L.: Isolation and characterization of an extracellular lipase from Mucor sp strain isolated from palm fruit. Enzym. Microb. Technol. 31, 968–975 (2002)
Ben Romdhane, I.B., Fendri, A., Gargouri, Y., Gargouri, A., Belghith, H.: A novel thermoactive and alkaline lipase from Talaromyces thermophilus fungus for use in laundry detergents. Biochem. Eng. J. 53, 112–120 (2010)
Wu, X.Y., Jäskeläinen, S., Linko, W.Y.: Purification and partial characterization of Rhizomucor miehei lipase for ester synthesis. Appl. Biochem. Biotechnol. 59, 145–158 (1996)
Sugihara, A., Shimada, Y., Tominaga, Y.: Purification and characterization of Aspergillus niger lipase. Agric. Biol. Chem. 52, 1591–1592 (1988)
Chopra, A.K., Chander, H., Singh, J.: Lipolytic activity of Syncephalastrum racemosum. J. Dairy Sci. 65, 1890–1894 (1982)
Simons, J.-W.F.A., Hendrik, A., Ruud, C.C., Niek, D., Friedrich, G., Arend, J.S.: The lipase from Staphylococcus aureus. Eur. J. Biochem. 242, 760–769 (1996)
Gray, C.J.: Stabilization of enzymes with soluble additives. In: Gupta, M.N. (ed.) Thermostability of Enzymes in Stabilization of Enzymes with Soluble Additives, pp. 124–143. Springer, New Delhi (1993)
Aoki, J., Inoue, A., Makide, K., Saiki, N., Arai, H.: Structure and function of extracellular phospholipase A1 belonging to the pancreatic lipase gene family. Biochimie 89, 197–204 (2007)
Jallouli, R., Khrouf, F., Fendri, A., Mechichi, T., Gargouri, Y., Bezzine, S.: Purification and biochemical characterization of a novel alkaline (phospho)lipase from a newly isolated Fusarium solani strain. Appl. Biochem. Biotechnol. 168, 2330–2343 (2012). https://doi.org/10.1007/s12010-012-9940-0
Knothe, G.: Improving biodiesel fuel properties by modifying fatty ester composition. Energy Environ. Sci. 2, 759–766 (2009)
Acknowledgements
This work is a part of a doctoral thesis by Ameni KTATA. Whose research was supported financially by “Ministère de l’enseignement supérieur, de la recherche scientifique et de la technologie-Tunisia” through a grant to “Laboratoire de Biochimie et Génie Enzymatique des Lipases-ENIS”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ktata, A., Bezzine, S., Sayari, A. et al. Newly Isolated Lipolytic and Oleaginous Fungal Strain, Production, Optimization and Biochemical Characterization of the Extracellular (phospho)lipase. Waste Biomass Valor 11, 6677–6687 (2020). https://doi.org/10.1007/s12649-019-00907-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12649-019-00907-3