Abstract
l-serine is a naturally occurring dietary amino acid that has recently received renewed attention as a potential therapy for the treatment of amyotrophic lateral sclerosis (ALS), Alzheimer’s disease (AD), hereditary sensory autonomic neuropathy type I (HSAN1), and sleep induction and maintenance. We have previously reported l-serine functions as a competitive inhibitor of l-BMAA toxicity in cell cultures and have since progressed to examine the neuroprotective effects of l-serine independent of l-BMAA-induced neurotoxicity. For example, in a Phase I, FDA-approved human clinical trial of 20 ALS patients, our lab reported 30 g l-serine/day was safe, well-tolerated, and slowed the progression of the disease in a group of 5 patients. Despite increasing evidence for l-serine being useful in the clinic, little is known about the mechanism of action of the observed neuroprotection. We have previously reported, in SH-SY5Y cell cultures, that l-serine alone can dysregulate the unfolded protein response (UPR) and increase the translation of the chaperone protein disulfide isomerase (PDI), and these mechanisms may contribute to the clearance of mis- or unfolded proteins. Here, we further explore the pathways involved in protein clearance when l-serine is present in low and high concentrations in cell culture. We incubated SH-SY5Y cells in the presence and absence of l-serine and measured changes in the activity of proteolytic enzymes from the autophagic-lysosomal system, cathepsin B, cathepsin L, and arylsulfatase and specific activities of the proteasome, peptidylglutamyl-peptide hydrolyzing (PGPH) (also called caspase-like), chymotrypsin, and trypsin-like. Under our conditions, we report that l-serine selectively induced the activity of autophagic-lysosomal enzymes, cathepsins B and L, but not any of the proteasome-hydrolyzing activities. To enable comparison with previous work, we also incubated cells with l-BMAA and report no effect on the activity of the autophagic lysosomes or the proteasomes. We also developed an open-source script for the automation of linear regression calculations of kinetic data. Autophagy impairment or failure is characteristic of many neurodegenerative disease; thus, activation of autophagic-lysosomal proteolysis may contribute to the neuroprotective effect of l-serine, which has been reported in cell culture and human clinical trials.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
l-serine is a naturally occurring, conditional (Metcalf et al. 2018) amino acid that has recently become the focus of renewed research since it was proposed as a potential treatment for ALS (Levine et al. 2016) and AD (ClinicalTrials.gov identifier: NCT03062449). Recent studies have also shown that L-serine has potential to treat neuropathy in hereditary sensory autonomic neuropathy type I (HSAN1) (Fridman et al. 2019; Garofalo et al. 2011), improve sleep induction and maintenance (Ito et al. 2014), and may be useful as a supplement in type I (Holm et al. 2018) and type II diabetes (Holm and Buschard 2019). A Phase I FDA-approved clinical trial for ALS has already been completed (Levine et al. 2016), and two more are currently underway for the use of l-serine in early-stage Alzheimer’s disease (AD) (ClinicalTrials.gov Identifier: NCT03062449) and in a larger group of patients with ALS (ClinicalTrials.gov Identifier: NCT03580616). In a randomized, placebo-controlled (dextrose hydrose powder), parallel-group trial with open-label extension, l-serine 400 mg/kg/day was shown to be effective at reducing a form of hereditary neuropathy in patients with HSAN1 over a 12-month period (Fridman et al. 2019). This was a similar dose to the ALS Phase I trial of 30 g/day (Levine et al. 2016), but the dosing for each patient in the latter trial only lasted for 3 months. l-Serine was shown to improve sleep in a randomized, placebo-controlled (trehalose), double-blind, crossover trial with 53 subjects who consumed 3 g of l-serine 30 min prior to sleep and reported it both induced and maintained sleep better than placebo, and the effect persisted 1 month after the participants ceased taking l-serine (Ito et al. 2014). A recent review article, citing data showing that patients with type I and II diabetes have reduced serum concentrations of l-serine, suggested that l-serine supplementation might be beneficial in type I and II diabetes (Holm and Buschard 2019).
Owing to our preliminary positive clinical findings for l-serine in ALS (Levine et al. 2016), we hypothesized l-serine might be neuroprotective; however, the mechanisms by which it imparts this protection have not been completely elucidated. We have previously reported that l-serine is a competitive inhibitor of l-BMAA—a non-protein amino acid implicated in neurodegenerative disease (Dunlop and Guillemin 2019)—and that coadministration of l-serine to cell cultures incubated with l-BMAA prevents incorporation, apoptosis, and autofluorescence (Dunlop et al. 2013). We are now interested in elucidating the mechanisms of the neuroprotective effects of l-serine independent of l-BMAA-mediated neurotoxicity. For example, we have previously shown in vitro that l-serine alone upregulates genes involved in endoplasmic reticulum (ER) protein homeostasis (proteostasis) and the unfolded protein response (UPR) (Dunlop et al. 2018a), and increases translation of the ER chaperone protein disulfide isomerase (PDI) (Dunlop et al. 2018b), which has been implicated in ALS (Perri et al. 2016). A recent study in HT22 mouse hippocampal neuronal cells showed that l-serine blocks the formation of reactive oxygen species generated by treatment with 2,3 dimethoxy-1,4-naphthoquinone (DMNQ), via a mechanism that contributes to the synthesis of glutathione, thus preserving mitochondrial homeostasis and imparting neuroprotection (Kim et al. 2019).
Our observations that l-serine reduces autofluorescence in cells treated with the non-protein amino acid, l-BMAA (Dunlop et al. 2013), could be a result of l-serine reducing the formation of misfolded/damaged proteins and/or increasing the metabolism of these substrates once they form. The two main mechanisms for protein degradation in eukaryotic cells are the autophagic-lysosome system and the ubiquitin-proteasome system. The 26S proteasome consists of multiple subunits with 3 active sites; the peptidylglutamyl-peptide hydrolyzing (PGPH) (caspase-like), the tryptic, and the chymotryptic sites contained inside the 20S catalytic core, which is capped by the 19S regulatory subunit. The 19s subunit recognizes polyubiquitin tags and linearizes and shuttles proteins into the 20S core for degradation (Tanaka 2009). Substrates that are too large, cross-linked, or otherwise unable to fit in the narrow core of the 20S subunit are shuttled for autophagic-lysosomal degradation (for a review of the ubiquitin-proteasome system, see (Schneider and Cuervo 2014)).
Lysosomes are large acidic (pH ~ 5) organelles that process macromolecules (such as mitochondria), cytosol fractions, plasma membrane components, aggregated proteins, lipids, and other long half-life proteins, primarily via autophagy (there are three types of autophagy: macro-, micro-, and chaperone-mediated). Autophagy plays a key role in the degradation of damaged or misfolded proteins, including some implicated in neurodegeneration. Impaired chaperone-mediated autophagy (CMA) has been implicated in the accumulation of aggregated tau in AD (Wang et al. 2009), and α-synuclein in Parkinson’s disease (PD) (Cuervo et al. 2004). Like the proteasome, CMA also requires linearization of substrates; thus, cross-linked/aggregated proteins will be processed by macroautophagy.
To determine if l-serine has any impact on the proteolytic activities of the autophagy-lysosomal system or the proteasomes, we incubated human neuroblastoma (SH-SY5Y) cells with low (100 μM) and high (1000 μM) concentrations of l-serine or l-BMAA and measured the activity of cathepsins B and L and the hydrolytic activities of the proteasome (tryptic, chymotryptic, and PGPH). We report that l-serine selectively induced the activity of cathepsins B and L, but not the proteasome. l-BMAA had no significant impact on the proteolytic activity, either in the lysosomes or in the proteasome. We included d,l-m-tyrosine as a negative control for proteasome activity, since we have previously shown it does not increase proteasome-hydrolyzing activity (Dunlop et al. 2008). The selective increase in autophagic-lysosomal proteolytic activity by l-serine may constitute a mechanism that contributes to its observed neuroprotection.
Materials
SH-SY5Y human neuroblastoma was from ATCC (ATCC® CRL-2266™) and was used at between population doubling (PD) 22 and 25. Dulbecco’s Modified Eagle’s Medium/Nutrient F-12 Ham (Cat. No. D8437), Minimum Essential Medium Eagle deficient in l-serine (Cat. No. M2279), l-serine ReagentPlus® ≥ 99% (TLC) (Cat. No. S4500), d,l-m-tyrosine crystalline (Cat. No. T3629), and adenosine 5′-triphosphate (ATP) disodium salt hydrate, grade I, ≥ 99%, from microbial (Cat. No. A2383), were purchased from Sigma-Aldrich. l-BMAA was synthesized by Irvine Chemistry Lab (Anaheim, CA, USA) and the structure and purity confirmed using nuclear magnetic resonance (NMR) spectroscopy, as described previously (Cox et al. 2016). Cathepsin L substrate (Z-Phe-Arg-AMC) was from AnaSpec Inc. (Cat. No. AS-24096) and Cathepsin B substrate (Z-Arg-Arg-AMC hydrochloride salt) was from Chem-Impex (Cat. No. 32723). 4-Nitrocatechol sulfate dipotassium salt (p-NCS) was from United States Biological (Cat. No. 433242). Hydroquinone (S4580) was from Selleck Chemicals. Nicotinamide adenine dinucleotide (NADH) disodium salt (Cat. No. 2735–1000) was from BioVision. Sodium pyruvate (Cat. No. 11 465 007 001) was from Roche. Proteasome substrates, Boc-Leu-Ser-Thr-Arg-Amido-4-Methylcoumarin (Boc-LSTR-AMC, “trypsin-like”), Ac-Nle-Pro-Nle-Asp-7-Amido-4-Methylcoumarin (Ac-nLPnLD-AMC) (PGPH or “caspase-like”), and digitonin (Cat. No. 11024-24-1) were purchased from Cayman Chemical Company, and N-Succinyl-Leu-Leu-Val-Tyr-7-Amido-4-Methylcoumarin (Suc-LLVY-AMC, “chymotrypsin-like”) was purchased from BioVision. Epoxomicin (Cat. No. A2606) was from ApexBio. All other chemicals were of reagent or tissue culture grade unless otherwise indicated.
Methods
Cell culture: SH-SY5Y human neuroblastoma (ATCC® CRL-2266™, PD18-20) were recovered from cryogenic storage into T-75 flasks in Dulbecco’s Modified Eagle’s Medium/Nutrient F-12 Ham (Sigma-Aldrich, Cat. No. D8437) containing 10% fetal bovine serum (FBS), 4 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. At between 70 and 80% confluence, the cells were trypsinized with warm (37 °C) 0.5% Trypsin-EDTA (Thermo Fisher, Cat. No. 15400054) and passaged into T-175 flasks, where they were left to multiply until they reached 80–90% confluence.
When cells were between PD22 and 25, they were plated into 6-well plates (1 × 106 cells/well) for cathepsin and arylsulfatase assays, 12-well plates for lactate dehydrogenase (LDH) assays (5 × 105 cells/well), and 96-well plates (2.4 × 104 cells/well) for MTT assays, and allowed to adhere overnight. The following day, the cell culture medium was changed to Minimum Essential Medium Eagle deficient in l-serine (Sigma-Aldrich Cat. No. M2279), and then supplemented with the following: untreated controls (UTC), l-serine 100 μM, l-serine 1000 μM, l-BMAA 100 μM, l-BMAA 1000 μM, or d,l-m-tyrosine 500 μM, for 24 or 48 h. The medium was either changed (for MTT assays) or supplemented (for cathepsins and LDH) at 24 h; then, incubations continued for 48 h. Sterile concentrated stocks of l-serine, l-BMAA, and d,l-m-tyrosine were dissolved in 20 mM HCl and stored at − 20 °C.
LDH Assay
Lactate dehydrogenase (LDH) is a cytosolic enzyme released by cells into the culture medium when the plasma membrane becomes compromised. A ratio of its activity in the medium versus in the cytosol is used to determine cell viability. LDH protocol for SH-SY5Y has been described previously (Dunlop et al. 2018b) with the following modifications. Following treatment, cultures were removed from the incubator and placed on ice. The media was aliquoted into Eppendorf tubes, and the tubes spun at 2500×g, 5 min, 4 °C. The supernatant was aliquoted into a fresh tube and designated M (medium). The remaining pellet was washed 1x in ice-cold PBS by resuspending the pellet, then centrifuging at 2500×g, 5 min, 4 °C. The cell monolayer was washed 1x in ice-cold PBS, then 500 μL of 0.1% Triton X-100 added to each well to lyse, and the plate placed on ice on a plate shaker for 5 min. The resulting lysate was pooled with the pellet generated above. The combined cell lysates were designated CL (cell lysate).
Lysates were diluted 1/10 into 0.1% Triton X-100 then 10 μL of each sample added to a 96-well plate. 100 μL LDH buffer (0.15 mg/mL NADH, 2.5 mM sodium pyruvate in PBS) was added to each well, then absorbance (340 nm) read on the Molecular Devices FlexStation 3 and acquired in the SoftMax® Pro Microplate Data Acquisition and Analysis Software (version 7.0.2) every 5 min, for 30 min. Viability was calculated from the change in absorbance in CL divided by the combined absorbance change of the M and CL fractions; % viability = (λ340 nm CL/λ340 nm CL + λ340 nm M) × 100. All incubations were conducted in triplicate wells (technical replicates) and in triplicate, independent incubations (n = 3). Data were converted to a percentage of control in Microsoft Excel for Mac (version 16.28) and graphed in GraphPad Prism for MacOS (version 8.2.1).
MTT Assay
Cells that are metabolically active cleave MTT (which appears as a yellow tetrazolium salt) to purple formazan crystals, and the change in absorbance is a measure of cell viability. Cells were plated at 2.4 × 104 cells/well in 96-well plates then allowed to adhere overnight. The following day, cells were washed 3x with warm PBS, then 100 μL treatment medium (UTC, l-serine 100 μM, l-serine 1000 μM, l-BMAA 100 μM, or l-BMAA 1000 μM) added. The medium was changed every 24 h. MTT activity was measured using the Roche Cell Proliferation Kit I (MTT) (Cat. No. 11465007001) according to the manufacturer’s instructions. Briefly, after the incubation period (24 or 48 h), 10 μL of the MTT labeling reagent was added to each well to a final concentration of 0.5 mg/mL, and the plates returned to the incubator for a further 4 h. One hundred microliter of solubilization solution was then added, and the cells were returned to the incubator overnight. The next morning, the absorbance was read at 550 nm on a Molecular Biosciences FlexStation 3 and acquired in SoftMax® Pro Microplate Data Acquisition and Analysis Software (version 7.0.2). All incubations were conducted in triplicate wells (technical replicates) and in triplicate, independent incubations (n = 3). Data was converted to a percentage of control in Microsoft Excel for Mac (version 16.28) and graphed in GraphPad Prism for MacOS (version 8.2.1).
Cathepsin Assays
Following incubation (24 h or 48 h), cells were placed on ice and scraped using a rubber policeman, then the slurry aliquoted into Eppendorf tubes. Tubes were spun at 2500×g, 5 min, 4 °C, and the supernatant aspirated. Cell pellets were washed 3x in ice-cold PBS, by gently resuspending the pellet, then centrifuged as described above and aspirating the supernatant. Pellets were placed at − 20 °C until required for assays.
Protein Assays
Proteasome assays
Protein concentrations were measured using the Quant-iT Protein Assay Kit adapted for microplates (Thermo Fisher, Cat. No. Q33210) and calculated from a standard curve (400 ng/μL to 0 ng/µL).
Arylsulfatase and cathepsin assays
Owing to the presence of interfering concentrations of Triton X-100 in the cell lysate, protein concentrations were measured using a Bicinchoninic Acid (BCA) Kit for Protein Determination (Sigma-Aldrich, Cat. No. B9643). Concentrations of samples were determined using a standard curve, where the standards were bovine serum albumin (BSA) 2 to 0.0625 mg/mL.
Measurement of Arylsulfatase Activity
Arylsulfatase activity was determined by measuring the enzymatic hydrolysis of the ester sulfate bond of p-NCS based on the methods of Orange and Moore (Orange and Moore 1976) and as described previously (Dunlop et al. 2008). Absorbance was measured at 540 nm on a Molecular Devices FlexStation 3 and acquired in SoftMax® Pro Microplate Data Acquisition and Analysis Software (version 7.0.2). Results were normalized to protein concentration and expressed as a percentage of control.
Cathepsin Activity Assays
Measurement of cathepsins B and L activities has been described previously (Dunlop et al. 2008). Briefly, cell pellets were removed from − 20 °C, 500 μL 0.1% Triton X-100 added, and vortexed to mix. Pellets were lysed by freeze/thawing 3 times (− 80 °C, and 37 °C water bath) with vortexing in between. Tubes were spun at 13,000×g for 10 min at 4 °C to remove cell debris, then supernatant transferred to a fresh tube. Cathepsin activity was measured by the change in fluorescence generated by cleavage of AMC from the peptide substrate Z-Arg-Arg-AMC (cathepsin B) or Z-Phe-Arg-AMC (cathepsin L). Changes in fluorescence (ex 360 nm, em 460 nm) were measured every 2 min for 30 min, in triplicate, on a Molecular Devices Flexstation 3, and acquired in SoftMax® Pro Microplate Data Acquisition and Analysis Software (version 7.0.2). Data was exported as .CSV and analyzed using Ruby (see “Data Analysis” section) and activity quantitated as change in fluorescence, per minute, per milligram protein and expressed as percentage of control. Plots were graphed in GraphPad Prism, version 8.2.1.
Proteasome Activity Assays
Following treatment of cells, plates were removed from the incubator and placed on ice. Cells were scraped using a rubber policeman and aliquoted into Eppendorf tubes. Tubes were spun in a benchtop centrifuge at 2000 x g, 5 mins, 4 °C, then the supernatant aspirated. The resulting pellet was washed by gentle resuspension (3 times in ice-cold PBS then centrifugation as above) and stored at −80 °C until required for proteasome activity assays. The proteasome was extracted by permeabilization of the cell membrane using digitonin and “squeezing out” with centrifugation, as described previously (Dunlop et al. 2008) and based on the method of Kisselev (Kisselev and Goldberg 2005). Proteasome chymotryptic, tryptic and PGPH (caspase-like) activities were measured by the initial linear rates of cleavage of the fluorescently labeled peptide substrates N-Suc-Leu-Leu-Val-Tyr-AMC, Boc-Leu-Ser-Thr-Arg-AMC and Ac-Nle-Pro-Nle-Asp-AMC respectively, as described previously (Dunlop et al. 2008) with the following modifications –– samples were diluted to equal protein concentrations in homogenization buffer (50 mM Tris/HCl pH 7.5, 250 mM sucrose, 5 mM MgCl2, 2 mM ATP, 1 mM DTT, 0.5 mM EDTA, 0.025% digitonin) then aliquoted into tubes where one sample contained 20 μM epoxomicin, and incubated at 37 °C for 30 mins. Following incubation, 140 μL reaction buffer (50 mM Tris/HCl pH 7.5, 40 mM KCl, 5 mM MgCl2, 0.5 mM ATP, 1 mM DTT and 0.2 mg/mL BSA) was added to each well of a black/clear bottom 96-well plates in triplicate, then 20 μL lysate of each sample, (plus or minus epoxomicin) was added. The reaction was initiated by the addition of 50 μL of 4 x concentrated substrate in reaction buffer. Fluorescence (ex 360 nm, em 460 nm) was read every 2 mins for 30 mins at 37 °C on a Molecular Devices FlexStation 3, using SoftMax® Pro Microplate Data Acquisition and Analysis Software (version 7.0.2). Data was exported as .CSV and analyzed using Ruby (see “Data Analysis” section) and activity quantitated as change in fluorescence, per minute, per milligram protein, and expressed as a percentage of control. Plots were graphed in GraphPad Prism, version 8.2.1.
Data Analysis
We developed an open-source script in Ruby (version 2.5.4) for the automation of linear regression calculations using kinetic data acquired with SoftMax® Pro Microplate Data Acquisition and Analysis Software (version 7.0.2) on the Molecular Devices FlexStation 3. Data was output from SoftMax® Pro to a .CSV file then, a program written in Ruby imported the data, computed the mean R-squared, slope, and intercept, and output the results in .CSV form. From these results, mean slope values were used to calculate the change in fluorescence per minute, per milligram of protein for each sample. R-squared was monitored to ensure we were collecting data from the linear part of the curve. Results were expressed as a percentage of control. The Ruby code is available on GitHub at https://github.com/johncarney/flex-station-data.
Statistical Analysis
Data are representative of at least three independent experiments and expressed as mean + standard error of the mean (SEM) or standard deviation (SD). Statistics were conducted in GraphPad Prism for MacOS (version 8.2.1) using 1-way ANOVA with Kruskal-Wallis post-hoc analysis, where p < 0.05.
Results
Data Analysis
To reduce processing time and to minimize the risk of human error, a program to integrate kinetic data was written in Ruby (version 2.5.4). We compared results obtained from conducting linear regression manually in Microsoft Excel with the results from the Ruby program and report both methods returned exactly the same numbers (Table 1). As a result, all linear regression of kinetic data to obtain slope (m) and R-squared required for the calculation of the change in fluorescence, per minute, per milligram protein, was conducted using Ruby.
Assays for Cell Viability
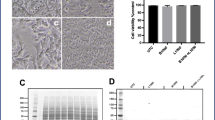
To ensure we were working at sub-toxic concentrations of both l-serine and l-BMAA, we conducted proliferation and viability assays using MTT and LDH assays, respectively. MTT is a yellow tetrazolium salt that is reduced to purple formazan crystals by metabolically active cells, which can be used to quantitate viability and proliferation. LDH is a cytosolic enzyme that leaks across the plasma membrane when the cells are damaged. LDH activity can be measured by the change in absorbance when NADH is oxidized to NAD+ by LDH (see “Materials” section), and the ratio of activity in the media versus in the cells constitutes cell viability. We report no significant difference in MTT or LDH activity across any time points (Fig. 1a, b, respectively), indicating there was no change in cell viability. Thus, we were working in sub-toxic concentrations of l-serine and l-BMAA.
Cell death was not induced by l-serine or l-BMAA. In order to determine if we were working at sub-toxic concentrations of l-serine and l-BMAA, cell viability and proliferation assays were conducted using MTT (a) and LDH (b). We report no significant differences between any conditions. Results are mean + SEM of at least 3 independent incubations. Statistics were conducted in GraphPad Prism for MacOS (version 8.2.1) using 1-way ANOVA with Kruskal-Wallis post hoc analysis, where p < 0.05
Activity Studies
Since we were interested in the impact of l-serine on proteolytic activity, we measured changes to key proteolytic enzymes from the autophagic-lysosomes and the proteasomes. In the lysosomes, we measured the activity of cathepsins B and L, and for the 26S proteasome, the tryptic, chymotryptic, and PGPH (caspase)-hydrolyzing activities. We report selective upregulation of cathepsin L in cells treated with l-serine 100 and 1000 μM at 24 h, but not at 48 h (Fig. 2a, b). The activity of cathepsin B was also increased at 24 h in cultures treated with l-serine 100 μM or 1000 μM, and this increase persisted at 48 h only in cells that had been treated with 1000 μM l-serine (Fig. 2c, d).
l-serine but not l-BMAA induced the activity of lysosomal cathepsins. Activities of cathepsin L (a, b) and cathepsin B (c, d) were measured in SH-SY5Y cells incubated with l-serine or l-BMAA for 24 h (a, c) or 48 h (b, d). l-serine, but not l-BMAA, selectively induced the activity of cathepsins L and B at 24 h. This effect persisted with cathepsin B at 48 h. Results are mean + SEM of at least 3 independent incubations. Linear regression was calculated using a custom program written in Ruby (version 2.5.4). Activity was expressed as change in fluorescence, per minute, per milligram protein and expressed as percentage of control. Statistics were conducted in GraphPad Prism for MacOS (version 8.2.1) using 1-way ANOVA with Kruskal-Wallis post hoc analysis, where p < 0.05
Evidence that these effects were selective and not just a result of an increase in the size or number of lysosomes is provided by the lack of change to the activity of arylsulfatase—a lysosomal enzyme not involved in proteolysis (Fig. 3).
Induction of cathepsins was selective as indicated by no change to arylsulfatase activity. There was no change in the activity of the non-proteolytic lysosomal enzyme, arylsulfatase, in response to any conditions, suggesting that any observed changes in proteolytic enzymes were selective and substrate dependent. a t = 24 h. b t = 48 h. Results are mean + SEM of at least 3 independent incubations. Statistics were conducted in GraphPad Prism for MacOS (version 8.2.1) using 1-way ANOVA with Kruskal-Wallis post hoc analysis, where p < 0.05
l-BMAA had no effect on the activity of autophagy-lysosomal enzymes under any of the conditions examined (Figs. 2 and 3).
We report no changes to the activity of any of the three specific activities of the proteasome under any of the conditions examined (Table 2) for either l-serine, l-BMAA, or the negative control, d,l-m-tyrosine. Evidence that we measured proteolysis specific to the proteasome was provided by simultaneous incubations that contained epoxomicin—an irreversible and specific proteasome inhibitor of the chymotryptic-hydrolyzing activity (epoxomicin also inhibits the trypsin and PGPH/caspase-hydrolyzing activities but at 100- and 1000-fold slower rates, respectively (Meng et al. 1999)). We report a mean slope of 21.308 for samples absent of 20 μM epoxomicin versus 0.728 for samples incubated in the presence of epoxomicin (Fig. 4), indicating an almost complete inhibition of the chymotryptic-hydrolyzing activity by epoxomicin.
Cleavage of Suc-LLVY-AMC was highly specific to the proteasome. Identical crude lysate samples were incubated in the absence (a) or presence (b) of 20 μM epoxomicin, a highly specific and irreversible inhibitor of the proteasome and chymotryptic activity measured using Suc-LLVY-AMC. Epoxomicin almost completely inhibited (mean slope 21.308 versus 0.728 for epoxomicin) cleavage of the substrate, indicating that virtually all proteolytic activity could be attributed to the proteasome. Seventy microliters of 0.191 mg/mL protein lysate was incubated at 37 °C for 30 min in the presence or absence of 20 μM epoxomicin, then 20 μL assayed in triplicate for cleavage of the chymotryptic substrate, Suc-LLVY-AMC. The mean slope of the uninhibited lysate was 21.308 versus 0.728 for the epoxomicin control. RFU = relative fluorescence units
Discussion
The two main pathways involved in the clearance of damaged or misfolded proteins in eukaryotic cells are the autophagic-lysosomal system and the ubiquitin proteasomes. Age-related increases in protein deposition are the result of an imbalance between the rate of formation of damaged or aberrant proteins and a decrease in the ability of the proteolytic machinery to remove these proteins. Thus, the ability of a cell to maintain proteostasis declines with age, and this is exacerbated in neurodegeneration, particularly in postmitotic cells such as neurons that lack the ability to dilute proteolytic-resistant substrates by mitosis.
It is well-established that autophagy is more efficient in young, healthy neurons, and this may in part be because aging contributes to a decline in the abundance of autophagy-related genes, such as autophagy-related protein 5 (ATG5), autophagy-related protein 7 (ATG7), and beclin-1 (Lipinski et al. 2010). Chaperone-mediate autophagy (CMA) is responsible for the degradation of many disease-related or pathogenic proteins, but it declines with age, as well as being impaired in many neurodegenerative diseases. Abnormal aggregation of α-synuclein is a hallmark of PD, and although it is normally degraded via CMA, mutations in the α-synuclein protein impair efficient passage into the lysosomes thereby contributing to accumulation (Alfaro et al. 2018). Lysosome-associated membrane protein 2 (LAMP2A) is the rate-limiting enzyme for CMA, and although the rate of transcription of LAMP2A remains unchanged with aging, the dynamics and stability of the lysosomal receptor are altered (Kiffin et al. 2007). A decline in the activity of chaperones and cofactors required for efficient autophagy might also contribute to accumulating damage. For example, in the substantia nigra pars compacta (SNpc) and amygdala of PD brains, heat shock cognate protein 71 kDa (Hsc70) and LAMP2A protein levels are reduced, and this corresponds to deposits of Lewy bodies that contain autophagy-related proteins (Alvarez-Erviti et al. 2010). A well-established neuropathologic feature of AD is the accumulation of autophagic vesicles such as lysosomes, specifically within damaged neuritic processes and synaptic terminals (Nixon et al. 2005). This phenotype is also observed in animal and cell-based models of AD when exposed to β-amyloid peptide (Lipinski et al. 2010). Additionally, in PD, Huntington’s disease (HD), and ALS, inclusion bodies contain abnormal amounts of ubiquitin, suggesting a link between dysfunction of CMA, the proteasome, and neurodegeneration (Alves-Rodrigues et al. 1998).
On the other hand, modulation of autophagy, either genetically, pharmacologically (for example, using rapamycin, metformin, resveratrol, or spermidine), or via caloric restriction, appears to extend longevity in animal models and can protect against neurodegeneration (Kaushik and Cuervo 2015). For example, in flies and mouse models of AD, activation of autophagy by rapamycin treatment increased clearance of tau and β-amyloid aggregates as well as rescued memory deficits in the mice (Berger et al. 2006; Caccamo et al. 2014). Likewise, in a human neuroblastoma cell line (differentiated SH-SY5Y) incubated with preformed fibrils to model α-synuclein accumulation, activation of autophagy by rapamycin treatment for 8 days increased clearance of α-synuclein aggregates (Gao et al. 2019). Also in SH-SY5Y over-expressing wild-type and mutant (A30P and A53T) α-synuclein, rapamycin increased clearance by stimulating autophagy (Webb et al. 2003). Expression of the autophagy-inducing protein beclin-1 is reduced in the brains of AD patients, but in amyloid precursor protein (APP) transgenic mice, increasing the expression of beclin-1 reduced amyloid pathology (Pickford et al. 2008). In fly models of HD, pharmacological activation of autophagy using rapamycin and a derivative of rapamycin (CCl-779) was shown to improve motor neuron functioning and, in a mouse model of HD, decrease aggregate formation (Berger et al. 2006; Ravikumar et al. 2004).
Whilst our initial hypothesis for one mechanism of l-serine-mediated neuroprotection was out-competing l-BMAA for incorporation into proteins (Dunlop et al. 2013), subsequent studies from our lab and others have revealed that l-serine has a neuroprotective effect independent of l-BMAA toxicity. For example, in a Phase I trial of l-serine for ALS patients, oral admission of l-serine 30 g/day reduced the progression of the disease by 85% (Levine et al. 2016), and this was independent of l-BMAA neurotoxicity. In human neuroblastoma (SH-SY5Y) cell cultures, we reported that l-serine alone differentially regulated proteins and genes in the UPR and ER stress pathways, again independent of l-BMAA (Dunlop et al. 2018a). In the same model system, we reported that l-serine selectively increased PDI protein, leading us to propose l-serine as a candidate for a small molecule proteostasis regulator (Dunlop et al. 2018b). Hence, l-serine-mediated neuroprotection occurs independent of l-BMAA-mediated neurotoxicity and comprises multiple mechanisms.
The selective activation of lysosomal cathepsins by l-serine reported here is consistent with our previous findings that l-serine upregulates protective mechanisms in the cell.
Since autophagy is impaired in neurodegenerative disease (Kaushik and Cuervo 2015), and therapies that modulate autophagy have the potential to reduce aggregate-mediated toxicity (Lipinski et al. 2010), our observations that l-serine selectively activates lysosomal cathepsins might position it as a useful pharmacological intervention for the regulation of autophagy.
In conclusion, we report that l-serine selectively activates proteolytic enzymes from the autophagy-lysosome pathway but not the ubiquitin-proteasome system. Investigations into l-serine’s observed neuroprotective effects continue to unearth fascinating mechanisms that contribute to the accumulating body of evidence as l-serine as a potentially useful neuroprotective molecule.
Abbreviations
- AD:
-
Alzheimer’s disease
- ALS:
-
Amyotrophic lateral sclerosis
- ATP:
-
Adenosine 5′-triphosphate
- l-BMAA:
-
Beta-methylamino-l-alanine
- BSA:
-
Bovine serum albumin
- ER:
-
Endoplasmic reticulum
- HSAN1:
-
Hereditary sensory autonomic neuropathy type I
- FBS:
-
Fetal bovine serum
- HD:
-
Huntington’s disease
- LDH:
-
Lactate dehydrogenase
- p-NCS:
-
4-Nitrocatechol sulfate dipotassium salt
- PDI:
-
Protein disulfide isomerase
- PGPH:
-
Peptidylglutamyl-peptide hydrolyzing
- UPR:
-
Unfolded protein response
References
Alfaro IE, Albornoz A, Molina A, Moreno J, Cordero K, Criollo A, Budini M (2018) Chaperone mediated autophagy in the crosstalk of neurodegenerative diseases and metabolic disorders. Front Endocrinol (Lausanne) 9:778. https://doi.org/10.3389/fendo.2018.00778
Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper JM, Caballero C, Ferrer I, Obeso JA, Schapira AHV (2010) Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol 67:1464–1472. https://doi.org/10.1001/archneurol.2010.198
Alves-Rodrigues A, Gregori L, Figueiredo-Pereira ME (1998) Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends Neurosci 21:516–520. https://doi.org/10.1016/S0166-2236(98)01276-4
Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ, Rubinsztein DC (2006) Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet 15:433–442. https://doi.org/10.1093/hmg/ddi458
Caccamo A, De Pinto V, Messina A, Branca C, Oddo S (2014) Genetic reduction of mammalian target of rapamycin ameliorates Alzheimer’s disease-like cognitive and pathological deficits by restoring hippocampal gene expression signature. J Neurosci 34:7988–7998. https://doi.org/10.1523/JNEUROSCI.0777-14.2014
Cox PA, Davis DA, Mash DC, Metcalf JS, Banack SA (2016) Dietary exposure to an environmental toxin triggers neurofibrillary tangles and amyloid deposits in the brain. Proc Biol Sci 283:20152397. https://doi.org/10.1098/rspb.2015.2397
Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D (2004) Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science 305:1292–1295
Dunlop RA, Guillemin GJ (2019) The cyanotoxin and non-protein amino acid β-methylamino-l-alanine (l-BMAA) in the food chain: incorporation into proteins and its impact on human health. Neurotox Res 36:601–611. https://doi.org/10.1007/s12640-019-00089-9
Dunlop RA, Dean RT, Rodgers KJ (2008) The impact of specific oxidized amino acids on protein turnover in J774 cells. Biochem J 410:131–140. https://doi.org/10.1042/BJ20070161
Dunlop RA, Cox PA, Banack SA, Rodgers KJ (2013) The non-protein amino acid BMAA is incorporated into human proteins in place of l-serine causing protein misfolding and aggregation. PLoS One 8:e75376. https://doi.org/10.1371/journal.pone.0075376
Dunlop RA, Powell J, Guillemin GJ, Cox PA (2018a) Mechanisms of l-serine neuroprotection in vitro include ER proteostasis regulation. Neurotox Res 33:123–132. https://doi.org/10.1007/s12640-017-9829-3
Dunlop RA, Powell JT, Metcalf JS, Guillemin GJ, Cox PA (2018b) l-serine-mediated neuroprotection includes the upregulation of the ER stress chaperone protein disulfide isomerase (PDI). Neurotox Res 33:113–122. https://doi.org/10.1007/s12640-017-9817-7
Fridman V, Suriyanarayanan S, Novak P, David W, Macklin EA, McKenna-Yasek D, Walsh K, Aziz-Bose R, Oaklander AL, Brown R, Hornemann T, Eichler F (2019) Randomized trial of l-serine in patients with hereditary sensory and autonomic neuropathy type 1. Neurology 92:e359–e370. https://doi.org/10.1212/WNL.0000000000006811
Gao J, Perera G, Bhadbhade M, Halliday GM, Dzamko N (2019) Autophagy activation promotes clearance of α-synuclein inclusions in fibril-seeded human neural cells. J Biol Chem 294:14241–14256. https://doi.org/10.1074/jbc.ra119.008733
Garofalo K, Penno A, Schmidt BP, Lee H-J, Frosch MP, von Eckardstein A, Brown RH, Hornemann T, Eichler FS (2011) Oral l-serine supplementation reduces production of neurotoxic deoxysphingolipids in mice and humans with hereditary sensory autonomic neuropathy type 1. J Clin Invest 121:4735–4745. https://doi.org/10.1172/JCI57549
Holm LJ, Buschard K (2019) L-serine: a neglected amino acid with a potential therapeutic role in diabetes. APMIS 127:655–659. https://doi.org/10.1111/apm.12987
Holm LJ, Haupt-Jorgensen M, Larsen J, Giacobini JD, Bilgin M, Buschard K (2018) L-serine supplementation lowers diabetes incidence and improves blood glucose homeostasis in NOD mice. PLoS One 13:e0194414. https://doi.org/10.1371/journal.pone.0194414
Ito Y, Takahashi S, Shen M, Yamaguchi K, Satoh M (2014) Effects of L-serine on human sleep. Springerplus 3:456. https://doi.org/10.1159/000119485
Kaushik S, Cuervo AM (2015) Proteostasis and aging. Nat Med 21:1406–1415. https://doi.org/10.1038/nm.4001
Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey AC, Martinez-Vicente M, Cuervo AM (2007) Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci 120:782–791. https://doi.org/10.1242/jcs.001073
Kim KY, Hwang S-K, Park SY, Kim MJ, Jun DY, Kim YH (2019) l-serine protects mouse hippocampal neuronal HT22 cells against oxidative stress-mediated mitochondrial damage and apoptotic cell death. Free Radic Biol Med 141:447–460. https://doi.org/10.1016/j.freeradbiomed.2019.07.018
Kisselev AF, Goldberg AL (2005) Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol 398:364–378
Levine TD, Miller RG, Bradley WG, Moore DH, Saperstein DS, Flynn LE, Katz JS, Forshew DA, Metcalf JS, Banack SA, Cox PA (2016) Phase I clinical trial of safety of L-serine for ALS patients. Amyotroph Lateral Scler Front Degener:1–5. https://doi.org/10.1080/21678421.2016.1221971
Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J (2010) Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A 107:14164–14169. https://doi.org/10.1073/pnas.1009485107
Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM (1999) Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci U S A 96:10403–10408. https://doi.org/10.1073/pnas.96.18.10403
Metcalf JS, Dunlop RA, Powell JT, Banack SA, Cox PA (2018) L-serine: a naturally-occurring amino acid with therapeutic potential. Neurotox Res 33:213–221. https://doi.org/10.1007/s12640-017-9814-x
Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM (2005) Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol 64:113–122. https://doi.org/10.1093/jnen/64.2.113
Orange RP, Moore EG (1976) Functional characterization of rat mast cell arylsulfatase activity. J Immunol 117:2191–2196
Perri ER, Thomas CJ, Parakh S, Spencer DM, Atkin JD (2016) The unfolded protein response and the role of protein disulfide isomerase in neurodegeneration. Front Cell Dev Biol 3:80. https://doi.org/10.3389/fcell.2015.00080
Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T (2008) The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid β accumulation in mice. J Clin Invest 118:2190–2199. https://doi.org/10.1172/JCI33585
Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC (2004) Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 36:585–595. https://doi.org/10.1038/ng1362
Schneider JL, Cuervo AM (2014) Autophagy and human disease: emerging themes. Curr Opin Genet Dev 26:16–23. https://doi.org/10.1016/j.gde.2014.04.003
Tanaka K (2009) The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci 85:12–36. https://doi.org/10.2183/pjab.85.12
Wang Y, Martinez-Vicente M, Krüger U, Kaushik S, Wong E, Mandelkow EM, Cuervo AM, Mandelkow E (2009) Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet 18:4153–4170. https://doi.org/10.1093/hmg/ddp367
Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC (2003) α-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem 278:25009–25013. https://doi.org/10.1074/jbc.M300227200
Funding
This research was supported by The William Stamps Farish Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dunlop, R.A., Carney, J.M. Mechanisms of l-Serine-Mediated Neuroprotection Include Selective Activation of Lysosomal Cathepsins B and L. Neurotox Res 39, 17–26 (2021). https://doi.org/10.1007/s12640-020-00168-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12640-020-00168-2