Abstract
Methamphetamine (METH) administration alters gene expression in the nucleus accumbens (NAc). We recently demonstrated that an acute METH injection produced prolonged increases in the expression of immediate early genes in the NAc of HDAC2-deficient mice, suggesting that HDAC2 might be an important regulator of gene expression in the rodent brain. Here, we tested the possibility that HDAC2 deletion might also impact METH-induced changes in the expression of various HDAC classes in the NAc. Wild-type (WT) and HDAC2 knockout (KO) mice were given a METH (20 mg/kg) injection, and NAc tissue was collected at 1, 2, and 8 h post treatment. We found that METH decreased HDAC3, HDAC4, HDAC7, HDAC8, and HDAC11 mRNA expression but increased HDAC6 mRNA levels in the NAc of WT mice. In contrast, the METH injection increased HDAC3, HDAC4, HDAC7, HDAC8, and HDAC11 mRNA levels in HDAC2KO mice. These observations suggest that METH may induce large-scale transcriptional changes in the NAc by regulating the expression of several HDACs, in part, via HDAC2-dependent mechanisms since some of the HDACs showed differential responses between the two genotypes. Our findings further implicate HDACs as potential novel therapeutic targets for neurotoxic complications associated with the abuse of certain psychostimulants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Methamphetamine (METH) use-disorders are serious international health problems with an estimated 25 million METH users worldwide (Chomchai and Chomchai 2015). METH causes an acute sense of euphoria, increased alertness, and hypersexuality in humans (Cruickshank and Dyer 2009). These clinical effects last several hours because of the long elimination half-life (10–12 h) of METH (Segal and Kuczenski 2006). In rodents, METH increases locomotor activity and produces stereotypic behaviors depending on the dose administered (Jing et al. 2014). In addition, an acute METH injection has been shown to alter gene expression in various brain regions (Cadet et al. 2001, 2010) including the nucleus accumbens (NAc), an important structure involved in the acute and chronic effects of psychostimulants (Kalivas and Volkow 2005). Moreover, these METH-induced transcriptional responses are accompanied by time-dependent post-translational histone modifications in the brain (Godino et al. 2015; Jayanthi et al. 2014; Martin et al. 2012).
Gene expression is regulated, in part, by dynamic interactions between chromatin, post-translational histone modifications, and transcription factors (TFs) (Bannister and Kouzarides 2011). In eukaryotic cells, DNA exists in chromatin whose basic subunits are called nucleosomes (Erdel et al. 2011). Each nucleosome is composed of 4 core histones including histones H2A, H2B, H3, and H4 that form an octomeric structure (2 of each core histone) (Morrison et al. 2007). Histones have N-terminal tails that contain lysine residues that can be reversibly acetylated by histone acetyltransferases (HATs) or deacetylated by histone deacetylases (HDACs) (Zhang et al. 2008). HDACs are divided into four known classes based on sequence similarities (Morrison et al. 2007). These include Class I (HDAC1, HDAC2, HDAC3, and HDAC8), Class IIA (HDAC4, HDAC5, HDAC7, and HDAC9), Class IIB (HDAC6 and HDAC10), Class III (Sirtuin1-7), and Class IV (HDAC11) (Gregoretti et al. 2004). Acetylation of histones allows for increased accessibility of TFs onto gene promoters with secondary increases in transcriptional activity (Verdone et al. 2006). In contrast, deacetylation of histones by repressor complexes containing HDACs results in condensed chromatin and suppressed gene expression (Kelly and Cowley 2013). While Sirtuins can also mediate transcription, they will not be the focus of this work as they are structurally and mechanistically distinct because they require NAD(+) for activation (Dali-Youcef et al. 2007), whereas the other HDACs require zinc as co-factors (de Ruijter et al. 2003; Wagner et al. 2013).
Because HDACs appear to be involved in psychostimulant-induced behaviors (Cadet 2014; Godino et al. 2015), they might provide important targets for the development of pharmacological therapeutics against the negative effects of psychostimulant exposure (Kalda and Zharkovsky 2015). For example, administration of the non-specific HDAC inhibitors, butyric acid, or valproic acid, was shown to potentiate amphetamine-induced behavioral sensitization in mice (Kalda et al. 2007). Similarly, administration of the histone deacetylase inhibitor, sodium butyrate, enhanced METH-induced locomotor sensitization in mice (Harkness et al. 2013). The latter effects may be secondary, in part, to METH-induced permissive epigenetic processes since psychostimulants are known to cause global hyperacetylation of histones (Godino et al. 2015). It is also important to note that HDACs may have roles beyond just the behaviors observed after injections of amphetamine-like drugs which are known neurotoxic agents that induced oxidative stress (Cadet et al. 2007). For example, inhibition of Class I and II HDACs by Trichostatin A (TSA) has neuroprotective effects against EtOH-induced overproduction of reactive oxygen species in human neuronal cells (Agudelo et al. 2011). METH-induced deacetylation of α-tubulin, an indicator of cellular structural loss, in endothelial cells was also shown to be preventable by treatment with the HDAC inhibitor, TSA (Fernandes et al. 2015). The suggestion for a role of HDACs in psychostimulant-induced biochemical effects is supported by the demonstration that an acute injection of METH produced time-dependent HDAC2 protein accumulation in the rat NAc (Martin et al. 2012). Our laboratory has also recently reported that binge toxic METH injections can influence the mRNA levels of several HDACs in the brain (Omonijo et al. 2014). Moreover, decreases in the striatal expression of glutamate receptors produced by chronic METH administration were mediated, in part, by increased HDAC1 and HDAC2 protein abundance on the promoters of these genes (Jayanthi et al. 2014).
In order to further test the role of specific HDACs in the effects of METH in the brain, we have used transgenic mice with a conditional deletion of HDAC2 to test if this protein was involved in the regulation of METH-induced changes in the expression of other HDACs. Here, we report that a single injection of METH decreased HDAC3, HDAC4, HDAC7, HDAC8, and HDAC11 mRNA levels in the NAc of wild-type (WT) mice. In the absence of HDAC2, however, the METH injection increased their expression. These results support the notion that HDAC2 might be an important regulator of gene expression after acute METH treatment.
Materials and Methods
Animals and Drug Treatment
We used male HDAC2flox/flox Camk2a Cre+ mice (HDAC2KO) and age-matched HDAC2flox/flox Camk2a Cre- (WT) littermates weighing 30–40 g (Total N = 48) in this study. HDAC2KO mice were generated by cross-breeding HDAC2loxP/loxP transgenic mice containing loxP sites between exon 2 and exon 4 of the mouse HDAC2 gene (Montgomery et al. 2007) with the C57BL/6 Camk2a-Cre transgenic mouse line (B6.Cg-Tg(Camk2a-cre)T29-1Stl/J) purchased from the Jackson Laboratory (Bar Harbor, ME). Mouse breeding was conducted by the National Institute of Drug Abuse (NIDA) Biomedical Research Center (BRC) Breeding Facility Core in Baltimore, MD. Genotyping for the specific deletion of HDAC2 was conducted by the Charles River’s Laboratory Testing Management® (LTM) division. All mice were housed in a temperature-controlled (22.2 ± 0.2 °C) vivarium with ad libitum access to food and water. After 14 weeks of age, mice of both genotypes received a single intraperitoneal (i.p.) injection of METH (20 mg/kg) and were subsequently euthanized at three different time points (1, 2, or 8 h post injection). Control mice of both genotypes received a single saline injection and were euthanized at either the 1- or 8-h time points. Six animals per genotype per time point were used in this study.
The time course of tissue collection was chosen based on our previous experiments showing HDAC2 involvement in the regulation of METH-induced IEG expression (Torres et al. 2015). The dose of METH was chosen because it produced robust changes in gene expression and increased HDAC2 protein levels within the NAc (Martin et al. 2012). All animal use procedures were conducted in adherence to the NIH Guide for the Care and Use of Laboratory Animals and were approved by the NIDA Intramural Research Program (IRP) Animal Care and Use Committee (ASP #12-CNRB-50).
RNA Isolation
NAc tissue was dissected, immediately placed on dry ice, and stored at −80 °C. Total RNA was isolated using Qiagen RNeasy® Mini kits, and genomic DNA contamination was eliminated using Qiagen RNase-free DNase (Qiagen, Valencia, CA) following the manufacturer’s protocol. Analysis of total RNA integrity was conducted using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA) and showed no degradation based on RIN values. Quantification of individual RNA was then assessed using a Nanodrop 2000 spectrophotometer system (Thermo Scientific, Waltham, MA).
Quantitative Real-Time Reverse Transcription-PCR Analysis
Un-pooled total RNA (0.5 μg) from each mouse was reverse-transcribed to cDNA using oligo dT primers from the Advantage RT for PCR kit (Clontech, Mountain View, CA). Specific gene primers for members of the HDAC family and PCR amplification were designed using the LightCycler Probe Design software (Roche, Indianapolis, IN). Primer sequences are listed in Table 1. Each custom-designed primer was then manufactured and HPLC-purified by the Synthesis and Sequencing Facility of the Johns Hopkins University (Baltimore, MD). The expression levels of each amplicon were examined by qRT-PCR using a LightCycler 480 II system (Roche, Indianapolis, IN) with iQ SYBR Green Supermix (BioRad, Hercules, CA). Purity of each qRT-PCR product was verified by separate melting curve analyses. Quantitative PCR values were normalized to clathrin (Clta) based on the observation that this reference gene had the most stable expression across both genotypes at various time points after the acute METH injection. Results are reported as fold changes calculated as the normalized gene expression value of each group relative to controls.
Statistical Analyses
Quantitative PCR data were analyzed using two-way analysis of variance (ANOVA) followed by Fisher’s LSD where appropriate (SPSS 20, IBM, Armonk, NY). All data are presented as mean ± SEM and considered statistically significant when p < 0.05.
Results
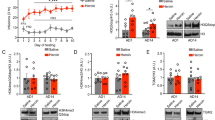
To test the potential role of HDAC2 in the regulation of METH-induced changes in Class I HDACs, we measured HDAC1, HDAC2, HDAC3, and HDAC8 mRNA levels in the NAc of WT and HDAC2KO mice (for a summary of results see Table 2). The METH injection did not cause significant alterations in HDAC1 [F(3,40) = 1.51, p = 0.22] mRNA levels in WT or HDAC2KO mice (Fig. 1a). There was a trend toward significant changes in HDAC2 [F(3,20) = 3.04, p = 0.05] mRNA levels in WT mice (Fig. 1b). HDAC2 mRNA was not measured in the HDAC2KO mice. In contrast, there were significant main effects of genotype [F(1,40) = 25.83, p < 0.001] and significant genotype*METH interactions [F(3,40) = 4.58, p < 0.01] in HDAC3 mRNA expression (Fig. 1c). HDAC3 mRNA levels were decreased at the 1- and 8-h time points after the METH injection in WT mice relative to saline controls (Fig. 1c). However, HDAC3 mRNA levels were significantly increased at the three time points in HDAC2KO mice compared to WT littermates (Fig. 1c). HDAC3 mRNA was also increased at the 8-h time point in comparison to saline-injected HDAC2KO mice (Fig. 1c). Moreover, there were significant main effects of genotype [F(1,38) = 10.9, p < 0.001] and genotype*METH interactions [F(3,38) = 3.19, p < 0.05] in HDAC8 mRNA levels (Fig. 1d). HDAC8 mRNA expression was comparable in both genotypes at the 1-h time point. However, HDAC8 expression was significantly increased (2.4-fold) in HDAC2KO at the 2-h time point relative to saline controls and WT mice (Fig. 1d). HDAC8 mRNA levels returned to normal values at the 8-h time point in HDAC2KO mice but were decreased in WT littermates.
METH-induced transcriptional responses in members of the Class I HDAC family: a HDAC1, b HDAC2, c HDAC3, and d HDAC8. WT and HDAC2KO mice were injected with a single dose of METH (20 mg/kg) and euthanized 1, 2, and 8 h later. Total RNA was extracted from the NAc and used in qRT-PCR assays. The relative amounts of transcripts were normalized to Clta. Data are presented as mean ± SEM (n = 6 animals per genotype per time point). Key to statistics: *p < 0.05, **p < 0.01, in comparison to respective saline-treated controls; # p < 0.05, ## p < 0.01, in comparison to respective METH-treated WT mice
The effects of METH on Class IIA HDACs are shown in Fig. 2. The METH injection caused significant main effects of genotype [F(1,40) = 29.67, p < 0.001] and significant genotype*METH interactions [F(3,40) = 16.17, p < 0.01] on HDAC4 mRNA levels (Fig. 2a). Decreased levels were observed at the 1-h time point in both WT and HDAC2KO mice relative to saline controls. HDAC4 mRNA levels remained decreased in WT mice, at both the 2- and 8-h time points, whereas they were significantly increased in HDAC2KO mice (Fig. 2a). METH treatment did not alter HDAC5 mRNA levels in the NAc of either WT or HDAC2KO mice [F(3,40) = 1.27, p = 0.29] (Fig. 2b). Figure 2c shows significant main effects of genotype [F(1,40) = 13.83, p < 0.005] and genotype*METH interactions [F(3,40) = 7.63, p < 0.01] in HDAC7 mRNA levels. HDAC7 mRNA levels decreased at 1 and 2 h, but were normalized at the 8-h time point after the METH injection in WT mice compared to saline controls. In contrast, HDAC7 mRNA levels increased at the 1-h time point in HDAC2KO mice compared to their respective saline-treated controls and to WT mice (see Table 2). HDAC7 mRNA levels increased at the 8-h time point in HDAC2KO mice in comparison to WT littermates. There were no significant changes [F(3,39) = 1.48, p = 0.23] in HDAC9 mRNA levels at any time point after the METH injection in WT or HDAC2KO mice (Fig. 2d).
Differential effects of acute METH treatment on Class IIA HDAC mRNA levels: a HDAC4, b HDAC5, c HDAC7, and d HDAC9. The graphs show results from qRT-PCR analyses. Data are presented as mean ± SEM (n = 6 animals per genotype per time point). Key to statistics: *p < 0.05, **p < 0.01, in comparison to respective saline-treated controls; ## p < 0.01, ### p < 0.001, in comparison to respective METH-treated WT mice
Figure 3 illustrates the effects of METH on Class IIB HDACs. We observed significant main effects of genotype [F(1,40) = 17.25, p < 0.001] and genotype*METH interactions [F(3,40) = 3.51, p < 0.05] on HDAC6 mRNA expression (Fig. 3a). In contrast to the observations with the other HDACs, there were significant increases in HDAC6 mRNA levels at the three time points in WT mice (Fig. 3a). However, significant increases in HDAC6 mRNA levels were only observed at the 8-h time point in HDAC2KO mice. There were no significant METH-induced changes in HDAC10 mRNA levels in either WT or HDAC2KO mice [F(3,39) = 0.70, p = 0.55] (Fig. 3b). Finally, METH caused significant main effects of genotype [F(1,38) = 24.58, p < 0.001] and significant genotype*METH interactions [F(3,38) = 5.19, p < 0.01] on HDAC11 mRNA expression (Fig. 3c). METH caused significant decreases in HDAC11 mRNA levels at the three time points post treatment in WT mice compared to controls. HDAC11 mRNA showed increased levels in HDAC2KO mice compared to WT littermates at all time points (Fig. 3c).
Effects of acute METH on the expression of Class IIB and Class IV members of the HDAC family: a HDAC6, b HDAC10, and c HDAC11. RNA extraction, qRT-PCR, and statistical analyses are as described in the methods section. Data are presented as mean ± SEM (n = 6 animals per genotype per time point). Key to statistics: *p < 0.05, **p < 0.01, ***p < 0.001, in comparison to respective saline-treated controls; # p < 0.05, ## p < 0.01, ### p < 0.001, in comparison to respective METH-treated genotype
Discussion
In the present study, we assessed the potential role of HDAC2 in mediating METH-induced transcriptional changes in all the members of zinc-dependent HDACs by using conditional HDAC2KO mice. The main findings of this paper are that (1) acute METH caused time-dependent decreases in HDAC3, HDAC4, HDAC7, HDAC8, and HDAC11 mRNA in the NAc of WT mice; (2) there were increased HDAC6 mRNA levels after acute METH; and (3) relative to WT mice, HDAC2KO mice showed increased HDAC3, HDAC4, HDAC7, HDAC8, and HDAC11 mRNA levels after a single METH injection. These findings suggest that acute METH causes down-regulation of HDACs in the NAc and implicate HDAC2 as a regulator of the expression of some HDACs following METH treatment.
Our laboratory had shown previously that binge METH injections did not cause significant changes in HDAC1 or HDAC2 mRNA levels measured at 16 h post treatment (Omonijo et al. 2014). In the present study, we also show that acute METH exposure does not alter HDAC1 or HDAC2 mRNA levels in the NAc of WT mice. However, following a similar dose of METH, HDAC2 protein levels were increased in the rat (Martin et al. 2012) and mouse (Torres et al. 2015) NAc. The differences between these reports and our current findings might be related to the fact that there is not a one-to-one correspondence between mRNA and protein expression (Wang 2008). More work is needed to explore these differences further. Nevertheless, the notion that HDAC2 might not be directly involved in the regulation of HDAC1 mRNA expression is not surprising given that the enzymatic activity of HDAC1 and HDAC2 is redundant (Montgomery et al. 2007). Among members of Class I HDACs, HDAC1 and HDAC2 are the most comparable with an estimated 83 % amino acid identity (Brunmeir et al. 2009). Given that deletion of HDAC2 does not alter HDAC1 expression in mouse embryonic stem cell lines (Winter et al. 2013), it is not far-fetched to suggest that HDAC1 is not regulated by HDAC2. Furthermore, HDAC1 might not compensate for the lack of HDAC2 in our transgenic mouse line since HDAC1 expression was not different between the two genotypes.
We also show, in addition, that acute METH decreased the mRNA expression of another Class I HDAC, HDAC3 in WT mice. Along with the various biological roles of HDAC3 (Emiliani et al. 1998), this enzyme is a negative regulator of transcription through the formation of SMRT and NCoR protein complexes (Karagianni and Wong 2007). Considering the regulatory role of HDAC3 on gene expression, the down-regulation of HDAC3 mRNA after acute METH exposure might also be involved in METH-induced transcriptional activation through increased histone acetylation. This notion is consistent with findings that cocaine exposure produces decreased HDAC3 enrichment on the promoters of c-fos and Nr4a2 (Rogge et al. 2013), two genes known to be up-regulated following acute cocaine (Hope et al. 1992) and METH treatment (Akiyama et al. 2008). We also showed that HDAC2KO mice display increases in HDAC3 mRNA levels after a single METH injection relative to WT mice. Because HDAC3 is essential for drug-induced memory consolidation (Malvaez et al. 2013), it is possible that HDAC3 might be involved in the improved memory observed after the loss of HDAC2 in rodents (Morris et al. 2013). Another Class I HDAC affected by the acute METH injection was HDAC8. Its molecular cloning was reported by two research groups (Buggy et al. 2000; Hu et al. 2000). HDAC8 was later shown to mediate transcriptional repression independent of protein complexes (Haider et al. 2011). Here, we show that HDAC8 mRNA expression was decreased in the NAc of WT mice at the 8-h time point after acute METH exposure. These results are in agreement with a previous report from our laboratory (Omonijo et al. 2014) and suggest that METH indeed does impact HDAC8 transcription. We also showed that, in HDAC2KO mice, HDAC8 mRNA levels were up-regulated at the 2-h time point and returned to baseline at the 8-h time point following acute METH treatment. Because overexpression of HDAC8 decreases pCREB activity (Gao et al. 2009) and METH prolongs expression of pCREB-responsive immediate early genes (IEGs) in the absence of HDAC2 (Torres et al. 2015), the transient up-regulation of HDAC8 might serve as a compensatory mechanism to repress pCREB-dependent transcription in HDAC2KO mice. Nevertheless, further studies are needed to fully explore this possibility.
The METH injection also impacted HDAC4 and HDAC7 mRNA expression. Unlike the Class I HDACs, members of the Class IIA subgroup are unique in that they shuttle between the nucleus and cell cytoplasm (Chawla et al. 2003). Herein, we observed that acute METH decreased HDAC4 mRNA levels in the NAc of WT mice. These findings are comparable to our observations for HDAC3 and suggest that METH might down-regulate HDAC4 expression by similar molecular mechanisms since HDAC4 is enzymatically inactive when not associated with HDAC3 (Guenther et al. 2001). We also observed that, relative to WT mice, HDAC2KO mice displayed increased HDAC4 mRNA levels at the 2- and 8-h time points after the acute METH injection. These observations suggest that HDAC2 is indeed involved in regulation of HDAC4 transcription. METH also caused decreased HDAC7 mRNA levels in WT mice. The METH-induced down-regulation of HDAC7 is interesting given that HDAC7 inhibits NR4a1 (Dequiedt et al. 2005), an orphan nuclear receptor known to be overexpressed after METH treatment (McCoy et al. 2011). Thus, decreases in HDAC7 mRNA might account for the METH-induced transcriptional activation of NR4a family members that play a key role in cell survival (Akiyama et al. 2008). METH also increased HDAC7 mRNA in HDAC2KO mice. Because HDAC7 also combines with protein complexes containing HDAC3 (Fischle et al. 2001), it is tempting to speculate that HDAC2 might also co-regulate these two enzymes to exert better control of their epigenetic effects.
Of note, HDAC6 mRNA was up-regulated in WT mice at all time points following the acute METH injection. HDAC6 is unique among other HDACs in that it has a zinc finger ubiquitin-binding domain able to identify misfolded proteins for lysosome degradation (Lee et al. 2010; Ouyang et al. 2012). HDAC6 also possesses tubulin deacetylase activity (Haggarty et al. 2003). Acetylation of microtubules plays a critical role in cell stability, whereas deacetylation results in cytoskeleton disarrangement (Li et al. 2007, 2012). Given that METH is known to increase blood–brain barrier (BBB) permeability (Ramirez et al. 2009), the up-regulation of HDAC6 mRNA might be involved in METH-induced disruption of cytoskeletal viability. This idea is consistent with the fact that endothelial cells treated with METH show α-tubulin degradation secondary to HDAC6 activation (Fernandes et al. 2015). Indeed, overexpression of HDAC6 in mammalian cells leads to hypoacetylation of tubulin (Hubbert et al. 2002), while inactivation of HDAC6 leads to tubulin hyperacetylation (Zhang et al. 2003). Another possible mechanism for the structural breakdown of actin filaments after METH exposure might also involve the expression of matrix metalloproteinases (MMPs). This idea is supported by the fact that METH-induced activation of MMP-9 was shown to produce BBB degradation via dysfunction of neurovascular matrix components and tight junctions (Martins et al. 2011; Ramirez et al. 2009). These effects appear to involve the action of HDACs that are involved in regulating MMP-9 expression (Poljak et al. 2014; Wang et al. 2013). On the other hand, acetyl-l-carnitine, an HDAC inhibitor (Huang et al. 2012), was shown to attenuate METH-induced activation of MMP-9 (Fernandes et al. 2014) and to prevent METH-induced deacetylation of tubulin (Fernandes et al. 2015). Thus, together, these results suggest that METH might compromise BBB integrity via increased HDAC2-dependent increased HDAC6 and MMP9 expression.
It needs to be pointed out that the expression of some HDACs was not altered by the acute METH injection. For example, there were no changes in HDAC9 or HDAC10 mRNA levels in WT or HDAC2KO mice. In contrast, Omonijo et al. (2014) showed that toxic METH doses produce decreases in both HDAC9 and HDAC10 mRNA levels in the rat striatum. The differences in response are attributable to the METH dosing regimen, the brain regions (NAc against dorsal striatum), and the species used (mice against rats). In fact, different schedules of METH exposure are known to cause distinct patterns of transcriptional responses in the rat brain (McCoy et al. 2011). Moreover, our results suggest that HDAC2 might not play a pivotal role in the regulation of HDAC9 or HDAC10 mRNA expression.
It should also be noted that the acute METH injection caused down-regulation of HDAC11 mRNA in WT mice. Given that HDAC11 also suppresses transcriptional activity (Sahakian et al. 2015; Gao et al. 2002), decreases in this enzyme, in addition to the decreased expression of other HDACs, might serve to potentiate METH-induced global changes in gene expression. We also found that, similar to the other HDACs, HDAC2 deletion resulted in overexpression of HDAC11 mRNA at the three time points after the METH injections. Because overexpression of HDAC11 decreases the acetylation of H3K9 and H3K14 (Liu et al. 2009), METH-induced HDAC11 overexpression may also decrease the abundance of these histone marks in HDAC2KO mice.
In summary, we report that acute METH treatment represses the mRNA expression of several members of the HDAC family. We have also shown, for the first time, that conditional deletion of HDAC2 produced METH-induced up-regulation in HDAC3, HDAC4, HDAC8, HDAC7, and HDAC11 mRNA levels in NAc. In contrast, acute METH treatment did not affect HDAC1, HDAC5, HDAC9, or HDAC10 mRNA levels in the NAc of HDAC2KO mice. These findings add further evidence for the notion that HDAC2 might act as a negative regulator of METH-induced transcriptional changes in the brain. Moreover, our work shows that HDAC2 is an important repressor of some, but not all, members of zinc-dependent HDAC classes. Future studies will focus on investigating whether there are direct associations between METH-induced repression of the transcription of HDAC family members and METH-induced global increases in gene expression.
References
Agudelo M, Gandhi N, Saiyed Z, Pichili V, Thangavel S, Khatavkar P, Yndart-Arias A, Nair M (2011) Effects of alcohol on histone deacetylase 2 (HDAC2) and the neuroprotective role of trichostatin A (TSA). Alcohol Clin Exp Res 35(8):1550–1556
Akiyama K, Isao T, Ide S, Ishikawa M, Saito A (2008) mRNA expression of the Nurr1 and NGFI-B nuclear receptor families following acute and chronic administration of methamphetamine. Prog Neuropsychopharmacol Biol Psychiatry 32(8):1957–1966
Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21(3):381–395
Brunmeir R, Lagger S, Seiser C (2009) Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol 53(2–3):275–289
Buggy JJ, Sideris ML, Mak P, Lorimer DD, McIntosh B, Clark JM (2000) Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem J 350(Pt 1):199–205
Cadet JL (2014) Epigenetics of stress, addiction, and resilience: Therapeutic implications. Mol Neurobiol. doi:10.1007/s12035-014-9040-y
Cadet JL, Jayanthi S, McCoy MT, Vawter M, Ladenheim B (2001) Temporal profiling of methamphetamine-induced changes in gene expression in the mouse brain: evidence from cDNA array. Synapse 41(1):40–48
Cadet JL, Krasnova IN, Jayanthi S, Lyles J (2007) Neurotoxicity of substituted amphetamines: molecular and cellular mechanisms. Neurotox Res 11(3–4):183–202
Cadet JL, Jayanthi S, McCoy MT, Beauvais G, Cai NS (2010) Dopamine D1 receptors, regulation of gene expression in the brain, and neurodegeneration. CNS Neurol Disord 9(5):526–538
Chawla S, Vanhoutte P, Arnold FJ, Huang CL, Bading H (2003) Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem 85(1):151–159
Chomchai C, Chomchai S (2015) Global patterns of methamphetamine use. Curr Opin Psychiatry 28(4):269–274
Cruickshank CC, Dyer KR (2009) A review of the clinical pharmacology of methamphetamine. Addiction 104(7):1085–1099
Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J (2007) Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann Med 39(5):335–345
de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 370:737–749
Dequiedt F, Van Lint J, Lecomte E, Van Duppen V, Seufferlein T, Vandenheede JR, Wattiez R, Kettmann R (2005) Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J Exp Med 201(5):793–804
Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E (1998) Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci USA 95(6):2795–2800
Erdel F, Krug J, Längst G, Rippe K (2011) Targeting chromatin remodelers: signals and search mechanisms. Biochim Biophys Acta 1809(9):497–508
Fernandes S, Salta S, Bravo J, Silva AP, Summavielle T (2014) Acetyl-l-Carnitine prevents methamphetamine-induced structural damage on endothelial cells via ILK-related MMP-9 activity. Mol Neurobiol. doi: 10.1007/s12035-014-8973-5
Fernandes S, Salta S, Summavielle T (2015) Methamphetamine promotes α-tubulin deacetylation in endothelial cells: the protective role of acetyl-l-carnitine. Toxicol Lett 234(2):131–138
Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, Verdin E (2001) Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J Biol Chem 276(38):35826–35835
Gao J, Siddoway B, Huang Q, Xia H (2009) Inactivation of CREB mediated gene transcription by HDAC8 bound protein phosphatase. Biochem Biophys Res Commun 379(1):1–5
Gao L, Cueto MA, Asselbergs F, Atadja P (2002) Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem 277(28):25748–25755
Godino A, Jayanthi S, Cadet JL (2015) Epigenetic landscape of amphetamine and methamphetamine addiction in rodents. Epigenetics 10(7):574–580
Gregoretti IV, Lee YM, Goodson HV (2004) Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol 338(1):17–31
Guenther MG, Barak O, Lazar MA (2001) The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol 21(18):6091–6101
Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL (2003) Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA 100(8):4389–4394
Haider S, Joseph CG, Neidle S, Fierke CA, Fuchter MJ (2011) On the function of the internal cavity of histone deacetylase protein 8: R37 is a crucial residue for catalysis. Bioorg Med Chem Lett 21(7):2129–2132
Harkness JH, Hitzemann RJ, Edmunds S, Phillips TJ (2013) Effects of sodium butyrate on methamphetamine-sensitized locomotor activity. Behav Brain Res 239:139–147
Hope B, Kosofsky B, Hyman SE, Nestler EJ (1992) Regulation of immediate early gene expression and AP-1 binding in the rat nucleus accumbens by chronic cocaine. Proc Natl Acad Sci USA 89(13):5764–5768
Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang GF, Johanson K, Sung CM, Liu R, Winkler J (2000) Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem 275(20):15254–15264
Huang H, Liu N, Guo H, Liao S, Li X, Yang C, Liu S, Song W, Liu C, Guan L, Li B, Xu L, Zhang C, Wang X, Dou QP, Liu J (2012) l-carnitine is an endogenous HDAC inhibitor selectively inhibiting cancer cell growth in vivo and in vitro. PLoS ONE 7(11):e49062
Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417(6887):455–458
Jayanthi S, McCoy MT, Chen B, Britt JP, Kourrich S, Yau HJ, Ladenheim B, Krasnova IN, Bonci A, Cadet JL (2014) Methamphetamine downregulates striatal glutamate receptors via diverse epigenetic mechanisms. Biol Psychiatry 76(1):47–56
Jing L, Zhang M, Li JX, Huang P, Liu Q, Li YL, Liang H, Liang JH (2014) Comparison of single versus repeated methamphetamine injection induced behavioral sensitization in mice. Neurosci Lett 560:103–106
Kalda A, Zharkovsky A (2015) Epigenetic mechanisms of psychostimulant-induced addiction. Int Rev Neurobiol 120:85–105
Kalda A, Heidmets LT, Shen HY, Zharkovsky A, Chen JF (2007) Histone deacetylase inhibitors modulates the induction and expression of amphetamine-induced behavioral sensitization partially through an associated learning of the environment in mice. Behav Brain Res 181(1):76–84
Kalivas PW, Volkow ND (2005) The neural basis of addiction: a pathology of motivation and choice. Am J Psychiatry 162(8):1403–1413
Karagianni P, Wong J (2007) HDAC3: taking the SMRT-N-CoRrect road to repression. Oncogene 26(37):5439–5449
Kelly RD, Cowley SM (2013) The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem Soc Trans 41(3):741–749
Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, Taylor JP, Cuervo AM, Yao TP (2010) HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J 29(5):969–980
Li G, Jiang H, Chang M, Xie H, Hu L (2007) HDAC6 α-tubulin deacetylase: a potential therapeutic target in neurodegenerative diseases. J Neurol Sci 304(1–2):1–8
Li Y, Shin D, Kwon SH (2012) Histone deacetylase 6 plays a role as a distinct regulator of diverse cellular processes. FEBS J 280(3):775–793
Liu H, Hu Q, D’ercole AJ, Ye P (2009) Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia 57(1):1–12
Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA (2013) HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc Natl Acad Sci USA 110(7):2647–2652
Martin TA, Jayanthi S, McCoy MT, Brannock C, Ladenheim B, Garrett T, Lehrmann E, Becker KG, Cadet JL (2012) Methamphetamine causes differential alterations in gene expression and patterns of histone acetylation/hypoacetylation in the rat nucleus accumbens. PLoS One 7(3):e34236
Martins T, Baptista S, Gonçalves J, Leal E, Milhazes N, Borges F, Ribeiro CF, Quintela O, Lendoiro E, López-Rivadulla M, Ambrósio AF, Silva AP (2011) Methamphetamine transiently increases the blood-brain barrier permeability in the hippocampus: role of tight junction proteins and matrix metalloproteinase-9. Brain Res 1411:28–40
McCoy MT, Jayanthi S, Wulu JA, Beauvais G, Ladenheim B, Martin TA, Krasnova IN, Hodges AB, Cadet JL (2011) Chronic methamphetamine exposure suppresses the striatal expression of members of multiple families of immediate early genes (IEGs) in the rat: normalization by an acute methamphetamine injection. Psychopharmacology 215(2):353–365
Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN (2007) Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev 21(14):1790–1802
Morris MJ, Mahgoub M, Na ES, Pranav H, Monteggia LM (2013) Loss of histone deacetylase 2 improves working memory and accelerates extinction learning. J Neurosci 33(15):6401–6411
Morrison BE, Majdzadeh N, D’Mello SR (2007) Histone deacetylases: focus on the nervous system. Cell Mol Life Sci 64(17):2258–2269
Omonijo O, Wongprayoon P, Ladenheim B, McCoy MT, Govitrapong P, Jayanthi S, Cadet JL (2014) Differential effects of binge methamphetamine injections on the mRNA expression of histone deacetylases (HDACs) in the rat striatum. Neurotoxicology 45:178–184
Ouyang H, Ali YO, Ravichandran M, Dong A, Qiu W, MacKenzie F, Dhe-Paganon S, Arrowsmith CH, Zhai RG (2012) Protein aggregates are recruited to aggresome by histone deacetylase 6 via unanchored ubiquitin C termini. J Biol Chem 287(4):2317–2327
Poljak M, Lim R, Barker G, Lappas M (2014) Class I to III histone deacetylases differentially regulate inflammation-induced matrix metalloproteinase 9 expression in primary amnion cells. Reprod Sci 21(6):804–813
Ramirez SH, Potula R, Fan S, Eidem T, Papugani A, Reichenbach N, Dykstra H, Weksler BB, Romero IA, Couraud PO, Persidsky Y (2009) Methamphetamine disrupts blood-brain barrier function by induction of oxidative stress in brain endothelial cells. J Cereb Blood Flow Metab 29(12):1933–1945
Rogge GA, Singh H, Dang R, Wood MA (2013) HDAC3 is a negative regulator of cocaine-context-associated memory formation. J Neurosci 33(15):6623–6632
Sahakian E, Powers JJ, Chen J, Deng SL, Cheng F, Distler A, Woods DM, Rock-Klotz J, Sodre AL, Youn JI, Woan KV, Villagra A, Gabrilovich D, Sotomayor EM, Pinilla-Ibarz J (2015) Histone deacetylase 11: a novel epigenetic regulator of myeloid derived suppressor cell expansion and function. Mol Immunol 63(2):579–585
Segal DS, Kuczenski R (2006) Human methamphetamine pharmacokinetics simulated in the rat: single daily intravenous administration reveals elements of sensitization and tolerance. Neuropsychopharmacology 31(5):941–955
Torres OV, McCoy MT, Ladenheim B, Jayanthi S, Brannock C, Tulloch I, Krasnova IN, Cadet JL (2015) CAMKII-conditional deletion of histone deacetylase 2 potentiates acute methamphetamine-induced expression of immediate early genes in the mouse nucleus accumbens. Sci Rep 5:13396
Verdone L, Agricola E, Caserta M, Di Mauro E (2006) Histone acetylation in gene regulation. Brief Funct Genom Proteom 5(3):209–221
Wagner FF, Weïwer M, Lewis MC, Holson EB (2013) Small molecule inhibitors of zinc-dependent histone deacetylases. Neurotherapeutics 10(4):589–604
Wang D (2008) Discrepancy between mRNA and protein abundance: insight from information retrieval process in computers. Comput Biol Chem 32(6):462–468
Wang F, Qi Y, Li X, He W, Fan QX, Zong H (2013) HDAC inhibitor trichostatin A suppresses esophageal squamous cell carcinoma metastasis through HADC2 reduced MMP-2/9. Clin Invest Med 36(2):E87–E94
Winter M, Moser MA, Meunier D, Fischer C, Machat G, Mattes K, Lichtenberger BM, Brunmeir R, Weissmann S, Murko C, Humer C, Meischel T, Brosch G, Matthias P, Sibilia M, Seiser C (2013) Divergent roles of HDAC1 and HDAC2 in the regulation of epidermal development and tumorigenesis. EMBO J 32(24):3176–3191
Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, Matthias P (2003) HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J 22(5):1168–1179
Zhang Y, Fang H, Jiao J, Xu W (2008) The structure and function of histone deacetylases: the target for anti-cancer therapy. Curr Med Chem 15(27):2840–2849
Acknowledgments
The authors wish to thank Ezekiell Mouzon at Mount Sinai School of Medicine for graciously donating HDAC2 floxed mice.
Funding
This study was supported by funds of the Intramural Research Program of the DHHS/NIH/NIDA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Torres, O.V., Ladenheim, B., Jayanthi, S. et al. An Acute Methamphetamine Injection Downregulates the Expression of Several Histone Deacetylases (HDACs) in the Mouse Nucleus Accumbens: Potential Regulatory Role of HDAC2 Expression. Neurotox Res 30, 32–40 (2016). https://doi.org/10.1007/s12640-015-9591-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12640-015-9591-3