Abstract
Animal and human fascioliasis is a health and economic problem in few of tropical and subtropical areas of the world, including Iran. The present study aimed to determine the genotype diversity of Fasciola isolates in different hosts from Gilan province, northern Iran, and compare it with those isolates from southwestern Iran. Forty-eight adult Fasciola spp. were collected from cattle, sheep, and goats from slaughterhouse in Talesh, north of Iran. DNA was extracted from each fluke and PCR-RFLP was used to find out the species of the isolates. The ribosomal ITS1 and ITS2, and mitochondrial genes of NDI and COI from individual Fasciola isolates of each host were PCR-amplified and the PCR products were sequenced. Genetic variation within and between the isolates was evaluated by comparing the sequences of ribosomal and mitochondrial genes. For analysis of phylogenetic diversity of the flukes, phylogenetic trees were constructed, using ITS1, ITS2, NDI, and COI sequences of the isolates. Based on PCR-RFLP profile, 5 (22.7%) of the total of sheep isolates and 18 (90%) of cattle isolates were identified as F. gigantica and other remaining samples from sheep, cattle and goats were identified as F. hepatica. Based on ITS1 and ITS2 sequences, six and seven nucleotide polymorphism were respectively noted in the isolates. On the other hand, CO1 region sequences showed considerable variation, which laid Talesh (north) isolates in a separate cluster. Findings of the study showed that the sequences of CO1 isolates from north and southwest have substantial differences mainly in CO1 region.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fasciola, is one of the most important food and water-borne parasitic zoonoses and a major public health problem in some of tropical and subtropical countries (Ashrafi et al. 2015; Hosseini et al. 2015; Mas-Coma et al. 2009; Sarkari et al. 2012). The two species F. hepatica and F. gigantica are considered as causative agents of human and animal fascioliasis. While F. hepatica is dominantly in temperate areas, F. gigantica is usually in tropical areas (Dar et al. 2012). Moreover, presence of intermediate forms of the parasite has been recognized in some Asian endemic areas including Iran, Thailand, Myanmar, Japan, Vietnam and Korea (Shoriki et al. 2014).
Human and animal fascioliasis is serious health and veterinary problems in Iran (Ashrafi et al. 2015; Hosseini et al. 2015; Moazeni et al. 2012; Sarkari et al. 2012; Mohammadi-Ghalehbin et al. 2012). Northern Province of Gilan in Iran is one of the most important endemic regions of the disease where two massive outbreaks of human fascioliasis occurred in 1987 and in 1997 and affected more than 10,000 people (Ashrafi et al. 2015; Rokni 2008).
The evaluation of Fasciola genotypes and increasing of the knowledge of genetic diversity in the northern part of Iran is necessary due to the presence of intermediate form and both species of Fasciola (Ashrafi et al. 2006). The most frequently used methods for taxonomical studies and differentiation of two Fasciola species have been morphological approaches, although differences in isoenzymatic and also in the electrophoretic patterns of two species are reported (Abdolahi Khabisi and Sarkari 2016; Sarkari et al. 2016). Nowadays, morphometric methods considered inappropriate for differentiation of the two Fasciola species. Molecular techniques, using mitochondrial markers including ND1 and CO1 and ribosomal DNA markers including ITS1 and ITS2, are recently utilized to find out the genetic variation of Fasciola species (Itagaki et al. 2005; Peng et al. 2009; Shafiei et al. 2013, 2014; Shoriki et al. 2014). The present study was conducted to determine the genotype diversity of Fasciola isolates in different hosts from Gilan province, northern Iran, and compare it with those isolates from southwestern Iran (Shafiei et al. 2014), which has a different geographical and ecological features.
Materials and methods
Study area
Fasciola isolates were collected from Talesh abattoir located in Gilan province, 140 km north-west of Rasht, the capital of Gilan province, on the southwest coast of the Caspian Sea (Fig. 1). Talesh is a mountainous wonderful region covering an area of 2373 square kilometers, about one quarter of surface area of Gilan province.
Parasites collection
Adult Fasciola (n = 48) were collected from liver of sheep (n = 22), goats (n = 6) and cattle (n = 20) from Talesh in Gilan province. The worms were washed four times, in PBS, and then preserved in 70% ethanol until use.
DNA extraction and PCR amplification
DNA extraction and PCR amplification was performed as previously described (Shafiei et al. 2014). Briefly, a portion of the apical and lateral zone of adult flukes was removed and homogenized, using 500 μL of lysis buffer (50 mL of Tris–HCl (100 Mm), pH 8; 1 mM of EDTA; 0.1% of SDS) in tissue grinder. 8 μL of proteinase K (20 mg/mL) was added and the sample was incubated overnight at 37 °C. DNA was extracted with phenol–chloroform method. PCR was performed to amplify the target genes, CO1, ND1, ITS1 and ITS2, using four pairs of primers as shown in Table 1 (Itagaki et al. 2005).
PCR-PFLP
PCR-RFLP was used to specifically determine the Fasciola species by fast digestion of ITS1 region, using RsaI enzyme. To do that, in 10 µl of PCR products, 17 µl of water nuclease free, 2 µl of fast digest green buffer (10×) and 1 µl of fast digest enzyme were mixed and incubated at 37 °C for 10 min. The product was loaded on 2.5% agarose gel.
DNA sequencing
PCR products of ITS1, ITS2, COI, and NDI of isolates from each host (cattle, sheep, and goat) were purified, using Vivantis purification kit (Vivantis Technologies Sdn. Bhd., Malaysia) and sequenced from both directions using the same primers which were used in the PCR.
Phylogenetic analysis
The sequences were aligned and compared with those of previously available sequences from our previous study in southwest of Iran and from other regions of the world, available in the GenBank, using the BLAST program of GenBank. Maximum likelihood tree was constructed, using the MEGA 5.0 software.
Results
PCR-RFLP analysis
Based on PCR-PRFLP profile, five (22.7%) of sheep isolates and 18 (90%) of cattle isolates were F. gigantica whereas 77.3% of sheep isolates, 10% of cattle isolates and all of the goat isolates were F. hepatica.
Genotype analysis based on ITS1 sequences
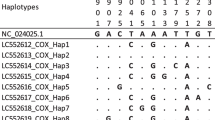
Sequences of ITS1 region (600 bp) of F. hepatica and F. gigantica were aligned with those of available sequences of both F. gigantica and F. hepatica from southwestern area of Iran. Between these isolates, six DNA variable sites were observed in which single-base substitution at different sites was occurred.
Genotype analysis based on ITS2 sequences
Multiple alignments between sequences of F. gigantica and F. hepatica isolates showed five variable sites. Aligned sequences of ITS2 region (505 bp) of Fasciola isolates form north of the country with those from southwest area showed seven variable sites which single-base substitution were occurred.
Genotype analysis based on CO1 sequences
Alignment of CO1 region (438 bp) sequences of F. gigantica and F. hepatica isolates showed 34 variable sites. Substitutions were observed at different nucleotide sites. Isolates from north area showed considerable nucleotide site substitutions in comparison with southwest isolates.
Genotype analysis based on ND1 sequences
Alignment between partial ND1 (535 bp) sequences of F. hepatica isolate showed only one variable site. However, 33 variable nucleotide sites were detected between north and southwest isolates.
Phylogenetic analysis
Phylogenetic trees were constructed, using ITS1, ITS2, NDI, and COI sequences of F. gigantica and F. hepatica from the present study along with sequences from the southwest region in our previous study, and also from other regions of the world. Phylogenetic analysis of ITS1 and ITS2 of northern isolates in comparison to southwest isolates and other identified isolates from different geographical regions showed two clusters of Fasciola species (Figs. 2, 3).
In CO1, north and southwest isolates separated into two different clusters (Fig. 4) demonstrating considerable differences between Iranian Fasciola isolates from north and south of the country. In ND1, highly relationship was observed between north isolates and southwest isolates (Fig. 5).
Discussion
In the present study, molecular characterization of Fasciola isolates was determined and the findings was compared with those available data from the isolates from southwest of the country. Based on phylogenetic analysis of the two species, ITS1 region showed six variable nucleotide sites which are compatible with the findings of previous studies (Itagaki et al. 2009; Shafiei et al. 2014).
F. hepatica and F. gigantica isolates were in separate clusters, based on ITS1 region. Nevertheless, close relationship between two species of north isolates and isolates from other geographical regions was observed.
Comparison of sequences of ITS2 region of north isolates and other isolates showed seven variable nucleotide sites among Fasciola species. Such variation was reported in previous studies as well (Choe et al. 2011; Erensoy et al. 2009; Shafiei et al. 2013; 2014).
Findings of the current study demonstrated a close association between north and southwest isolates regarding the ND1 region. As already shown in phylogenetic tree, north and southwest isolates are sit in one cluster.
Fasciola isolates of southwest of Iran and Asian zones showed genetic variation in mitochondrial genes including ND1 and CO1 regions in a few studies (Itagaki et al. 2005; Semyenova et al. 2009). Considering the findings of current and other related studies, ND1 region is a suitable marker for evaluation of genetic diversity of Fasciola. Having said that, in our study, this region showed only one nucleotide variation among north isolates and high relationship was observed between the ND1 sequence of north and southwest isolates.
CO1 sequences of F. hepatica isolated from goat and sheep from north showed 100% identity with KF992217.1 (from southwest Iran), AB553810.1 (from Egypt), GQ398051.1 (from Iran) and FJ895604.1 (from north of Iran) isolates. In addition, samples isolated from cattle showed 100% identity with HQ857101 (from Mauritania) and AB553784 (from Egypt).
CO1 mitochondrial gene has been suggested as an approperiate region for evaluation of genetic diversity in different hosts and geographical areas for Fasciola and also other helminthes including Echinococcus (Sarkari et al. 2015; Shafiei et al. 2014). Our results showed that the sequences of CO1 isolates from north and southwest have substantial differences. Genetic diversity in north and south isolates of Fasciola might be linked to the host species, including intermediate and final hosts. In keeping with these findings, previous studies on Iranian isolates of Fasciola showed high level of diversity in CO1 region (Moazeni et al. 2012).
Taken together, findings of the present study based on RFLP-PCR and sequencing of ribosomal and mitochondrial DNA, presented only two species of Fasciola in the studied area. However, in this study, Iranian isolates presented less variation in ITS1, ITS2 and ND1 regions. Based on CO1 marker, high genetic variation was noted in north isolates in comparison with isolates from southwest of Iran and also other geographical regions of the world.
References
Abdolahi Khabisi S, Sarkari B (2016) Detection of Fasciola hepatica and Fasciola gigantica common and uncommon antigens, using rabbit hyper immune serum raised against their excretory-secretory and somatic antigens. J Parasit Dis 40(4):1552–1557
Ashrafi K, Valero M, Panova M, Periago M, Massoud J, Mas-Coma S (2006) Phenotypic analysis of adults of Fasciola hepatica, Fasciola gigantica and intermediate forms from the endemic region of Gilan, Iran. Parasitol Int 55(4):249–260
Ashrafi K, Valero MA, Peixoto RV, Artigas P, Panova M, Mas-Coma S (2015) Distribution of Fasciola hepatica and F. gigantica in the endemic area of Guilan, Iran: relationships between zonal overlap and phenotypic traits. Infect Genet Evol 31:95–109
Choe S-E, Nguyen TT-D, Kang T-G, Kweon C-H, Kang S-W (2011) Genetic analysis of Fasciola isolates from cattle in Korea based on second internal transcribed spacer (ITS-2) sequence of nuclear ribosomal DNA. Parasitol Res 109(3):833–839
Dar Y, Amer S, Mercier A, Courtioux B, Dreyfuss G (2012) Molecular identification of Fasciola spp. (Digenea: Fasciolidae) in Egypt. Parasite: journal de la Société Française de Parasitologie 19(2):177
Erensoy A, Kuk S, Ozden M (2009) Genetic identification of Fasciola hepatica by ITS-2 sequence of nuclear ribosomal DNA in Turkey. Parasitol Res 105(2):407–412
Hosseini G, Sarkari B, Moshfe A, Motazedian MH, Abdolahi Khabisi S (2015) Epidemiology of human fascioliasis and intestinal helminthes in rural areas of Boyer-Ahmad Township, Southwest Iran; a population based study. Iran J Public Health 44(11):1520–1525
Itagaki T et al (2005) Genetic characterization of parthenogenic Fasciola sp. in Japan on the basis of the sequences of ribosomal and mitochondrial DNA. Parasitology 131(05):679–685
Itagaki T, Sakaguchi K, Terasaki K, Sasaki O, Yoshihara S, Van Dung T (2009) Occurrence of spermic diploid and aspermic triploid forms of Fasciola in Vietnam and their molecular characterization based on nuclear and mitochondrial DNA. Parasitol Int 58(1):81–85
Mas-Coma S, Valero MA, Bargues MD (2009) Chapter 2. Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv Parasitol 69:41–146
Moazeni M, Sharifiyazdi H, Izadpanah A (2012) Characterization of Fasciola hepatica genotypes from cattle and sheep in Iran using cytochrome C oxidase gene (CO1). Parasitol Res 110(6):2379–2384
Mohammadi-Ghalehbin B, Chinifroush-Asl MM, Ramzi F (2012) Extra-hepatic fascioliasis with peritoneal malignancy tumor feature. J Parasit Dis 36(1):78–80
Peng M, Ichinomiya M, Ohtori M, Ichikawa M, Shibahara T, Itagaki T (2009) Molecular characterization of Fasciola hepatica, Fasciola gigantica, and aspermic Fasciola sp. in China based on nuclear and mitochondrial DNA. Parasitol Res 105(3):809–815
Rokni M (2008) The present status of human helminthic diseases in Iran. Ann Trop Med Parasitol 102(4):283–295
Sarkari B, Ghobakhloo N, Moshfea A, Eilami O (2012) Seroprevalence of human fasciolosis in a new-emerging focus of fasciolosis in Yasuj district, southwest of Iran. Iran J Parasitol 7(2):15–20
Sarkari B, Mansouri M, Khabisi SA, Mowlavi G (2015) Molecular characterization and seroprevalence of Echinococcus granulosus in wild boars (Sus scrofa) in south-western Iran. Ann Parasitol 61(4):269–273
Sarkari B, Sedaghat B, Hatam G (2016) Comparative study on isoenzyme patterns of Fasciola hepatica and Fasciola gigantica. Trop Biomed 33(3):462–468
Semyenova SK et al (2009) Genetic differentiation in eastern European and western Asian populations of the liver fluke, Fasciola hepatica, as revealed by mitochondrial nad1 and cox1 genes. J Parasitol 92(3):525–530
Shafiei R, Sarkari B, Moshfe A (2013) A consistent PCR-RFLP assay based on ITS-2 ribosomal DNA for differentiation of Fasciola species. Iran J Basic Med Sci 16(12):1266
Shafiei R, Sarkari B, Sadjjadi SM, Mowlavi GR, Moshfe A (2014) Molecular and morphological characterization of Fasciola spp. isolated from different host species in a newly emerging focus of human fascioliasis in Iran. Vet Med Int 2014:405740
Shoriki T, Ichikawa-Seki M, Devkota B, Rana HB, Devkota SP, Humagain SK, Itagaki T (2014) Molecular phylogenetic identification of Fasciola flukes in Nepal. Parasitol Int 3(6):758–762
Acknowledgements
The study was financially supported by the office of vice-chancellor for research of Shiraz University of Medical Sciences (Grant No. 90-01-43-3855). The study was the subject of M. Parhoode MD thesis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Sarkari, B., Parhoode, M., Abdolahi Khabisi, S. et al. Genetic diversity of Fasciola spp. isolates from northern part of Iran: comparison with southwestern isolates. J Parasit Dis 41, 768–772 (2017). https://doi.org/10.1007/s12639-017-0886-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12639-017-0886-6