Abstract
Today, resistance of microorganisms to antibiotics has become a major challenge. To overcome this problem, development of new drugs, besides research on their antibacterial activity, is essential. Among chemical components, antimicrobial peptides (AMPs) exhibit antibacterial activity and can be selected as suitable antimicrobial candidates. In this study, a novel antimicrobial peptide, called dendrocin-ZM1, with a molecular weight of ~3716.48 Da, was isolated from Zataria multiflora Boiss (ZM) and purified via precipitation with ammonium sulfate and reverse-phase HPLC chromatography; it was then sequenced via Edman degradation. The in silico method was used to examine the physicochemical properties of dendrocin-ZM1. In this study, four reference strains (gram-positive and gram-negative) and one clinical vancomycin-resistant Staphylococcus aureus strain were used to survey the antimicrobial activities. Moreover, to examine cytotoxicity and hemolytic activity, a HEK-293 cell line and human red blood cells (RBCs) were used, respectively. Evaluation of the physicochemical properties of dendrocin-ZM1, as an AMP, indicated a net charge of + 7 and a hydrophobicity percentage of 54%. This peptide had an amphipathic alpha-helical conformation. It exhibited broad-spectrum antibacterial activities against the tested strains at minimum inhibitory concentrations (MICs) of 4–16 μg/mL. Besides, this peptide showed negligible hemolysis and cytotoxicity in the MIC range. It also exhibited heat stability at temperatures of 20 to 80 °C and was active in a broad pH range (from 6.0 to 10.0). Overall, the present results suggested dendrocin-ZM1 as a remarkable antimicrobial candidate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Escherichia coli, as an intestinal and extra-intestinal pathogen, is responsible for various diseases, ranging from urinary tract infection (UTI) and diarrhea to neonatal meningitis and bacteremia. Recent evidence suggests a dramatic increase in hospital-acquired E. coli infection and its heavy burden around the world, causing significant morbidity and mortality [1]. Besides, Staphylococcus aureus is a major cause of skin and soft tissue infections, bacteremia, and life-threatening diseases in both community and healthcare settings [2]. Similarly, a dramatic increase has been recently reported in the prevalence of S. aureus infections worldwide, associated with significant morbidity, mortality, and hospital costs. In the USA, medical costs related to S. aureus infections range from $18,588 to $29,069 per patient [3].
Generally, infections caused by bacteria are intensified by the emergence of drug-resistant strains [4]. Recent studies have reported an increase in the prevalence of multidrug-resistant (MDR) bacteria, which has complicated the availability and use of antibiotics for the treatment of patients and management of diseases [3, 4]. In the past few decades, many studies have indicated an increase in the clinical isolates of vancomycin-resistant S. aureus (VRSA) and vancomycin-intermediate S. aureus (VISA) in different areas. The increased prevalence of these isolates has become an emerging global concern and a public health challenge [3]. Besides, these resistant bacteria have posed a major challenge to the treatment of infections; consequently, discovery of new alternatives for the treatment of these infections is necessary [5].
Among different compounds with antibacterial activities, the use of antimicrobial peptides (AMPs) in resistance-free therapies against infectious diseases has shown the most promising results; therefore, by increasing our knowledge of these peptides, we can address the issue of resistant bacteria [6]. AMPs are known as peptides that contribute to the host defense. They are small-molecular-weight proteins (2–10 kDa), which can be found in living organisms and act as first-line defense against a wide range of pathogens [6, 7]. AMPs are molecules with short amino acid residues (10–50). They are amphipathic and positively charged, with strong thermal stability, broad-spectrum antimicrobial activity, and low immunogenicity [7]. They are well known for having multiple targets in the plasma membrane, low relative cytotoxicity, rapid killing of MDR microorganisms, effectiveness at low concentrations, and low antimicrobial resistance of pathogens. Besides, they exhibit antioxidant, anti-inflammatory, wound-healing, anticancer, and immunomodulatory properties [6].
Although AMPs are isolated from a wide spectrum of sources, plants are one of the most important natural sources of AMPs with numerous therapeutic potentials. AMPs are isolated from different parts of plants, including fruits, seeds, leaves, flowers, and roots [6, 7]. In the past several decades, many studies have been conducted on medicinal plants [2, 3, 5].
Zataria multiflora Boiss (ZM), known as “Shirazi thyme” in Iran, is one of the popular plants with medicinal properties. This plant, which belongs to the Lamiaceae family, is a valuable medicinal plant, which grows in different geographic areas, such as India, Pakistan, and Afghanistan, as well as central and southern parts of Iran. It has a broad range of biological properties, including antimicrobial, antinociceptive, spasmolytic, and anti-inflammatory effects [6, 7]. However, given the scarcity of information, in this study, we aimed to isolate, purify, and characterize novel AMPs from ZM, with effective antimicrobial activities against E. coli ATCC, S. aureus ATCC, and clinical VRSA strains.
Materials and Methods
Test Organisms and Plant Collection
E. coli ATCC 25,922 and ATCC 35,218, S. aureus ATCC 25,923, and ATCC43300 strains were used in the present study. A VRSA clinical strain was kindly provided by the Department of Microbiology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran. All strains were cultured in Trypticase Soy Broth (Merck, Darmstadt, Germany) containing 25% glycerol and stored at −70 °C. The bacterial strains were grown on Nutrient agar (Merck, Darmstadt, Germany) medium. All were incubated aerobically for 24 h at 37 °C, accompanied by the control strain [8]. In the current study, the sampling collected mature and fresh leaves of ZM in March 2020 from Shiraz (Fars province, South of Iran). The taxonomic identities of plants were authenticated in the herbarium of the Research Institute of Forests and Rangelands, Tehran, Iran.
Extraction and Purification of the Peptide
Isolation of antibacterial peptides from the ZM leaves was performed based on the method explained by Seyedjavadi et al. [8]. At first, approximately 400 g of ZM leaves were frozen and grounded to a fine flour in liquid nitrogen using a porcelain mortar then stored in a − 80 °C freezer for further investigation. The powder was treated with two liters of extraction buffer containing 10 mM Na2HPO4, 100 mM KCl, 15 mM NaH2PO4, and 1.5% EDTA (Sigma-Aldrich, St. Louis, MO, USA) at 4 °C for 4 h in physiological pH under continuous stirring to extract the peptide leaves. Afterward, the crude extract was centrifuged at 3900 g at 4 °C for 20 min, and subsequently, the supernatant was filtered. The resultant supernatant was saturated with 85% ammonium sulfate ((NH4)2SO4) (Sigma-Aldrich, St. Louis, MO, USA) and stirred continuously at 4 °C for 24 h. The resulting precipitated peptides were clarified by centrifugation at 6900 g at 4 °C for 20 min. In the next step, after dissolving the pellets in distilled water, the residual (NH4)2SO4 was removed by dialysis against de-ionized water using benzoylated membrane performance (MWCO 2000 Da) (Sigma-Aldrich, St. Louis, MO, USA) at 4 °C within 12 h. Insoluble debris was also eliminated by centrifugation of the dialyzed suspension at 6900 g at 4 °C for 10 min; then, the resultant supernatant was accumulated for the purification of the peptide. The protein extract was subjected to an ultracentrifugal filter with a 10 kDa cutoff (Millipore, Bedford, MA, USA) for screening peptides with low molecular weight, and the resultant filtrated solution was concentrated using a 1 kDa ultra-membrane and then lyophilized by freeze-drying [9].
In order to purify the antibacterial peptides, 15 mg lyophilized extract was dissolved in 1 mL distilled water and 250 μL of which was injected to reverse phase-high performance liquid chromatography (RP-HPLC) (C18 column, 7.8 × 300 mm; Tosoh, Tokyo, Japan) using the solution A (0.1% TFA in water) combined with the gradient of 5 to 65% (v/v) solution B (0.098% TFA in acetonitrile). All chromatographic steps were implemented at the flow rate of 1 mL/min for 85 min. The resulting peptide from the column was investigated at a wavelength of 220 nm. Based on the fractions obtained during HPLC, each fraction was manually collected then lyophilized by the freeze-dryer. Fractions were evaluated to determine antibacterial activity, and those that had the highest activity were collected. Fractions were re-injected onto HPLC under the aforementioned conditions to ensure the purity of the fractions with antibacterial activity [8, 9].
SDS-PAGE
Analysis of molecular weight and purity of individual fractions was carried out by tricine-sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions on a Bio-Rad electrophoresis apparatus as described by Schagger and Von Jagow with minor modifications [10]. Products were run on a 12% Tris-tricine gel with tricine-SDS running buffer for 65 min in 210 V. After electrophoresis, protein bands were visualized by silver staining then the molecular mass of novel peptide was approximately determined by comparing of its electrophoretic mobility with molecular mass of protein ladder (2–250 kDa) [8].
Measurement of In vitro Antibacterial Activity
The effect of the collected peptide on the growth of the E. coli ATCC 25,922 and S. aureus ATCC 25,923 strains was investigated as previously stated by Tam et al. [11]. For the first step, the bacterial inoculum was prepared based on the clinical and the laboratory standards institute (CLSI) guideline (CLSI, 2019). In the study for the first step, a suspension of overnight bacterial culture with turbidity adjusted to the 0.5 McFarland standard (equivalent to 108 colony forming units (CFU)/mL) using a spectrophotometer (optical density (OD) 625 nm of ~ 0.2) was prepared in Müller-Hinton broth medium (MHB) (Merck, Darmstadt, Germany). Afterward, 50 μL of suspension was added to10 mL of MHB, resulting in an inoculum of 106 CFU/mL used in the earlier methods. For antibacterial activity assays first, bacterial inoculum containing approximately 106 cells/mL was incubated in MHB containing different concentrations of the fractions obtained from ZM, and the final assay volume was adjusted to 200 μL. The assay was performed in 96-well flat bottomed microtiter polystyrene plates for 24 h at 30 °C. The cell growth was determined by OD, monitored every 6 h, using a microplate reader at a wavelength of 620 nm. Every experiment was performed in triplicate.
MIC Assay
According to the CLSI, broth microdilution method was done to study the minimal inhibitory concentration (MIC) value of the purified peptide against the bacteria (CLSI, 2019). In this experiment, susceptibility testing was assessed in 96-well flat bottomed microtiter polystyrene plates. The peptide solutions were prepared fresh in each run test and diluted twofold. The range of concentrations assayed for the purified peptide was 1–256 μg/mL. In brief, the pure colony of bacteria overnight culture was suspended in 2 mL of MHB and kept in an incubator at 37 °C to reach 4 × 106 CFU/mL. The suspension above was diluted with fresh MHB culture media to reach a concentration of ~106 CFU/mL and placed in the wells of a 96-well plate. Each well was contained with 90 μL of diluted bacterial suspension and 10 μL of serially diluted peptide and incubated for 18 h at 37 °C. Next, the absorbance was measured spectrophotometrically at a wavelength of 600 nm using an ELISA reader, and the findings were compared to the control sample. This experiment was done in triplicate. Pure broths with inoculum suspensions and pure broths without bacterial or peptides were considered positive and negative controls [12]. MIC assay refers to the lowest antimicrobial agent concentration at which the microorganism’s visible growth was inhibited.
MBC Assay
To determine the lowest concentration of peptide required to kill bacteria, 20 µL of the mixture from each well showing no visible bacterial growths plated on Mueller–Hinton agar (Merck, Darmstadt, Germany) and after overnight incubation, the concentration that caused no bacterial growth was considered as minimum bactericidal concentration (MBC) value [8].
Amino Acid Sequence Analysis
The active fraction was lyophilized and subjected to the amino acid sequence. Edman degradation method was employed to identify the amino acid sequence of the purified antimicrobial peptide. For this, an ABI Procise Edman Micro Sequencer (Model 492) was connected online to an ABI PTH Amino Acid Analyzer (Model 140 C) [8, 9].
Sequence Alignment and Phylogenetic Tree
The search was done in the antimicrobial peptide database to find similar peptides with the highest similarity to our new peptide (https://www.aps.unmc.edu/AP/main.php). After the search, 13 peptides with the highest similarity to new peptides were identified. The sequence of identified peptides with our new desired peptide was aligned using the primary local alignment search tool (BLAST) program (https://www.ncbi.nlm.nih.gov/BLAST). After manually adjusting alignment, a phylogenetic tree was generated by CLC main workbench software. The phylogenetic tree was assessed using bootstrap analysis with 100 replications to calculate the reproducibility of the tree topology [8].
Bioinformatics and Physicochemical Analysis
The measurement of physicochemical parameters was carried out using the Prot Param (ExPASy Proteomics Server: (http://www.expasy.org/tools/protparam HTML). Its AMP probability was evaluated by four machine learning algorithms such as support vector machine (SVM), random forest (RF), artificial neural network (ANN), and discriminant analysis (DA) from the CAMPR3 server (http://www.camp.bicnirrh.res.in/ 2019). The threshold of each algorithm is between 0.5 and 1. The peptide is AMP if this threshold number is > 0.5. The specific kind of antimicrobial activity (such as antibacterial, antifungal, antiviral) of the peptide, the prediction server iAMPpred (http://cabgrid.res.in:8080/amppred/server.php) have been used. The secondary structure of SM-985 was predicted using the PSIPRED server (http://bioinf.cs.ucl.ac.uk/psipred/). A helical wheel diagram was drawn using the online software to predict the position of amino acids in the peptide (http://lbqp.unb.br/NetWheels/). The three-dimensional model (3D) of the peptide was generated using I-TASSER (http://zhanglab.ccmb.med.umich.edu/ITASSER/). Accelrys, DS visualizer ve, assessed the model quality. 1.7. To predict the hemolytic property of the peptide, the HemoPI server (http://crdd.osdd.net/raghava/hemopi/) was used (SVM score ranges between 0 and 1, i.e., 1 very likely to be hemolytic, 0 very unlikely to be hemolytic) [13].
Cytotoxicity Assay
To evaluate the toxicity of the novel peptide against HEK293 cell line, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was employed. For this purpose, cultured HEK293 cell line in Dulbecco’s modified Eagle’s medium (DMEM) (supplemented with 10% fetal bovine serum) was added to 96-well plates with approximately 1 × 105 cells per well. After incubation for 18 h under optimal conditions, including 5% CO2 at 37 °C in a humidified incubator, the cells were treated with different peptide concentrations for 24 h. Subsequently, 10 µL of the MTT (0.5 mg/mL in phosphate-buffered saline, PBS) (Sigma-Aldrich, St. Louis, MO, USA) was added to each well and allowed to incubate for another 4 h under the same conditions. After incubation, the supernatants of each well were discarded, and then 100 μL of DMSO (dimethyl sulfoxide) was added to the wells with gentle shaking to dissolve the formazan crystals. The OD of each well was measured spectrophotometrically at a wavelength of 570 nm using an ELISA reader. Cell viability was estimated as follows:
1% Triton X-100 and PBS were used as the positive and negative controls for MTT assay [8, 9]. Each test was performed three times.
Hemolytic Assay
To investigate the hemolytic activities of the novel peptide, primarily obtained human RBCs from fresh whole blood, a person with blood type O was washed five times using sterile PBS, then centrifuged at 3900 g for 15 min. In the next step, RBCs were resuspended in PBS to reach a final concentration of 4% (v/v). Subsequently, 20 μL of serial dilutions of novel peptide and 80 μL of the RBC suspension were added to a 96-well microtiter plate and incubated for 1 h at 37 °C. After incubation, samples were centrifuged at 3900 g for 5 min, and soon afterward, the supernatant was transferred into a new 96-well plate, then absorbance of the supernatant indicating hemoglobin release was measured at 570 nm. In this experiment, RBCs treated with PBS were used as negative control and 0.1% Triton X-100 (Sigma-Aldrich, St. Louis, MO, US) was used as the positive control in each run test. All the experiments were performed in triplicate [14]. Hemolysis percent was estimated as follows:
Evaluation of the Influence of Temperature and pH on the Activity of Novel Peptide
In vitro thermal and pH activity of the peptide isolated from ZM was determined based on the standard method described by Chan and colleagues. For the thermostability analysis, 100 µL of the active peptide was heated at 20–100 °C for 1 h. The RDA assay estimated the antibacterial activity against S. aureus ATCC 25,923 as the indicator strain as previously described by Wang et al. As a control, a non-heated peptide at temperatures above was applied.
The pH was ranged from 2 to 12 to investigate the pH effect on the isolated peptide. pH stability was estimated by incubation of the peptide solution adjusted in different pH values (1 N NaOH or 1 N HCl) for 1 h at 25 °C. After incubation, the pH of a mixture containing the peptide was adjusted to 7.2 with a 0.5 M sodium citrate buffer. The antibacterial activity of peptide was evaluated using an RDA assay against S. aureus ATCC 25,923. The peptide dissolved in 100 µL of solution (pH of 7.2) was taken as control. All the experiments were performed in triplicate [8, 9].
Data and Statistical Analysis
GraphPad Prism 5 Statistical software (GraphPad Software, Inc., La Jolla, CA, USA) was applied for statistical analyses. All experiments were performed in triplicate on three separate occasions.
Results
Peptide Isolation and Purification
The low molecular weight peptides obtained from centrifugal filters were lyophilized and subjected to C18 RP-HPLC. Fourteen fractions were obtained (Fig. S1a) and all the fractions were collected as individual fractions. The antibacterial activity was tested for each fraction. Out of 14 fractions, fraction 5 had the best antibacterial activity. The active fraction was re-injected on the same column under the same elution conditions (Fig. S1b).
Growth Inhibition Assay and Antimicrobial Activity
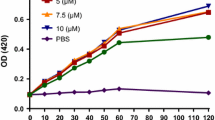
A growth inhibition assay was performed with the 14 fractions obtained by RP-HPLC against S. aureus ATCC 25,923 and E. coli ATCC 25,922 strains (data not shown), and just 4 fractions (F3, F5, F7, and F10) from Zataria multiflora demonstrated significant differences in comparison with the control (absence of peptide fractions) (Fig. 1). The F3 fraction 16.1% and 17.2%, the F7 fraction 13.5% and 15.6%, and the F10 fraction 18.7% and 19.4% showed growth inhibition against S. aureus and E. coli, respectively. The F5 fraction showed the highest inhibition activity with 64.8% and 69.3% against S. aureus and E. coli growth inhibition, respectively.
Assay MIC values of the peptide were determined using micro broth dilution against S. aureus and E. coli strains. As shown in Table 1, the MICs of peptide for S. aureus, MRSA, and VRSA were 8, 16, and 16 µg/mL, respectively. The MIC values of the peptide against the E. coli were 4–8 µg/mL, respectively. The MFC values were equal or two times higher than MIC values across bacterial strains.
Evaluation of Purity of the Peptide
The purity of fraction 5 on the SDSPAGE has been shown in Fig. S1c. After the SDS-PAGE analysis, the purified peptide indicated a single band with a molecular weight of about 3.7 kDa.
Peptide Identification
The peptide band of the F5 fraction was sequenced using the Edman degradation procedure to determine the amino acid sequence. As a result, a fragment of 33 amino acid residues was obtained (TTLRLNTLAYKVAWLVNVKAFWAAGRALKKVGR).
Sequence Alignment and Phylogenetic Tree
The sequence of this peptide demonstrated no complete homology to any known AMPs. After BLAST analysis, we observed that the peptide was a novel antimicrobial peptide from Zataria multiflora Boiss. Sequence alignment and phylogenetic tree showed that the new peptide had the highest sequence similarity with dendrocin obtained from bamboo shoots, Dendrocalamus latiflora Munro. The results are presented in Fig. 2a, b. Thus, this new peptide was named dendrocin-ZM1 according to the source of peptide purification (Zataria multiflora Boiss).
The alignment (a) and phylogenetic tree (b) of dendrocin-ZM1. a The alignment of dendrocin-ZM1 amino acid sequence as well as the sequences related to other antimicrobial peptides. b Phylogenetic tree of dendrocin-ZM1. Phylogenetic tree was acquired by CLC main workbench software. Each sequence name is written at the end of the relevant branch. The tree reliability was determined using bootstrap method with 100 replications
Bioinformatics Analysis
According to the online prediction servers CAMPR3 (Table 2) and iAMPpred, dendrocin-ZM1 is AMP and has antibacterial activity. The physicochemical parameters and sequences of dendrocin-ZM1 are given in Table 2. The calculated molecular weight of the peptide is 3716.48 Da. Dendrocin-ZM1 has 33 amino acids, and based on the physicochemical properties, dendrocin-ZM1 displayed AMP properties due to the high positive charge and hydrophobic ratio (+ 7 and 54%, respectively) and protein-binding potential (Boman index) value of 0.81 kcal/mol. The instability and aliphatic index value for dendrocin-ZM1 were 0.98 and 112.42, respectively. Online server PSIPRED was applied to predict the secondary structure of the dendrocin-ZM1 peptide, and the result showed that dendrocin-ZM1 contained an α-helix structure (Fig. 3a).
a Graphical result from secondary structure prediction of dendrocin-ZM1 using PSIPRED. b Helical wheel diagram of dendrocin-ZM1 showing the amphipathic α-helix conformation. c The dendrocin-ZM1 3D structure contains α-helix, as predicted by I-TASSER. The 3D structure model visualized using Accelrys discovery studio visualizer software
A helical wheel diagram indicated that the dendrocin-ZM1 peptide had amphipathic α-helix conformations, with both hydrophilic and hydrophobic faces (Fig. 3b). Results of the tertiary structure prediction by the server I-TASSER indicated that the dendrocin-ZM1 peptide contains an α-helix structure (Fig. 3c). The evaluation by I-TASSER also showed that C-score, a confidence score to calculate the models’ global accuracy (in the range of −5.0 to 2.0), for our new peptide was 1.16.
Hemolytic Activity and Cytotoxicity
The results of hemolytic activity of the peptide on RBCs demonstrated (Fig. 4a) that dendrocin-ZM1 had slight hemolytic activity (> 5%) at concentrations the MIC doses (4–16 μg/mL) on human RBCs. As the concentration of this antimicrobial peptide increases, the percentage of hemolysis increases. At the highest concentration (128 μg/mL), it showed 17% hemolytic activity. The results were the mean of 3 independent experiments. The cytotoxicity of the peptide against HEK293 cell line was further evaluated after 24 h of treatment by using MTT assay (Fig. 4b). Dendrocin-ZM1 demonstrated mild cytotoxicity (from 2.3 to 5.3%) at MIC doses (4–16 μg/mL). The cell death rates of dendrocin-ZM1 were 7.3 and 12.4% at concentrations of 64 and 128 μg/mL, respectively.
Hemolytic activity, cytotoxicity and stability of dendrocin-ZM1. a Hemolytic activity of dendrocin-ZM1 against human erythrocytes. b Cytotoxicity assay of dendrocin-ZM1 on HEK293 mammalian cell line using MTT assay. The effects of pH (c) and temperature (d) on the antibacterial activity of dendrocin-ZM1 against S. aureus ATCC 25,923 cells
Effects of pH and Temperature on Antimicrobial Activity
As shown in Fig. 4c, the stability of the antibacterial peptide was evaluated in different pH values. The results demonstrated that the dendrocin-ZM1 remained stable after 1 h at pH values ranging from 6 to 10. However, its activity remarkably decreased when the pH values were below 5. The effect of temperature on the dendrocin-ZM1 (Fig. 4c) displayed that this peptide was stable at different temperatures ranging from 20 to 80 °C during 1 h of treatment.
Discussion
The emergence of MDR bacterial strains has become a serious health challenge. Some studies have focused on a new generation of antimicrobial molecules to treat infections caused by MDR strains [4]. AMPs are a new generation of potential antibiotics, with an antimicrobial mechanism comparable to that of traditional antibiotics [6, 7]. Due to their importance, different studies have been conducted to investigate new AMPs [2, 5, 15,16,17]. In the present study, we succeeded to isolate a novel AMP, dendrocin-ZM1, from ZM with broad-spectrum activities against both gram-positive and gram-negative bacteria. According to the BLAST results for dendrocin-ZM1, no sequence of this peptide was completely homologous to other AMP sequences registered in scientific databases, which indicates its novelty as an AMP derived from ZM.
In this study, the predictive and physicochemical properties of the new AMP were investigated in silico. Before in vitro experiments, dendrocin-ZM1 was examined using some online tools. This peptide contained 33 amino acids and seven positively charged residues, including three arginine and four lysine residues, creating a net charge of + 7, which is important for electrostatic interactions with negatively charged components in the cell membranes of bacterial strains [18]. Dendrocin-ZM1 also contained ten 13-residue hydrophobic regions with a hydrophobic percentage of 53%, which facilitated interactions with the membrane and consequently disrupted the integrity of bacterial membrane [19, 20]. Studies show that hydrophobicity is one of the important parameters in the antimicrobial activity of AMPs, which creates some pores in the membrane of bacteria; these pores can cause degradation in the bacterial membrane and destroy the organism [20].
By using the Protein Structure Prediction Server (PSIPRED) to predict the secondary structure of dendrocin-ZM1, it was found that this peptide had an α-helical conformation; this conformation was related to the antimicrobial activity of AMPs [21]. In this regard, some earlier studies have reported some AMPs with a similar structure [22, 23], which plays an important role in antimicrobial activity. The helical wheel of dendrocin-ZM1 represents the amphipathic α-helical conformation of this AMP. It is known that peptides with α-helical structures are an important class of cytolytic AMPs [24]. Another study showed that α-helical AMPs with an amphipathic structure can dramatically increase the potential antibacterial activity, as they can interact with the membranes [25, 26]. Most of these peptides consist of a charged part for membrane interaction and a hydrophobic part for insertion into the lipid bilayer. This physical interaction and membrane lysis can be explained by the carpet model for AMPs [26, 27]. In this model, the α-helical peptide can change the bacterial membrane permeability, leading to the loss of membrane integrity, degradation, and disturbance of ionic balance. In peptides with α-helical structures, insertion of amphipathic parts and snorkel effects, along with pore formation in the membrane, are the mechanisms of action [28,29,30].
A previous study revealed that the polar side of peptides is associated with the binding of AMP phospholipid to the cell membrane, while the hydrophobic side is inserted into the membrane [31].
Different mechanisms have been defined to explain the effects of AMPs, including damage to the cell membrane, intracellular target interactions (inhibition of synthesis of nucleic acids, proteins, and cell wall), and apoptosis induction [32]. The prediction results and physiochemical properties of dendrocin-ZM1 suggest that this new peptide has antibacterial activities by affecting the membrane permeability. Therefore, it may interact with the microbial membrane and damage the integrity of bacterial cell membranes. However, further studies are needed to determine the exact antimicrobial function of dendrocin-ZM1.
To explore the therapeutic potential of dendrocin-ZM1, the MIC and MBC assays were performed against gram-positive and gram-negative bacteria (S. aureus and E. coli). Dendrocin-ZM1 showed the strongest antimicrobial activity to both standard microbial strains and resistant strains. Interestingly, it exhibited a higher antimicrobial activity against E. coli with MIC of 4–8 μg/mL, compared to S. aureus with MIC of 8–16 μg/mL, which might be related to the bacterial cell wall [33].
Some important challenges related to the use of chemical compounds in the healthcare setting are cytotoxicity and hemolysis; therefore, assessment of these factors is essential. As shown in Fig. 4a, b, dendrocin-ZM1 exhibits slight hemolytic and cytotoxic activities at concentrations exceeding the MIC range. Regarding cytotoxicity, another study demonstrated that hydrophobicity was correlated with peptide cytotoxicity [34].
In addition to cytotoxicity and hemolysis, another important factor is the stability of peptide. Two important factors in stability are pH and heat; therefore, we evaluated the effects of these variables on the antimicrobial activity of dendrocin-ZM1. Surprisingly, dendrocin-ZM1 showed significant stability in a wide pH range of 6–10. Moreover, it showed acceptable stability in different temperature ranges. In conclusion, the antimicrobial activity of dendrocin-ZM1 at 20–80 °C indicates the good thermal stability of this peptide. Our findings were in line with another previous study [35]. Moreover, we had previously calculated the stability of our AMP in the antimicrobial peptide database (APD), which revealed its acceptable stability (Table 2). On the other hand, based on our findings, heat could not change the primary structure of the peptide and destroyed it [36].
Present research has several limitations worth noting. Firstly, it was limited to in vitro findings, and thus there is a need for further in vivo study. Secondly, due to some of the technical limitations mode of action of AMP were not investigated in the current study. Throughout the study, we believe that AMPs have great potential as an alternative therapeutic option to conventional drugs to treat infections, a belief that is strongly supported by the many positive results shown in this study. The field of AMP research is constantly evolving, and AMP databases have amassed a large quantity of data on the subject. However, further researches are needed to determine the relationship between different physicochemical properties and the specificity and synergistic capacity of AMPs to obtain low-cost and highly safe AMPs with remarkable antimicrobial effects. More research on toxicity against animal or human cells, modes of action, in vivo effects, and negative and positive interactions with commonly used antibiotics is also required. The critical issue is to develop the most efficient methods for isolating and purifying newer and safer naturally occurring antimicrobial peptides that are effective against pathogens.
Conclusion
The present study investigated a novel AMP, called dendrocin-ZM1, with an α-helical structure isolated from ZM. Dendrocin-ZM1 showed very promising antimicrobial activities against VRSA and the tested strains, while it exhibited negligible cytotoxic and hemolytic activities. The results revealed that dendrocin-ZM1 could be used as a novel antimicrobial agent to combat bacterial infections. However, further research is needed to better understand the function of this new peptide in the bacterial cell.
Availability of Data and Material
The datasets generated and analyzed during this study were included in the main document of this manuscript.
References
MacKinnon MC, Sargeant JM, Pearl DL, Reid-Smith RJ, Carson CA, Parmley EJ, McEwen SA (2020) Evaluation of the health and healthcare system burden due to antimicrobial-resistant Escherichia coli infections in humans: a systematic review and meta-analysis. Antimicrob Resist Infect Control 9:1–22. https://doi.org/10.1186/s13756-020-00863-x
Cheung GY, Bae JS, Otto M (2021) Pathogenicity and virulence of Staphylococcus aureus. Virulence 12(1):547–569. https://doi.org/10.1080/21505594.2021.1878688
Shariati A, Dadashi M, Moghadam MT, van Belkum A, Yaslianifard S, Darban-Sarokhalil D (2020) Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci Rep 10:1–16. https://doi.org/10.1038/s41598-020-69058-z
Zaman SB, Hussain MA, Nye R, Mehta V, Mamun KT, Hossain N (2017) A review on antibiotic resistance: alarm bells are ringing. Cureus 9:e1403. https://doi.org/10.7759/cureus.1403
Ebbensgaard A, Mordhorst H, Overgaard MT, Nielsen CG, Aarestrup FM, Hansen EB (2015) Comparative evaluation of the antimicrobial activity of different antimicrobial peptides against a range of pathogenic bacteria. PLoS One 10:e0144611. https://doi.org/10.1371/journal.pone.0144611
Lei J, Sun L, Huang S, Zhu C, Li P, He J, Mackey V, Coy DH, He Q (2019) The antimicrobial peptides and their potential clinical applications. Am J Transl Res 11:3919–3931
Sajed H, Sahebkar A, Iranshahi M (2013) Zataria multiflora Boiss. (Shirazi thyme)—an ancient condiment with modern pharmaceutical uses. J Ethnopharmacol 145:686–698. https://doi.org/10.1016/j.jep.2012.12.018
Seyedjavadi SS, Khani S, Zare-Zardini H, Halabian R, Goudarzi M, Khatami S, Imani Fooladi AA, Amani J, Razzaghi-Abyaneh M (2019) Isolation, functional characterization, and biological properties of MCh-AMP1, a novel antifungal peptide from Matricaria chamomilla L. Chem Biol Drug Des 93:949–959. https://doi.org/10.1111/cbdd.13500
Khani S, Seyedjavadi SS, Zare-Zardini H, Hosseini HM, Goudarzi M, Khatami S, Amani J, Fooladi AA, Razzaghi-Abyaneh M (2019) Isolation and functional characterization of an antifungal hydrophilic peptide, Skh-AMP1, derived from Satureja khuzistanica leaves. Phytochemistry 164:136–143. https://doi.org/10.1016/j.phytochem.2019.05.011
Schägger H, Von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166:368–379. https://doi.org/10.1016/0003-2697(87)90587-2
Tam JP, Wang S, Wong KH, Tan WL (2015) Antimicrobial peptides from plants Pharmaceuticals 8:711–757. https://doi.org/10.3390/ph8040711
Lee K-S, Yun E-Y, Goo T-W (2020) Evaluation of the antimicrobial activity of an extract of Lactobacillus casei-infected Hermetia illucens larvae produced using an automatic injection system. Animals 10:2121. https://doi.org/10.3390/ani10112121
Chaudhary K, Kumar R, Singh S, Tuknait A, Gautam A, Mathur D, Anand P, Varshney GC, Raghava GP (2016) A web server and mobile app for computing hemolytic potency of peptides. Sci Rep 6:1–13. https://doi.org/10.1038/srep22843
Asoodeh A, Zardini HZ, Chamani J (2012) Identification and characterization of two novel antimicrobial peptides, temporin-Ra and temporin-Rb, from skin secretions of the marsh frog (Rana ridibunda). J Pept Sci 18:10–16. https://doi.org/10.1002/psc.1409
Aguilar-Toalá JE, Deering AJ, Liceaga AM (2020) new insights into the antimicrobial properties of hydrolysates and peptide fractions derived from chia seed (Salvia hispanica L.). Probio Antimicrob Proteins 12:1571–1581. https://doi.org/10.1007/s12602-020-09653-8
Hu B, Pan Y, Li Z, Yuan W, Deng L (2019) EmPis-1L, an effective antimicrobial peptide against the antibiotic-resistant VBNC state cells of pathogenic bacteria. Probio Antimicrob Proteins 11:667–675. https://doi.org/10.1007/s12602-018-9446-3
Noll KS, Sinko PJ, Chikindas ML (2011) Elucidation of the molecular mechanisms of action of the natural antimicrobial peptide subtilosin against the bacterial vaginosis-associated pathogen Gardnerella vaginalis. Probio Antimicrob Proteins 3:41–47. https://doi.org/10.1007/s12602-010-9061-4
Dathe M, Nikolenko H, Meyer J, Beyermann M, Bienert M (2001) Optimization of the antimicrobial activity of magainin peptides by modification of charge. FEBS Lett 501:146–150. https://doi.org/10.1016/S0014-5793(01)02648-5
Kim H, Jang JH, Kim SC, Cho JH (2014) De novo generation of short antimicrobial peptides with enhanced stability and cell specificity. J Antimicrob Chemother 69:121–132. https://doi.org/10.1093/jac/dkt322
Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS (2007) Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob Agents Chemother 51:1398–1406. https://doi.org/10.1128/AAC.00925-06
Park CB, Yi K-S, Matsuzaki K, Kim MS, Kim SC (2000) Structure–activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. Proc Natl Acad Sci U S A 97:8245–8250. https://doi.org/10.1073/pnas.150518097
Bonduelle C (2018) Secondary structures of synthetic polypeptide polymers. Polym Chem 9:1517–1529. https://doi.org/10.1039/C7PY01725A
Shen W, He P, Xiao C, Chen X (2018) From antimicrobial peptides to antimicrobial poly (α-amino acid) s. Adv Healthc Mater 7:1800354. https://doi.org/10.1002/adhm.201800354
Zelezetsky I, Tossi A (2006) Alpha-helical antimicrobial peptides—using a sequence template to guide structure–activity relationship studies. Biochim Biophys Acta 1758:1436–1449. https://doi.org/10.1016/j.bbamem.2006.03.021
Khara JS, Obuobi S, Wang Y, Hamilton MS, Robertson BD, Newton SM, Yang YY, Langford PR, Ee PL (2017) Disruption of drug-resistant biofilms using de novo designed short α-helical antimicrobial peptides with idealized facial amphiphilicity. Acta Biomater 57:103–114. https://doi.org/10.1016/j.actbio.2017.04.032
Ginsburg I (2004) Bactericidal cationic peptides can also function as bacteriolysis-inducing agents mimicking beta-lactam antibiotics?; it is enigmatic why this concept is consistently disregarded. Med Hypotheses 62:367–374. https://doi.org/10.1016/j.mehy.2003.11.017
Saini SS, Chopra AK, Peterson JW (1999) Melittin activates endogenous phospholipase D during cytolysis of human monocytic leukemia cells. Toxicon 37:1605–1619. https://doi.org/10.1016/S0041-0101(99)00110-5
Crovella S, Antcheva N, Zelezetsky I, Boniotto M, Pacor S, Falzacappa MV, Tossi A (2005) Primate β-defensins-structure, function and evolution. Curr Protein Pept Sci 6:7–21. https://doi.org/10.2174/1389203053027593
Zhao C, Nguyen T, Boo LM, Hong T, Espiritu C, Orlov D, Wang W, Waring A, Lehrer RI (2001) RL-37, an alpha-helical antimicrobial peptide of the rhesus monkey. Antimicrob Agents Chemother 45:2695–2702. https://doi.org/10.1128/AAC.45.10.2695-2702.2001
Huang Y, Huang J, Chen Y (2010) Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein Cell 1:143–152. https://doi.org/10.1007/s13238-010-0004-3
He J, Luo X, Jin D, Wang Y, Zhang T (2018) Identification, recombinant expression, and characterization of LGH2, a novel antimicrobial peptide of Lactobacillus casei HZ1. Molecules 23:2246. https://doi.org/10.3390/molecules23092246
Moravej H, Moravej Z, Yazdanparast M, Heiat M, Mirhosseini A, Moosazadeh Moghaddam M, Mirnejad R (2018) Antimicrobial peptides: features, action, and their resistance mechanisms in bacteria. Microb Drug Resist 24(6):747–767. https://doi.org/10.1089/mdr.2017.0392
Silhavy TJ, Kahne D, Walker S (2010) The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. https://doi.org/10.1101/cshperspect.a000414
Wiradharma N, Sng MY, Khan M, Ong ZY, Yang YY (2013) Rationally designed α-helical broad-spectrum antimicrobial peptides with idealized facial amphiphilicity. Macromol Rapid Commun 34:74–80. https://doi.org/10.1002/marc.201200534
Wang G, Li X, Wang Z (2009) APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res 37:D933–D937. https://doi.org/10.1093/nar/gkn823
Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ (1992) Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111:229–233. https://doi.org/10.1016/0378-1119(92)90691-H
Funding
This work is financially supported by a research grant from Deputy of Research, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran [Grant No. 20359]. The funding agency has no role in the design of the project, work execution, analyses, interpretation of the data, and manuscript writing and submission as well.
Author information
Authors and Affiliations
Contributions
SSS, MG, and MRA designed the study. MG, SSS, MRA, MH, and MJN reviewed the literature. SSS and MG performed the laboratory investigations. SSS, MG, AH, MH, MD, and HZZ participated in data interpretation. MG, SSS, BP, and MH drafted the manuscript. SSS, MG, MRA, and BP critically reviewed the manuscript and approved the final version of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
The entire procedures were approved by the Ethics Committee of the Shahid Beheshti University of Medical Sciences in Tehran, Iran (IR. SBMU. MSP.REC. 1398. 788). A written informed consent was obtained from participants.
Consent to Participate
Not.
Consent for Publication
Not.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Seyedjavadi, S.S., Razzaghi-Abyaneh, M., Nasiri, M.J. et al. Isolation and Chemical Characterization of an Alpha-Helical Peptide, Dendrocin-ZM1, Derived from Zataria multiflora Boiss with Potent Antibacterial Activity. Probiotics & Antimicro. Prot. 14, 326–336 (2022). https://doi.org/10.1007/s12602-022-09907-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-022-09907-7