Abstract
This study aimed to select beneficial strains from the oral cavity of healthy volunteers and to evaluate these as potential oral probiotic candidates. The selection process was based on the isolation, differentiation, identification, and safety assessment of LAB strains, followed by a series of experiments for the selection of appropriate candidates with beneficial properties. In the screening procedure, 8 isolates from the oral cavity of a Caucasian volunteers were identified as Streptococcus (Str.) salivarius ST48HK, ST59HK, ST61HK, and ST62HK; Lactiplantibacillus plantarum (Lb.) (Lactobacillus plantarum) ST63HK and ST66HK; Latilactobacillus sakei (Lb.) (Lactobacillus sakei) ST69HK; and Lactobacillus (Lb.) gasseri ST16HK based on 16S rRNA sequencing. Physiological and phenotypic tests did not show hemolytic, proteinase, or gelatinase activities, as well as production of biogenic amines. In addition, screening for the presence of efaA, cyt, IS16, esp, asa1, and hyl virulence genes and vancomycin-resistant genes confirmed safety of the studied strains. Moreover, cell-to-cell antagonism indicated that the strains were able to inhibit the growth of tested representatives from the genera Bacillus, Enterococcus, Streptococcus, and Staphylococcus in a strain-specific manner. Various beneficial genes were detected including gad gene, which codes for GABA production. Furthermore, cell surface hydrophobicity levels ranging between 1.58% and 85% were determined. The studied strains have also demonstrated high survivability in a broad range of pH (4.0–8.0). The interaction of the 8 putative probiotic candidates with drugs from different groups and oral hygiene products were evaluated for their MICs. This is to determine if the application of these drugs and hygiene products can negatively affect the oral probiotic candidates. Overall, antagonistic properties, safety assessment, and high rates of survival in the presence of these commonly used drugs and oral hygiene products indicate Str. salivarius ST48HK, ST59HK, ST61HK, and ST62HK; Lb. plantarum ST63HK and ST66HK; Lb. sakei ST69HK; and Lb. gasseri ST16HK as promising oral cavity probiotic candidates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to the World Health Organization, probiotics are defined as “live microorganisms which, when administered in adequate amounts, confer benefits to the health of the host” [1]. With positive effects on human health, probiotics can contribute to the prevention or treatment of different clinical conditions [2]. Positive health benefits of bacterial species on humans were initially suggested by Ilya Ilyich Mechnikov and his collaborator Stamen Grigorov. This concept has since been developed over the last century [3,4,5] and became well established as a powerful tool and even as an adjunct to western medicine in the prevention and treatment of some diseases [6,7,8]. Although the initial concept for the application of probiotics targeted the gastro-intestinal tract (GIT) microbial balance [9], their role and impact on the immune system, skin health, brain, kidney, and liver functions has also been progressively explored in the past decades [10, 11].
The oral cavity, serving as a principal access to the digestive tract, plays an essential role in the wellness of humans and other animals [12]. The oral cavity is a complex structure composed of several elements, such as the teeth, tongue, palate, and buccal mucosa, that are involved in a functional complex. These serve as principal habitats for diverse microorganisms involved in and supporting the normal functional status quo of macroorganisms [13]. It was suggested that more than 700-1000 bacterial species are permanently or transiently colonize the human oral cavity and play a role in the functional balance. Additionally, the biodiversity in the human oral cavity also includes representatives of some viruses, fungi, and even members of the Archaea [2, 14]. The oral cavity of humans and other animals is a habitat for different representatives of lactic acid bacteria (LAB), which comprise a large group of bacteria from the genera Lactobacillus, Lactococcus, Streptococcus, Leuconostoc, Enterococcus, and Weissella, some of which plays a significant role in oral health [12]. LAB are Gram-positive bacteria belonging to the phylum Firmicutes, class Bacilli, and order Lactobacillales, with a low G + C content (less than or equal to 55 mol %) in their DNA. In the human oral cavity, LAB are mainly from the genera Lactobacillus (now subdivided into 25 new genera) and Streptococcus, which account for 29.2% of the whole oral microbiome. LAB are widely spread in nature and are known to be autochthonous, but several representatives are specifically considered as beneficial microorganisms and are described and applied as probiotic cultures [8, 15].
The conditions in the human mouth provide an optimal setting for the growth and colonization of microorganisms. However, the specificity of the saliva composition and presence of different enzymes and bioactive peptides serve as selection pressure for microbial presence and dominance in the mouth [16, 17]. The environmental conditions in the oral cavity also allow successful niching in for various microorganisms due to the optimal temperature, ambient moisture, available nutrients, minerals, vitamins, and various tissue surfaces that enable successful microbial attachment, which ultimately facilitates colonization in this environment. This exemplifies the diverse ecological niches associated with living systems, which are innately equipped with deterrents and mechanisms for effective reduction and/or control of different pathogens. Furthermore, it has been confirmed that the saliva of several animals was found to be highly antiseptic [18, 19].
The role of LAB in the oral cavity needs to be evaluated as either beneficial or detrimental. Several LAB can be considered as beneficial, especially those that are adapted to the mouth, since they promote microbial balance, which subsequently results in good oral health [12, 15]. Some specific LAB or oral pathogens, however, may contribute to oral diseases such as dental caries and periodontitis [20]. Previous studies have shown the relationship between oral pathogens and precancerous lesions of gastric cancer (PLGC), rheumatoid arthritis, and type-2 diabetes, indicating that oral health is an important component of overall human health [8, 21, 22]. The beneficial properties of LAB (and generally for all probiotics) are strain-specific [23]. In the selection process of appropriate probiotic candidates, both for application in the oral cavity and/or other purposes, careful evaluation and consideration of their association with specific bacterial strain(s) is necessary, especially concerning the environment in which they are intended to be applied. Some of the key factors in the evaluation process of new probiotic strains are their interactions with microorganisms typical of the specific ecological niche, its safety, and appropriate identification.
LAB, particularly representatives of the former genus Lactobacillus, were evaluated and applied as potential probiotics in numerous studies [24, 25]. There have been reports of LAB with beneficial effects on the control of dental problems, such as dental caries, which is frequently associated with the oral pathogen Streptococcus mutans, and halitosis, which is commonly caused by Gram-negative bacteria such as Porphyromonas gingivalis [20, 26]. Oral probiotics strains, developed for dental health, have been investigated in clinical studies. Some of these include the application of Streptococcus salivarius strains M18 and K12 as reported by Wescombe et al. [27] and He et al. [29]. It was demonstrated that Str. salivarius M18 is effective in inhibiting the oral pathogen Str. mutans, thereby reducing the risk of dental caries [27, 28]. In the other study, Str. salivarius K12 was found to have a beneficial effect on dental health and supported halitosis treatment post-removal of tongue coating [29].
The aim of this study was to select possible beneficial strains from the oral cavity of healthy volunteers and to evaluate these as potential oral probiotic candidates. The selection process was based on isolation of LAB strains, differentiation, identification, safety assessment, and a series of experiments leading to the selection of potential candidates for the application as oral probiotics.
Materials and Methods
Isolation, Differentiation, Identification, and Selection of LAB Strains with Antimicrobial Activity
For the isolation of potential oral probiotic candidates with antagonistic properties against common oral cavity pathogens, saliva swabs from 26 healthy volunteers of Caucasian and Asian ethnicities were obtained and processed. The modified triple agar layer method was adapted from dos Santos et al. [30] and performed on M17-lactose (Difco, Detroit, MI, USA) and MRS (Difco) supplemented with 1% agar (Difco). Saliva swabs were surface spread on the solid media (M17-lactose and MRS) in triplicates and covered with 10 mL of 1% agar. Plates were incubated aerobically at 37 °C for 24–48 h and examined for bacterial growth. Plates with distinct individual colonies were selected and overlaid with an additional layer of BHI (Difco), supplemented with 1% agar and the test (indicator) organisms (Str. mutans KACC 16833, Streptococcus gordonii KACC 13829, and Listeria monocytogenes ATCC 15313) at 105 CFU/mL (final concentration). Plates were incubated for an additional 24 h at 37 °C and evaluated for the presence of inhibition zones around the previously formed colonies. Individual colonies were isolated and grown in M17-lactose or MRS broth. Preliminary identification of the isolates was done based on the morphology observed via phase contrast microscopy, catalase reaction, and reactions to other physiological and biochemical tests indicated in Bergey’s Manual of Systematic Bacteriology of Archaea and Bacteria [31]. Stock cultures were kept in M17-lactose or MRS broth supplemented with 20% (v/v) glycerol at −20 °C.
The isolates were initially screened for the production of antibacterial metabolites against Str. mutans KACC 16833, Str. gordonii KACC 13829, and L. monocytogenes ATCC 15313 by determining bacteriocin production and cell-to-cell interaction according to dos Santos et al. [30]. Cell-free supernatants (CFS) of the cultures (MRS, 24 h, 37 °C) obtained after centrifugation (12,000 × g, 10 min, 20 °C) were heat-treated for 10 min at 80 °C and spotted on the surface of previously prepared solid BHI medium, supplemented with 1% agar and 105 CFU/mL of Str. mutans KACC 16833, Str. gordonii KACC 13829, or L. monocytogenes ATCC 15313. Plates were incubated for 24 h at 37 °C and evaluated for the presence of inhibition zones. Zones larger than 2 mm were considered as positive. In a parallel experiment, deferred antagonism assay was carried out by spotting 10 μL of exponentially growing cultures of the LAB isolates on the surface of similarly prepared plates. Plates were incubated following the same conditions and examined for the presence of clear inhibition zones. Isolates confirmed to be potentially antagonistic were selected for subsequent experiments.
Putative LAB isolates were cultured in MRS broth for 24 h at 37 °C and subjected to DNA extraction using ZR Fungal/Bacterial DNA Kit (Zymo Research, Irvine, CA, USA) following the manufacturer’s recommendations. The obtained DNA was quantified in SPECTROstar Nano nanodrop (BMG LABTECH, Ortenberg, Germany). Differentiation of the isolates was performed by rep-PCR DNA fingerprinting with the primer (GTG)5 (Table 1) according to de Castilho et al. [32]. PCR reactions were performed in a Veriti 96-well Thermal Cycler, Applied Biosystems (Thermo Scientific, Waltham, MA, USA). Profiling of the generated amplicons was done via gel electrophoresis, run on 1.5% (w/v) agarose gels stained with 0.02 μL/mL of SYBR®Safe (Thermo Scientific) in 1 × TAE buffer at 100 V for 1 h (GH-200 Genera Biosystems, Victoria, Australia; Elite 300 Plus Power Supply, Wealtec Bioscience Co., Ltd., Taiwan). The gel was visualized using Omega Lum™G gel documenter (Aplegen, Inc., Pleasanton, CA, USA). Isolates were grouped according to the generated profiles and representatives of each group were subjected to 16S rRNA sequencing with primers 8F and 1492R (SolGent Analysis Service, Daejeon, Republic of Korea) as commercial service.
Safety Evaluation of the Selected Strains
Phenotypic Safety Test
The investigated strains were subjected to selected phenotypic tests to determine their hemolytic activity and gelatinase production according to Colombo et al. [23] and biogenic amine production following the recommendations of Bover-Cid and Holzapfel [33]. For hemolytic activity, the selected strains were streaked on the surface of trypticase soy agar supplemented with 5% (v/v) defibrinated sheep blood (Synergy Innovation, Seongnam-si, Republic of Korea). Plates were incubated at 37 °C for 24 h and observed for evidence of possible hemolytic activity of each strain. Clear halos around the colonies were considered as β-hemolysis, greenish halos were noted as partial or α-hemolysis, and no change was recorded as γ-hemolysis. Cultures of Bacillus cereus ATCC 27438 was applied as β-hemolysis, Escherichia coli ATCC 25922 as α-hemolysis, and Lb. plantarum ATCC 14197 as γ-hemolysis controls [23].
Gelatinase production was performed according to Colombo et al. [23] with some modifications. Ten-microliter aliquots were deposited on the surface of Luria Bertani (LB) agar (Difco), supplemented with 3% (w/v) gelatin (Difco). Plates were incubated for 48 h at 37 °C and then maintained at 4 °C for 4 h. A positive result for gelatin hydrolysis was exhibited by transformation of the solid agar medium to liquid phase. Bacillus amyloliquefaciens ST109 and Lb. plantarum ATCC 14197 served as positive and negative controls, respectively.
The ability to produce biogenic amines was performed according to Bover-Cid and Holzapfel [33]. The strains were cultured at 37 °C for 24 h for at least five consecutive times in MRS broth supplemented with 0.1% (w/v) of the respective amino acid precursors for the production of biogenic amines. After the last transfer, the strains were streaked in duplicate on a modified MRS agar supplemented with the biogenic amine precursors described above (1%, w/v). The plates were incubated for up to 4 days at 37 °C and change in color from yellow to purple was considered as positive result for the production of a biogenic amine. E. coli ATCC 25922 and Lb. plantarum ATCC 14197 served as positive and negative controls, respectively.
Screening for Presence of Virulence, Biogenic Amines, and Vancomycin Resistance-Associated Genes
As a safety criterion, the DNA previously obtained from the studied strains were evaluated by PCR for the presence of the selected virulence genes (efaA, cyt, IS16, esp, asa1, and hyl) originally recommended by EFSA for Enterococcus spp. (Table 1) [34], Vankerckhoven et al. [35], Leavis et al. [36], and Martín-Platero et al. [37]. In addition, the presence of genes associated with vancomycin resistance (vanA, vanB, vanC, vanD, vanE, and vanG) was investigated according to Valledor et al. [38], as well as the genes involved in the production of biogenic amines (hdc1, hdc21, tdc, odc) (Table 1) as indicated by de la Rivas et al. [39]. PCR reactions were performed in a Veriti 96-well Thermal Cycler, Applied Biosystems (Thermo Scientific) and the products were separated on 2.0% (w/v) agarose gels in 1 × TAE and visualized as described before.
Antibiotic Resistance
The selected cultures were subjected to phenotypic analysis of antibiotic resistance by using antibiotic disks (Oxoid Ltd., Basingstoke, England). The following antibiotics were used: ampicillin (10 μg/disk), erythromycin (10 μg/disk), gentamicin (10 μg/disk), penicillin (1 U/disk), streptomycin (10 μg/disk), daptomycin (10 μg/disk), vancomycin (10 μg/disk), and tobramycin (10 μg/disk). The strains were supplemented to MRS agar at 105 CFU/mL (final concentration) and the disks placed on the surfaces. Plates were incubated at 37 °C for 24 h and inhibition zone diameters around antibiotic disks were measured in millimeters. The isolates were interpreted as either resistant (R) or sensitive (S) according to the recommendations of the European Committee on Antimicrobial Susceptibility Testing [40]. The presence of intermediate resistance was considered as resistant.
Evaluation of Beneficial Properties
β-Galactosidase Production
The expression of β-galactosidase was evaluated using ONPG disks (Sigma-Aldrich, St. Louis, MO, USA) following manufacturer’s instructions. The strains were grown on solid MRS for 24–48 h at 37 °C until the formation of distinct individual colonies. A suspension of each strain was prepared by homogenizing 3–4 colonies in 0.2 mL of sterile saline solution (0.85% NaCl, w/v) in a test tube with an ONPG disk. The tubes were incubated at 37 °C for 4–6 h and regularly observed for yellow color formation as evidence of ONPG hydrolysis via the action of β-galactosidase to ortho-nitrophenol. The experiment was performed in duplicates in at least 2 independent occasions.
Bile Salt Deconjugation
To evaluate the ability of the probiotic candidates to perform bile salt deconjugation, bacterial cultures previously grown in MRS broth at 37 °C for 24 h were streaked individually on MRS agar plates supplemented with 0.5% (w/v) sodium taurodeoxycholate hydrate, taurcholate acid sodium salt hydrate, glycocholic acid hydrate, sodium glycocholate hydrate, sodium glycochenodeoxycholate, or sodium taurochenodeoxycholate (Sigma-Aldrich). The presence of an opaque halo around the colonies after incubation at 37 °C for 72 h in anaerobic conditions (GasPak System, Oxoid) indicated a positive result for bile salt deconjugation following the protocol of de Maraes et al. [41]. The test was performed in two independent occasions, twice for each strain.
Screening for Presence of Beneficial Genes
The DNA previously obtained were evaluated for the presence of selected beneficial genes related to adhesion (msa, map, mub, and eftu), folate production (pabB, pabC, folKQ, and folPE), GABA production (gad), and bile salt hydrolase enzymes (bsh) (Table 1) by PCR according to de Moraes et al. [41], dos Santos [30], and Bajic et al. [42] (Table 1). PCR reactions were performed on a Veriti 96-well Thermal Cycler, Applied Biosystems (Thermo Scientific) and the amplicons were separated on 1.0–2.0% (w/v) agarose gels in 1 × TAE and visualized as described before.
Evaluation of Antagonistic Properties
The ability of the strains to produce antagonistic metabolites was explored in two directions: production of antimicrobial peptides (bacteriocins) and cell-to-cell interaction as described previously and according to dos Santos et al. [30]. Test organisms, including different oral cavity-related pathogens, opportunistic pathogens, and beneficial strains from culture collections of HEM Inc. (Human Effective Microbes, Pohang, Republic of Korea), ProBacLab (Handong Global University, Pohang, Republic of Korea), KCTC (Jeongeup, Republic of Korea), KACC (Jeollabuk-do, Republic of Korea), and ATCC (American Type Culture Collection, Manassas, VA, USA) (Table 2), were used in a test panel to evaluate the antagonistic properties of the strains. Inhibition zones greater than 2 mm were considered evidence of potential production of antimicrobial metabolites, including bacteriocins. All experiments were performed in triplicate in two independent occasions.
Hydrophobicity
The ability of the selected strains to adhere to hydrocarbons as a criterion of hydrophobicity was determined according to Todorov and Dicks [43]. The strains were grown in MRS broth at 37 °C until stationary phase and cells were harvested (12,000 × g, 5 min, 4 °C), washed twice in 50 mM potassium phosphate buffer (pH 6.5), and re-suspended in the same buffer. Cell suspensions were adjusted to OD560 nm 1.0 (UV/VIS Spectrophotometer, Optizen™ POP) using 50 mM potassium phosphate buffer (pH 6.5), and mixed with 0.6 mL of n-hexadecane (proportion 5:1) and vigorously vortexed for 2 min. The two phases were allowed to separate for 1 h at 37 °C and the OD of the aqueous phase was determined at 560 nm. The percentage cell surface hydrophobicity was calculated as [(A0 − A)/A0] × 100, where A0 and A are the optical density readings before and after extraction with n-hexadecane, respectively.
Growth in Different Initial pH of the Media
Fifteen microliters of the strains initially grown in MRS broth (37 °C, 24 h) was transferred to 135 μL modified MRS broth (Difco) adjusted to pH 2.0, 4.0, 6.0, 8.0 10.0, and 12.0 with 1 M HCl or 1 M NaOH before autoclaving. All tests were conducted in sterile 96-well flat-bottom microtiter plates (SPL, Pocheon-si, Gyeonggi-do, Republic of Korea). Optical density readings at 600 nm were recorded every hour for 24 h in SPECTROstar Nano (BMG LABTECH). Cultures grown in MRS broth without pH adjustment served as control. All experiments were performed in duplicate in two independent occasions.
Effect of Commercial Drugs on Bacterial Survival
The strains were tested for susceptibility to commercial drugs from different commonly used groups and oral hygiene products (Table 3). Strains were grown in MRS broth (Difco) at 37 °C for 24 h and imbedded into MRS soft agar (1.0%, w/v, Difco) at 105 CFU/mL final concentrations. One tablet from each of the commercial drugs was solubilized in 5 mL sterile distilled water under aseptic conditions, with the final concentration as specified in Table 3. Ten microliters of each drug solution was spotted onto the surface of the previously described MRS agar plates. The plates were incubated for 24 h at 37 °C and examined for the presence of inhibition zones. The drugs generating inhibition zones larger than 2 mm were considered for further determination of the minimal inhibition concentration (MIC) by applying two-fold serial dilutions of the drugs.
In addition, two commercial pharmaceutical oral cleaning powders, Cleaning Time Ocean Mint (Bareun, LLC, Chun-cheon, Gang-wondo, Republic of Korea) and Coolush powder (CO-FOODS, LLC, Suwanee GA, USA), and their individual ingredients were evaluated for possible inhibitory effects against the studied strains. One commercial sachet of Cleaning Time Ocean Mint and Coolush powder were diluted in 5 mL sterile distilled water under aseptic conditions. All ingredients were dissolved in sterile distilled water under aseptic conditions to final concentrations of 0.1 g/mL (Table 3). As with the test done on commercial drugs, 10 μL of the prepared solution was spotted on the surface of MRS plates, which were incubated at 37 °C for 24 h and evaluated for the presence of inhibition zones.
Results and Discussion
Isolation and Identification of Beneficial LAB
Numerous and diverse microorganisms colonize the oral cavity, including beneficial bacteria that could exert positive effects on human and oral health. Several LAB in the oral cavity contribute to the prevention of oral diseases and numerous species have been reported for their probiotic features [12]. In this study, putative beneficial LAB strains have been isolated from the oral cavity, and have been identified as Lactobacillus gasseri ST16HK; Streptococcus salivarius ST48HK, ST59HK, ST61HK, and ST62HK; Lactiplantibacillus plantarum (Lactobacillus plantarum) ST63HK and ST66HK; and Latilactobacillus sakei (Lactobacillus sakei) ST69HK. These showed potential for further development as oral probiotics based on the characterization of their antimicrobial activity and properties.
Presumptive LAB were isolated from the saliva of healthy volunteers of different ethnicities with the aim of evaluating them as potential probiotics for oral cavity application. More than 120 bacterial colonies showed inhibition zones in the triple-level approach and were isolated and selected as promising candidates based on their antimicrobial activity against the test microorganisms (Str. mutans KACC 16833, Str. gordonii KACC 13829, and L. monocytogenes ATCC 15313). The isolates were tested for antimicrobial activity, with the results serving as basis for further evaluation. Differentiation of the promising isolates based on rep-PCR revealed 9 unique clusters, with representatives from each group subjected to partial 16S rRNA gene sequencing. Grouping the isolates based on their antimicrobial activities, representatives from Cluster 1 revealed a 99% similarity to Str. mutans, a well-known opportunistic pathogen typically isolated from the oral cavity of humans [44]. Representatives from the other 8 clusters were identified as Lb. gasseri strain ST16HK; Str. salivarius strains ST48HK, ST59HK, ST61HK, and ST62HK; Lb. plantarum strains ST63HK and ST66HK; and Lb. sakei strain ST69HK. All 8 strains (Lb. gasseri ST16HK; Str. salivarius ST48HK, ST59HK, ST61HK, and ST62HK; Lb. plantarum ST63HK and ST66HK; and Lb. sakei ST69HK) showed inhibitory activity against the pathogens Str. mutans KACC 16833, Str. gordonii KACC 13829, and L. monocytogenes ATCC 15313 with cell-to-cell interaction. While LAB from the genera Lactobacillus, Lactococcus, Streptococcus, Leuconostoc, Enterococcus, and Weissella can be found in the oral cavity [45], Lactobacillus and Streptococcus were mostly isolated in this study.
As the gateway to the digestive system, the oral cavity is where mechanical and initial enzymatic digestion occurs. These enzymes, although primarily aid in the digestion processes, also play a crucial role in the elimination and serve as an initial barrier-function of the entry of potential pathogens to the rest of the digestive tract. In addition to this, the oral cavity is considered a rich ecological niche, with more than 700–1000 bacterial species that are permanently or temporarily associated with it [2, 14]. The dynamic state of the microbial population in the oral cavity is dependent of the age, social economic status of the individual, and the type of diet and lifestyle [2, 46]. It has been clearly demonstrated that during different life stages, specific bacterial groups may colonize the oral cavity [47], with Str. salivarius being typically associated with the human oral cavity. This was suggested by Andrewes and Horder [48] and this association has been frequently confirmed [49, 50]. Lb. plantarum, Lb. gasseri, and Lb. sakei have also been reported as inhabitants of the human oral cavity [47, 49]. However, the isolation of different LAB from the oral cavity cannot support the postulate that these species are part of the residual microbiota of this ecosystem. Several LAB isolated from the oral cavity may be temporally introduced by foods. However, their presence in the oral cavity should be evaluated, specifically their stability in this ecological niche while also investigating their potentially beneficial, negative, or a hitherto undefined role in host health.
Safety Evaluation of the Selected Strains
Antibiotic Resistance, Hemolysis, Gelatinase, and Biogenic Amine Production
The safety of the 8 selected strains was evaluated as part of the approach “safety is priority,” an essential step for the application of beneficial strains as oral probiotics. Antibiotic resistance, hemolytic activity, gelatinase production, and biogenic amine production were determined as part of physiological testing.
Str. salivarius ST48HK, ST59HK, ST61HK, and ST62HK; Lb. plantarum ST63HK and ST66HK; Lb. sakei ST69HK; and Lb. gasseri ST16HK responded in different ways when tested for antibiotic resistance (Table 4). Special attention should be given to tobramycin (primary antibiotic treatment for Gram-negative infections, Pseudomonas-related infections and tuberculosis) [51] resistance exhibited by Lb. gasseri ST16HK. Moreover, from all the tested strains, only Str. salivarius ST59HK was clearly inhibited by vancomycin, while all other strains were resistant. According to EFSA, vancomycin resistance is intrinsic to most LAB species, and strains of Lb. plantarum and Lb. sakei showing resistance can still be considered as safe. On the other hand, the resistance of Str. salivarius strains ST48HK, ST61HK, and ST62HK to some of tested antibiotics can be considered as a safety concern.
The antibiotic resistance of the strains evaluated as probiotic candidates is considered a key issue. The uncontrolled application of antibiotics, especially during the last 3 decades of the twentieth century in health care and farming practices, resulted in the wide distribution of antibiotic resistance, not only among pathogens, but also among beneficial organisms [52]. Moreover, a serious concern is the possible scenario of antibiotic resistance transfer from beneficial bacterial species to other inhabitants of the GIT, including opportunistic and effective pathogens [52]. EFSA (https://www.efsa.europa.eu/en/topics/topic/antimicrobial-resistance) is regularly updating information and recommendations for the safety criteria regarding antibiotic resistance in different bacterial groups.
Evaluation of the hemolytic activity of the 8 selected strains revealed that they all exhibited γ-hemolytic activity. The presence of α- and particularly β-hemolysis can be considered as virulence factors, and are observed in different pathogenic species, including E. coli, B. cereus, and some Enterococcus spp. This is of significance, especially to Str. salivarius strains since the genus Streptococcus is known as human and other animal pathogen [30, 53]. According to the Bulletin of the International Dairy Association [54], only a few species of the genus Streptococcus (Streptococcus gallolyticus subsp. macedonicus, Str. salivarius subsp. salivarius, Str. salivarius subsp. thermophilus, and Streptococcus thermophilus) are considered as safe. Although Str. salivarius has a disputable reputation, MacDonald et al. [55] and Wescombe et al. [27] suggested that strains belonging to this species can be applied as beneficial strains. On the contrary, Wilson et al. [56] reported that Str. salivarius can be associated with meningitis cases.
The evaluation for gelatinase production demonstrated that all the strains gave negative results. Gelatinase production, although not a common feature among representatives of the genus Lactobacillus, is frequently reported in some representatives of the genus Streptococcus, especially for strains with a clear pathogenic profile [57]. The assessment for biogenic amine (BA) production also generated negative results. The production of biogenic amines is considered crucial for the safety application of beneficial organisms, especially for those intended to be applied as starter or adjunct cultures in food fermentation processes. BAs may be produced by amino acid decarboxylation in diverse foods, especially during fermentation, and their accumulation may be considered as a potential health hazard. Specific thresholds for different BAs are regulated for different fermented food products and normally are subjected to strict control [58]. This characteristic has been shown to be a strain-specific property [33]. Different studies have demonstrated that BA productions such as histamine, recorded for a strain of Lb. plantarum [59], and tyramine, for Lb. curvatus strains, are typically observed [33] and thus require close monitoring. This has been further highlighted by Barbieri [60], who reviewed the link between biogenic amine production and the presence of LAB in different food products, and emphasized the significant role of different strains of Lactobacillus spp.
Detection of Virulence and Vancomycin-Resistant Genes
In addition to physiological tests, safety evaluation of the selected strains was also done based on PCR screening for virulence and vancomycin-resistant genes. In this study, although detection of virulence genes (asa1) and vancomycin-resistant genes (vanB and vanD) were observed in some of the strains evaluated, the majority of the selected beneficial strains are still considered safe to be applied as oral probiotics. However, some concerns about the presence of vanB and vanD need to be looked at as these can be potential risks [38]. Transferable antibiotic-resistant genes, especially to vancomycin, are considered to pose a threat on the safety of LAB strains. In this study, 7 out of the 8 evaluated strains showed resistance to vancomycin based on the performed antibiotic susceptibility tests. However, for all hetero-fermentative and some homo-fermentative lactobacilli, vancomycin resistance is considered by EFSA [34] as an intrinsic feature. The phenotypic demonstration of vancomycin resistance by three Str. salivarius strains (ST48HK, ST61HK, and ST62HK) is a crucial point for risk assessment for the possible applications of these strains. Based on PCR assays carried out for the detection of different vancomycin genes (vanA, vanB, vanC, vanD, vanE, and vanG), only vanD was detected in Str. salivarius ST48HK, while vanB was detected in Lb. plantarum ST63HK and ST66HK. The presence of vancomycin-resistant genes needs to be evaluated, as well as the localization of these associated genes, for possible horizontal gene transfer. It has been highlighted that the presence of vanA and vanB genes are much more concerning than the rest of the van genes. This is due to their plasmid localization, which favors horizontal gene transfer [38]. Thereby, it was suggested by EFSA that testing for presence of various key virulence genes, efaA, cyt, IS16, esp, asa1, and hyl, is necessary for safety evaluation of some LAB species. Screening for these aforementioned genes demonstrated their absence across the evaluated strains, except for Str. salivarius ST61HK and ST62HK, where asa1 was detected. This observation is a pivotal point for considerations for possible application of these strains. Screening for the presence of genes related to the production of biogenic amines is also considered a key test in the evaluation of LAB intended to be applied as a probiotic or starter cultures. The ingestion of excessive amounts of BA, especially the combinations of formed biogenic amines, can pose a serious health consequence for consumers [33]. In the detection assay conducted in this study for the detection of the crucial enzyme for BA production, only hdc1 was detected in Lb. gasseri ST16HK.
The molecular-based evaluation for the presence of virulence factors is one of the predictive approaches in the safety evaluation of potential beneficial bacteria [61]. However, the presence of specific genes needs to be discussed in the context of the presence of functional operons related to the effective expression of these virulence factors. It has previously been shown that some virulence genes can still be present, even though they may not be expressed. Thus, the expression of a specific factor should be further examined by determining whether a functional operon is present or not [62]. Possible horizontal gene transfer may occur between bacterial species and strains carrying the targeted virulence genes. It is likely that potential transfer occurred, involving only parts of the operon. Moreover, as suggested by Perin et al. [62], the expression of virulence genes (and other genes as well) can be related to environmental conditions, including temperature, pH, or specificity of the particular inductors. It must also be considered that these assessments are only carried out in vitro and in DNA-level screenings; thus, further evaluation of expression of the functionality of the virulence genes and the conditions required for their effective expression needs to be considered in future safety analysis.
Additional Beneficial Properties of the Selected Strains
β-Galactosidase Production and Hydrophobicity
Production of β-galactosidase is essential for bacterial growth when the available carbohydrate is limited to lactose [63]. From a probiotic point of view, the expression of β-galactosidase can be associated with the role of probiotics in the reduction of lactose intolerance in individuals [64]. However, the production of β-galactosidase is only considered an additional beneficial property of probiotics promoting oral health, since lactose digestion typically occurs in the small intestine, with little to no significance in the oral cavity. In this study, we have evaluated β-galactosidase production through ONPG disks. Among the evaluated strains, only Str. salivarius ST48HK showed a positive result by forming of a yellow color due to the action of β-galactosidase on ortho-nitrophenol.
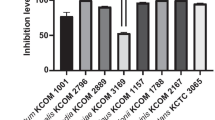
The strains demonstrated a wide range of cell-surface hydrophobicity as indicated in Fig. 1. According to Krausova et al. [65], hydrophobicity above 40% can be considered as high. This characteristic has been considered a predictive tool for potential probiotic application because it determines the ability to adhere to the intestinal mucosa [30, 65]. However, this can only be regarded as a preliminary indication of mucus adhesion characteristics as the entire process is much more complex in vivo, and involves different factors such as expression of specific adherence proteins, distribution of carbohydrates on the cell surface, and electrical charges [66]. While a simple method, cell surface hydrophobicity to hydrocarbons serves as a presumptive assay for the adherence properties of the strains under investigation.
Levels of the hydrophobicity, recorded for the evaluated in this study potential probiotic strains. SD were presented. Lactobacillus gasseri (ST16HK), Streptococcus salivarius (ST48HK ST59HK, ST61HK, and ST62HK), Lactiplantibacillus plantarum (ST63HK and ST66HK), and Latilactobacillus sakei (ST69HK)
Detection of Adhesion and Beneficial Genes
The presence and expression of adhesion genes can be regarded as a desirable property for a strain to colonize the GIT and, therefore, interact with the host. Colonizing the digestive system, including the oral cavity, needs to be discussed as a functional characteristic in the context of the biological role and application of a specific probiotic strain. Adhesion may support prolonged “domestication” and can be considered as beneficial for long-term presence in the oral cavity and the GIT, thereby improving interaction with other bacterial species and promoting immune responses in the host [67]. In the scenario, where probiotics will interact with toxic metabolites (toxins, heavy metals, etc.) and be involved in their removal, quick transit passage may be considered as beneficial [68]. Results indicated that adhesion genes map, mub, and eftu were detected in Str. salivarius ST61HK and ST62HK. On the other hand, eftu gene was detected in both Lb. gasseri ST16HK and Lb. sakei ST69HK, while only map gene was present in Lb. plantarum ST63HK.
It is interesting to note that some of the evaluated strains were positive for map, mub, and eftu genes, which are typically present in Lb. plantarum strains [69]. However, the presence of these genes was determined even in representatives of Enterococcus and Leuconostoc [43], possibly making them more universally distributed. Moreover, the other evaluated genes (EF1249, EF2380, EF2662, and prg) were not detected in the strains being studied. The presence of a variety of adhesion proteins is strain-specific and may serve as functional support for possible application as novel probiotics.
Screening for the deconjugation-associated gene (bsh) showed its presence in Str. salivarius ST61HK and ST62HK, along with Lb. sakei ST69HK. This observation points to the possibility that these strains can be involved in various physiological processes including the metabolism of bile salts. However, based on the phenotypic assays, no evidence of degradation of the tested salts was demonstrated by the strains. The ability to deconjugate bile salts in vitro is considered a beneficial property of the strains since they can reduce cholesterol levels and the toxicity of bile salts [70], particularly those that are intended to be applied as probiotics. This characteristic was previously reported in Lb. mucosae CNPC006 and Lb. mucosae CNPC007 [41], with their ability to deconjugate GDC salts and TDC salts in a strain-specific manner. Additionally, some Lb. reuteri strains have also been reported to have the ability to degrade GDC salts [71]. Benefits associated with this characteristic lead to the reduction of the toxicity, formation of insoluble forms, and control of the presence of less absorbable bile acids in the intestinal lumen. Additionally, this may have an effect on the reduction of serum cholesterol levels [72].
The production of specific beneficial metabolites may also serve as an additional support for determining a newly isolated probiotic candidate’s potential for its intended application. The presence of gad gene in all the strains may be considered as an additional beneficial characteristic. GABA, a known neurotransmitter, plays an important role in brain development. Aside from this, its other applications such as possible treatment for diabetes, positive influence and suppression inflammatory immune responses, and its ability to promote “regulatory” immune responses, especially concerning autoimmune diseases [73, 74], merit further evaluation, quantification, and exploitation.
Folate production is considered a beneficial property of LAB important in the formulation of functional fermented food products naturally enriched with vitamin B9. Laiño et al. [75] and Levit et al. [76] reported on different LAB strains with the ability to produce vitamin B9, and proposed the application of such strains in the formulation of dairy beverages for children to combat folate deficiency. In addition to previously identified beneficial genes, folate encoding genes were also screened. It was observed that pabB, pabC, folKQ, and folPE with the exception of folPE genes were detected in Str. salivarius ST48HK, but not in the other strains. In Str. salivarius ST61HK and ST62HK, the genes encoding for folate (pabB, pabC, folKQ, and folPE) and GABA (gad) production were detected in addition to all the adhesion genes. Only three genes (pabB, pabC, folKQ) were found to be associated with Str. salivarius ST48HK, suggesting strain-specific diversity in this species, and also suggesting a potential for selecting of appropriate strains as oral probiotic candidates.
Antagonistic Properties of the Selected Strains
Production of antimicrobial peptides (bacteriocins) and cell-to-cell interactions were determined through activity against the test organisms listed in Table 2. Lb. plantarum ST63HK showed activity against most of the test organisms via cell-to-cell interaction, whereas a few positive results were observed for bacteriocin activity. Lb. gasseri ST16HK showed activity against Lactobacillus paracasei, L. monocytogenes ATCC 15313, Staphylococcus delphini KACC 13,58, and Str. gordonii KACC 13829. Str. salivarius ST48HK showed inhibitory activity against L. monocytogenes ATCC 15,313, St. delphini KACC 13258, and Str. gordonii KACC 13829. The inhibition by Lb. plantarum ST63HK was detected against E. coli, St. delphini KACC 13258, and Str. gordonii KACC 13829, whereas Lb. plantarum ST66HK was active against St. delphini KACC 13258 (Table 2).
LAB can produce different antimicrobial metabolites as part of their defense mechanism. Organic acids, low molecular antimicrobials, diacetyl, H2O2, and antimicrobial peptides are just a few of the metabolites formed by LAB as a means to compete against other microorganisms present in the same ecological niche [77]. Moreover, Chikindas et al. [77] suggested that bacteriocins can be regarded as molecules of great complexity than just simple “killers,” since they are actively involved in the regulatory cell processes and quorum sensing interactions. In addition to cell-to-cell inhibitory interactions, evidence for possible production of bacteriocins was sparse.
Growth in Media With Various Initial pH
The growth rate of the selected strains was determined in MRS broth with varying initial pH values. At pH values below 6.0, only slight growth was detected, most strains showed notable growth above pH 6.0, suggesting that they can grow effectively in the human oral cavity, emphasizing their potential as oral probiotics. In this study, bacterial growth on MRS broth adjusted to higher pH levels demonstrated slow growth of Lb. gasseri ST16HK compared to the control MRS medium (with pH 6.0). Optimal growth for Lb. gasseri ST16HK was recorded at pH 8.0, compared to other pH conditions. Str. salivarius ST48HK showed the lowest growth rate at pH 2.0 and 4.0, while other pH conditions showed no significant differences. Str. salivarius ST59HK showed the lowest growth rate at pH 2.0, pH 4.0, and pH 12.0 while continuous log phase was observed at pH 6.0. Str. salivarius ST61HK showed a significantly lower growth rate in pH 2.0 compared to the standard MRS medium. Str. salivarius ST62HK exhibited the lowest growth rate in pH 4.0 compared to the control and showed maximum growth rate at pH 6.0 and 8.0. For Lb. plantarum ST63HK, pH conditions showed no significant difference between pH 6.0, pH 8.0, pH 10.0, and pH 12.0 compared to the control with the lowest growth rate at pH 2.0 and pH 4.0. For Lb. plantarum ST66HK, growth was lowest in pH 2.0 and pH 4.0 compared to the control, with no significant differences at pH 6.0 and pH 8.0. Growth rate of Lb. sakei ST69HK was maximal at pH 8.0 and 10.0 compared to the other conditions. At pH values 2.0, pH 4.0, and pH 12.0, the lowest growth rate was detected for Lb. sakei ST69HK (Fig. 2).
Comparison of the growth of Lactobacillus gasseri ST16HK, Streptococcus salivarius ST48HK ST59HK, ST61HK, and ST62HK, Lactiplantibacillus plantarum ST63HK and ST66HK, and Latilactobacillus sakei ST69HK in MRS (Difco) at different initial pH levels. The experiment was performed in triplicate. SD were lower than 2%, and for simplicity, were not presented
Survival of Selected Strains in Commercial Drugs and Oral Hygiene Products
The efficacy of probiotics can be influenced by the interference of general drugs and hygienic products. Previous research [78,79,80] reported on the possible inhibitory effects of commercial non-antibiotic drugs on the viability of probiotic strains. In this study, 13 commercial drugs (Table 3) inhibited the growth of the strains. In order to estimate the potential effect, minimal inhibitory concentrations were calculated to evaluate the effective concentration against the selected strains. Nurofen, Doloran, Nurofurantiona, and oral hygienic products are inhibited all 8 strains. Buscopan inhibited the growth of Str. salivarius ST59HK, Lb. plantarum ST63HK, and Lb. plantarum ST66HK; Analgin inhibited the growth of Lb. gasseri ST16HK, Lb. plantarum ST63HK, and Lb. plantarum ST66HK. Empeace reduced the growth of Str. salivarius ST48HK, Str. salivarius ST61HK, and Str. salivarius ST62HK, while Mortin/Ibuprofen reduced the growth of all the strains except Lb. plantarum ST63HK. Treda did not influence the growth of Lb. gasseri ST16HK and Lb. plantarum ST66HK. Sandrin inhibitory effect was observed to the growth of Lb. plantarum ST66HK and Str. salivarius ST61HK. Different samples of Listerine inhibited the strains in a strain-specific manner (Table 3). The different ingredients of the two hygienic oral products did not show inhibitory effects against the strains. MIC levels were also calculated for the commercial drugs and hygienic products (Table 3), an information relevant to the long-term application of the drugs and/or hygienic products and potential negative effect on the probiotics. The generated inhibitory effects against the studied strains and the calculated concentrations are listed in Table 3.
Conclusions
Eight beneficial strains with potential as oral probiotics were isolated from the saliva of healthy individuals. Their differentiation and identification were based on repPCR and 16S rRNA partial genome sequencing. These strains were further evaluated by screening for the presence of different safety and beneficial properties. Results suggest that the strains Lb. gasseri ST16HK; Str. salivarius ST48HK, ST59HK, ST61HK, and ST62HK; Lb. plantarum ST63HK and ST66HK; and Lb. sakei ST69HK can be considered as promising oral probiotic candidates. In order to be recommended as probiotics, additional tests to confirm their efficacy in an appropriate animal model, as well as in vivo toxicological studies, have to be performed, in accordance to national and international regulations.
Data Availability
All data generated or analyzed during this study are included in this published article and comply with research standards.
References
FAO/WHO (2006) Probiotics in food health and nutritional properties and guidelines for evaluation. FAO/WHO, Rome. https://www.fao.org/publications/card/en/c/7c102d95-2fd5-5b22-8faf-f0b2e68dfbb6/
Mahasneh SA, Mahasneh AM (2017) Probiotics: a promising role in dental health. Dent J 5(4):26. https://doi.org/10.3390/dj5040026
Schmalstieg FC, Goldman AS (2008) Ilya Ilich Metchnikoff (1845–1915) and Paul Ehrlich (1854–1915): the Centennial of the 1908 Nobel Prize in Physiology or Medicine. J Med Biogr 16(2):96–104. https://doi.org/10.1258/jmb.2008.008006
Fuller R (1991) Probiotics in human medicine Gut 32:439–442. https://doi.org/10.1136/gut.32.4.439
Vasiljevic T, Shah NP (2008) Probiotics — from Metchnikoff to bioactives. Int Dairy J 18:714–728. https://doi.org/10.1016/j.idairyj.2008.03.004
Metchnikoff E (1907) Lactic acid as inhibiting intestinal putrefaction. The prolongation of life: Optimistic studies. W Heinemann, London 1907:161–183
Rastogi P, Saini H, Dixit J, Singhal R (2011) Probiotics and oral health. Natl J Maxillofac Surg 2(1):6–9. https://doi.org/10.4103/0975-5950.85845
Chugh P, Dutt R, Sharma A, Bhagat N, Dhar MS (2020) A critical appraisal of the effects of probiotics on the oral health. J Funct Foods 70:103985. https://doi.org/10.1016/j.jff.2020.103985
Schrezenmeir J, de Vrese M (2001) Probiotics, prebiotics, and synbiotics — approaching a definition. Amer J Clin Nutr 73(2):361s–364s. https://doi.org/10.1093/ajcn/73.2.361s
Kober M-M, Whitney PB (2015) The effect of probiotics on immune regulation, acne, and photoaging. Int J Womens Dermatol 1(2):85–89. https://doi.org/10.1016/j.ijwd.2015.02.001
Day RL, Harper AJ, Woods RM, Davies OG, Heaney LM (2019) Probiotics: current landscape and future horizons. Future Sci OA 5(4):FSO391. https://doi.org/10.4155/fsoa-2019-0004
Lahtinen SJ, Forssten S, Aakko J, Granlund L, Rautonen N, Salminen S, Viitanen M, Ouwehand AC (2011) Probiotic cheese containing Lactobacillus rhamnosus HN001 and Lactobacillus acidophilus NCFM® modifies subpopulations of fecal lactobacilli and Clostridium difficile in the elderly. Age (Dordr) 34(1):133–143. https://doi.org/10.1007/s11357-011-9208-6
Bustamante M, Oomah BD, Oliveira WP, Burgos-Díaz C, Rubilar M, Shene C (2020) Probiotics and prebiotics potential for the care of skin, female urogenital tract, and respiratory tract. Folia Microbiol 65(2):245–264. https://doi.org/10.1007/s12223-019-00759-3
Yamashita Y, Takeshita T (2017) The oral microbiome and human health. J Oral Sci 59(2):201–206. https://doi.org/10.2334/josnusd.16-0856
Holzapfel WH, Wood BJB (2014) Lactic acid bacteria: biodiversity and taxonomy. pp 606. John Wiley & Sons, Ltd. https://doi.org/10.1002/9781118655252
Faran A, Syed M, Farzeen T (2012) Oral microbial habitat a dynamic entity. J Oral Biol Craniofacial Res 2(3):181–187. https://doi.org/10.1016/j.jobcr.2012.07.001
Marsh PD, Martin MV (2009) Mouth as a microbial habitat. In: Lewis MA (Ed.) Oral Microbiology Textbook. Churchill Livingstone Elsevier; Edinburgh, London, New York, Oxford: 2009. pp. 8–23
Hart BL, Powell KL (1990) Antibacterial properties of saliva: role in maternal periparturient grooming and in licking wounds. Physiol Behavior 48(3):383–386. https://doi.org/10.1016/0031-9384(90)90332-x
Vila T, Rizk AM, Sultan AS, Jabra-Rizk MA (2019) The power of saliva: antimicrobial and beyond. PLoS Pathog 15(11):e1008058. https://doi.org/10.1371/journal.ppat.1008058
Gao L, Liu Y, Kim D, Li Y, Hwang G, Naha PC, Cormode DP, Koo H (2016) Nanocatalysts promote Streptococcus mutans biofilm matrix degradation and enhance bacterial killing to suppress dental caries in vivo. Biomaterials 101:272–284. https://doi.org/10.1016/j.biomaterials.2016.05.051
Casarin RC, Barbagallo A, Meulman T, Santos VR, Sallum EA, Nociti FH, Duarte PM, Casati MZ, Gonçalves RB (2012) Subgingival biodiversity in subjects with uncontrolled type-2 diabetes and chronic periodontitis. J Periodontal Res 48(1):30–36. https://doi.org/10.1111/j.1600-0765.2012.01498.x
Kaur S, White S, Bartold M (2012) Periodontal disease as a risk factor for rheumatoid arthritis: a systematic review. JBI Libr Syst Rev 10(42 Suppl):1–12. https://doi.org/10.11124/jbisrir-2012-288
Colombo M, Castilho NPA, Todorov SD, Nero LA (2018) Beneficial properties of lactic acid bacteria naturally present in dairy production. BMC Microbiol 18:219. https://doi.org/10.1186/s12866-018-1356-8
Lebeer S, Vanderleyden J, De Keersmaecker SC (2008) Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev 72(4):728–764. https://doi.org/10.1128/MMBR.00017-08
Reid G (1999) The scientific basis for probiotic strains of Lactobacillus. Appl Environ Microbiol 65(9):3763–3766. https://doi.org/10.1128/AEM.65.9.3763-3766.1999
Porter SR, Scully C (2006) Oral malodour (halitosis). BMJ (Clinical research ed) 333(7569):632–635. https://doi.org/10.1136/bmj.38954.631968.AE
Wescombe PA, Hale JD, Heng NC, Tagg JR (2012) Developing oral probiotics from Streptococcus salivarius. Future Microbiol 7(12):1355–1371. https://doi.org/10.2217/fmb.12.113
Burton JP, Wescombe PA, Macklaim JM, Chai MH, Macdonald K, Hale JD, Tagg J, Reid G, Gloor GB, Cadieux PA (2013) Persistence of the oral probiotic Streptococcus salivarius M18 is dose dependent and megaplasmid transfer can augment their bacteriocin production and adhesion characteristics. PLoS One 8(6):e65991. https://doi.org/10.1371/journal.pone.0065991
He L, Yang H, Chen Z, Ouyang X (2020) The effect of Streptococcus salivarius K12 on halitosis: a double-blind, randomized, placebo-controlled trial. Probiotics Antimicrob Protein 12:1321–1329. https://doi.org/10.1007/s12602-020-09646-7
dos Santos KMO, de Matos CR, Salles HO, de Melo Franco BDG, Arellano K, Holzapfel WH, Todorov SD (2020) Exploring beneficial/virulence properties of two dairy-related strains of Streptococcus infantarius subsp. infantarius. Probiotics Antimicrob Proteins 12(4):1524–1541. https://doi.org/10.1007/s12602-020-09637-8
De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB (2009) Bergeys manual of systematic bacteriology: the Firmicutes. UK, Springer, London
de Castilho NPA, Colombo M, de Oliveira LL, Todorov SD, Nero LA (2019) Lactobacillus curvatus UFV-NPAC1 and other lactic acid bacteria isolated from calabresa, a fermented meat product, present high bacteriocinogenic activity against Listeria monocytogenes. BMC Microbiol 19:63–75. https://doi.org/10.1186/s12866-019-1436-4
Bover-Cid S, Holzapfel WH (1999) Improved screening procedure for biogenic amine production by lactic acid bacteria. Int J Food Microbiol 53(1):33–41. https://doi.org/10.1016/s0168-1605(99)00152-x
EFSA (2012) European Food Safety Authority. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. Parma, Italy: EFSA; Available from: https://www.efsa.europa.eu/en/efsajournal/pub/2740
Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, Jabes D, Goossens H (2004) Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol 42(10):4473–4479. https://doi.org/10.1128/JCM.42.10.4473-4479.2004
Leavis HL, Willems RJ, van Wamel WJ, Schuren FH, Caspers MP, Bonten MJ (2007) Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Path 3(1):e7. https://doi.org/10.1371/journal.ppat.0030007
Martín-Platero AM, Valdivia E, Maqueda M, Martínez-Bueno M (2009) Characterization and safety evaluation of enterococci isolated from Spanish goats’ milk cheeses. Int J Food Microbiol 132(1):24–32. https://doi.org/10.1016/j.ijfoodmicro.2009.03.010
Valledor SJD, Bucheli JEV, Holzapfel WH, Todorov SD (2020) Exploring beneficial properties of the bacteriocinogenic Enterococcus faecium ST10Bz strain isolated from boza, a Bulgarian cereal-based beverage. Microorganisms 8:1474. https://doi.org/10.3390/microorganisms8101474
de las Rivas B, Marcobal A, Muñoz R (2005) Improved multiplex-PCR method for the simultaneous detection of food bacteria producing biogenic amines. FEMS Microbiol Lett 244(2):367–372. https://doi.org/10.1016/j.femsle.2005.02.012
EUCAST (2016) European Committee on Antimicrobial Susceptibility Testing. http://www.eucast.org/ast_of_bacteria/
de Moraes GMD, de Abreu LR, do Egito AS, Salles HO, da Silva LMF, Nero LA, Todorov SD, Dos Santos KMO (2017) Functional properties of Lactobacillus mucosae strains isolated from Brazilian goat milk. Probiotics Antimicrob Proteins 9(3):235–245. https://doi.org/10.1007/s12602-016-9244-8
Bajic SS, Djokic J, Dinic M, Veljovic K, Golic N, Mihajlovic S, Tolinacki M (2019) GABA-producing natural dairy isolate from artisanal Zlatar cheese attenuates gut inflammation and strengthens gut epithelial barrier in vitro. Front Microbiol 10. https://doi.org/10.3389/fmicb.2019.00527
Todorov SD, Dicks LMT (2008) Evaluation of lactic acid bacteria from kefir, molasses and olive brine as possible probiotics based on physiological properties. Ann Microbiol 58(4):661–670. https://doi.org/10.1007/BF03175572
Loesche WJ (1986) Role of Streptococcus mutans in human dental decay. Microbiol Rev 50(4):353–380. https://doi.org/10.1128/mr.50.4.353-380.1986
Ahrné S, Nobaek S, Jeppsson B, Adlerberth I, Wold AE, Molin G (1998) The normal Lactobacillus flora of healthy human rectal and oral mucosa. J Appl Microbiol 85:88–94. https://doi.org/10.1046/j.1365-2672.1998.00480.x
Stamatova I, Jukka HM (2009) Probiotics: health benefits in the mouth. Amer J Dent 22(6):329–338
Deo PN, Deshmukh R (2019) Oral microbiome: unveiling the fundamentals. J Oral Maxillofacial Pathol: JOMFP 23(1):122–128. https://doi.org/10.4103/jomfp.JOMFP_304_18
Andrewes FW, Horder TJ (1906) A study of the streptococci pathogenic for man. Lancet 2:708
Abranches J, Zeng L, Kajfasz JK, Palmer SR, Chakraborty B, Wen ZT, Richards VP, Brady LJ, Lemos JA (2018) Biology of oral streptococci. Microbiol Spectr 6(5):10. https://doi.org/10.1128/microbiolspec.GPP3-0042-2018
Du Toit M, Huch M, Cho G-S, Franz CMAP (2014) The genus Streptococcus. In: Holzapfel WH, Wood BLB (eds) Lactic acid bacteria — biodiversity and taxonomy. John Wiley & Sons Ltd., Chichester, West Sussex, UK, pp 457–505
Lode H (1998) Tobramycin: a review of therapeutic uses and dosing schedules. Curr Therap Res 59(7):420–453. https://doi.org/10.1016/S0011-393X(98)85082-0
Sharma L, Nagpal R, Jackson CR, Patel D, Singh P (2021) Antibiotic-resistant bacteria and gut microbiome communities associated with wild-caught shrimp from the United States versus imported farm-raised retail shrimp. Sci Rep 11:3356. https://doi.org/10.1038/s41598-021-82823-y
Dekker JP, Lau AF (2016) An update on the Streptococcus bovis group: classification, identification, and disease associations. J Clin Microbiol 54(7):1694–1699. https://doi.org/10.1128/JCM.02977-15
Bulletin of the International Dairy Association 495/2018 (2018) Inventory of microbial food cultures with safety demonstration in fermented food products. Update of the Bulletin of IDF No455–2012. www.fil-idf.org
MacDonald KW, Chanyi RM, Macklaim JM, Cadieux PA, Reid G, Burton JP (2021) Streptococcus salivarius inhibits immune activation by periodontal disease pathogens. BMC Oral Health 21(1):245. https://doi.org/10.1186/s12903-021-01606-z
Wilson M, Martin R, Walk ST, Young C, Grossman S, McKean EL, Aronoff DM (2012) Clinical and laboratory features of Streptococcus salivarius meningitis: a case report and literature review. Clin Med Res 10(1):15–25. https://doi.org/10.3121/cmr.2011.1001
Patterson MJ (1996) Streptococcus. In: Baron S (Ed.) Medical Microbiology. Galveston (TX): University of Texas Medical Branch at Galveston. Available from: https://www.ncbi.nlm.nih.gov/books/NBK7611/
Özogul Y, Özogul F (2019) Biogenic amines formation, toxicity, regulations in food. In: Biogenic amines in food: Analysis, occurrence and toxicity. Saad B, Tofalo R (Eds). pp 1–17. Royal Society of Chemistry, London, UK. https://doi.org/10.1039/9781788015813
Yazgan H, Kuley E, Güven GT, Regenstein JM, Özogul F (2021) The antimicrobial properties and biogenic amine production of lactic acid bacteria isolated from various fermented food products. J Food Process Preserv 45:e15085. https://doi.org/10.1111/jfpp.15085
Barbieri F, Montanari C, Gardini F, Tabanelli G (2019) Biogenic amine production by lactic acid bacteria: a review. Foods 8(1):17. https://doi.org/10.3390/foods8010017
Todorov SD, Stojanovski S, Iliev I, Moncheva P, Nero LA, Ivanova IV (2017) Technology and safety assessment for lactic acid bacteria isolated from traditional Bulgarian fermented meat product “lukanka.” Braz J Microbiol 48(3):576–586. https://doi.org/10.1016/j.bjm.2017.02.005
Perin LM, Miranda RO, Todorov SD, Franco BDGDM, Nero LA (2014) Virulence, antibiotic resistance and biogenic amines of bacteriocinogenic lactococci and enterococci isolated from goat milk. Int J Food Microbiol 185:121–126. https://doi.org/10.1016/j.ijfoodmicro.2014.06.001
Hill C, Ross RP (1998) Starter cultures for the dairy industry. In: Roller S, Harlander S (Eds.) Genetic Modification in the Food Industry. Springer, Boston, MA. https://doi.org/10.1007/978-1-4615-5815-6_9
Masoumi SJ, Mehrabani D, Saberifiroozi M, Fattahi MR, Moradi F, Najafi M (2021) The effect of yogurt fortified with Lactobacillus acidophilus and Bifidobacterium sp. probiotic in patients with lactose intolerance. Food Sci Nutr 9:1704–1711. https://doi.org/10.1002/fsn3.2145
Krausova G, Hyrslova I, Hynstova I (2019) In vitro evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains. Fermentation 5(4):100. https://doi.org/10.3390/fermentation5040100
Monteagudo-Mera A, Rastall RA, Gibson GR, Charalampopoulos D, Chatzifragkou A (2019) Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl Microbiol Biotechnol 103(16):6463–6472. https://doi.org/10.1007/s00253-019-09978-7
Guan C, Chen X, Jiang X, Zhao R, Yuan Y, Chen D, Zhang CC, Lu M, Lu Z, Gu R (2020) In vitro studies of adhesion properties of six lactic acid bacteria isolated from the longevous population of China. RSC Adv 10(41):24234–24240. https://doi.org/10.1039/D0RA03517C
Zoghi A, Massoud R, Todorov SD, Chikindas ML, Popov I, Smith S, Khosravi-Darani K (2021) Role of the lactobacilli in food bio-decontamination: friends with benefits. Enzyme Microb Technol 150:109861. https://doi.org/10.1016/j.enzmictec.2021.109861
Ramiah K, van Reenen CA, Dicks LMT (2007) Expression of the mucus adhesion genes mub and mapA, adhesion-like factor ef-tu and bacteriocin gene plaA of Lactobacillus plantarum 423, monitored with real-time PCR. Int J Food Microbiol 116(3):405–409. https://doi.org/10.1016/j.ijfoodmicro.2007.02.011
Vizoso Pinto MG, Franz CM, Schillinger U, Holzapfel WH (2006) Lactobacillus spp. with in vitro probiotic properties from human faeces and traditional fermented products. Int J Food Microbiol 109(3):205–214. https://doi.org/10.1016/j.ijfoodmicro.2006.01.029
Taranto M, Sesma F, Font de Valdez G (1999) Localization and primary characterization of bile salt hydrolase from Lactobacillus reuteri. Biotechnol Lett 21:935–938. https://doi.org/10.1023/A:1005652501404
De Smet I, Van Hoorde L, De Saeyer N, Vande Woestyne M, Verstraete W (1994) In vitro study of bile salt hydrolase (BSH) activity of BSH isogenic Lactobacillus plantarum 80 strains and estimation of cholesterol lowering through enhanced BSH activity. Microb Ecol Health Dis 7:315–329. https://doi.org/10.3109/08910609409141371
Soltani N, Qiu H, Aleksic M, Glinka Y, Zhao F, Liu R, Li Y, Zhang N, Chakrabarti R, Ng T, Jin T, Zhang H, Lu WY, Feng ZP, Prud’homme GJ, Wang Q (2011) GABA exerts protective and regenerative effects on islet beta cells and reverses diabetes. Proc Natl Acad Sci USA 108(28):11692–11697. https://doi.org/10.1073/pnas.1102715108
Purwana I, Zheng J, Li X, Deurloo M, Son DO, Zhang Z, Liang C, Shen E, Tadkase A, Feng ZP, Li Y, Hasilo C, Paraskevas S, Bortell R, Greiner DL, Atkinson M, Prud’homme GJ, Wang Q, (2014) GABA promotes human β-cell proliferation and modulates glucose homeostasis. Diabetes 63(12):4197–4205. https://doi.org/10.2337/db14-0153
Laiño JE, LeBlanc J-G, Savoy de Giori G (2012) Production of natural folates by lactic acid bacteria starter cultures isolated from artisanal Argentinean yogurts. Can J Microbiol 58(5):581–588. https://doi.org/10.1139/w2012-026
Levit R, Savoy de Giori G, de Moreno de LeBlanc A, LeBlanc JG, (2018) Protective effect of the riboflavin-overproducing strain Lactobacillus plantarum CRL2130 on intestinal mucositis in mice. Nutr 54:165–172. https://doi.org/10.1016/j.nut.2018.03.056
Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LMT (2018) Functions and emerging applications of bacteriocins. Curr Opin Biotechnol 49:23–28. https://doi.org/10.1016/j.copbio.2017.07.011
Berginc K (2015) Pharmacokinetic interactions between drugs and dietary supplements: probiotic and lipid supplements. In: Dietary supplements: Safety, efficacy and quality. Berginc K, Kret S (Eds.) Elsevier, Amsterdam, NL. pp. 69–83. https://doi.org/10.1533/9781782420811.2.69.
Casarotti SN, Carneiro BM, Todorov SD, Nero LA, Rahal P, Penna ALB (2017) In vitro assessment of safety and probiotic potential characteristics of Lactobacillus strains isolated from water-buffalo mozzarella cheese. Ann Microb 67:289–301. https://doi.org/10.1007/s13213-017-1258-2
Nascimento LCS, Casarotti SN, Todorov SD, Penna ALB (2019) Probiotic potential and safety of enterococci strains. Ann Microbiol 69:241–252. https://doi.org/10.1007/s13213-018-1412-5
Charteris WP, Kelly PM, Morelli L, Collins JK (2001) Gradient diffusion antibiotic susceptibility testing of potentially probiotic Lactobacilli. J Food Prot 64:2007–2014. https://doi.org/10.4315/0362-028X-64.12.2007
Author information
Authors and Affiliations
Contributions
Concept: SDT; experimental work: HK, JIIF, SDT; data analysis: HK, JIIF, SDT; funds: WHH, SDT; writing of the manuscript: HK, JIIF, SDT; corrections and editing: WHH, SDT.
Corresponding author
Ethics declarations
Ethics Approval
This article does not contain any studies with human or animal subjects.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kim, H., Fugaban, J.I.I., Holzapfel, W.H. et al. Selection of Beneficial Bacterial Strains With Potential as Oral Probiotic Candidates. Probiotics & Antimicro. Prot. 14, 1077–1093 (2022). https://doi.org/10.1007/s12602-021-09896-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-021-09896-z