Abstract
Antimicrobial peptides (AMPs) have been an area of great interest, due to the high selectivity of these molecules toward bacterial targets over host cells and the limited development of bacterial resistance to these molecules through evolution. The peptides are known to selectively bind to bacterial cell surfaces through electrostatic interactions, and subsequently, the peptides insert into the cell membrane and cause local disruptions of membrane integrity leading to cell death. Previous experiments showed that replacing the Leu residues in the AMP C18G with other naturally occurring hydrophobic residues resulted in side-chain-dependent activities. This work extends the investigation to non-natural hydrophobic amino acids and the effect on peptide activity. Minimal inhibitory concentration (MIC) results demonstrated that amino acid substitutions containing long flexible carbon chains maintained or increased antimicrobial activity compared to natural analogues. In solution, the peptide showed aggregation only with the most hydrophobic non-natural amino acid substitutions. Binding assays using Trp fluorescence confirm a binding preference for anionic lipids while quenching experiments demonstrated that the more hydrophobic peptides are more deeply buried in the anionic lipid bilayers compared to the zwitterionic bilayers. The most effective peptides at killing bacteria were also those which showed some level of disruption of bacterial membranes; however, one peptide sequence exhibited very strong activity and very low levels of red blood cell hemolysis, yielding a promising target for future development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although the first case of antibiotic resistance was identified in 1940, resistance development has rapidly increased since the late 1990s [1]. Despite this, there has been a significant decrease in the number of new antibiotics developed and through clinical trials during this same period. The development of new antibiotics is primarily hampered due to the economic challenges of developing these compounds that will rapidly lose viability in the clinic due to resistance development [1, 2]. The World Health Organization and the US Centers for Disease Control have both identified antimicrobial resistance as a major concern for public health, with ~ 3 M antibiotic resistant infections per year in the USA alone.
The resistance phenomenon has fueled significant research in academic settings on the development and application of novel antimicrobial molecules, drug delivery platforms, and combinatorial drug interactions to enhance efficacy. Numerous groups have reported progress on natural products [3, 4], polymers [5,6,7], peptidomimetics [8,9,10], metals and metal nanoparticles [11,12,13], quorum sensing inhibitors [14], polysaccharides [15], ionic liquids [16,17,18], and peptides [19,20,21,22]. Each of these systems has inherent benefits and drawbacks regarding efficacy, cytotoxicity, cost, and resistance development [23]. Thus, none have effectively emerged as a front runner, driving the continued research into all of them.
Antimicrobial peptides (AMPs) have been the focus of tremendous research since the discovery of magainin in the 1980s [24]. Although originally isolated from the skin secretions of X. laevis, AMPs have since been found in all higher organisms [25]. AMPs have been classified by their amino acid composition, charge, secondary structural elements, intramolecular connections, among other characteristics (reviewed in [26]). Of these, by far the most widely studied class of AMPs are the amphiphilic, cationic, and helical peptides. This class of peptides are often characterized by the traditional facially amphiphilic structure that is adopted upon binding to bacterial cell surfaces in which the hydrophobic amino acid side chains are clustered on one face of the a-helix while the cationic residues are localized on the opposite face of the helix. AMP recognition and binding to the target bacteria is initially driven by favorable electrostatic interactions between the cationic peptide and the anionic bacterial cell surface. Once bound, the facially amphiphilic helix structure facilitates the peptide interaction with the non-polar core of the bacterial lipid bilayer while maintaining the electrostatically favorable interactions between cationic side chains and anionic lipid headgroups. Upon interacting with the bilayer, the peptides induce membrane permeabilization or destabilization, which is linked to the antibacterial mechanism [27]. Despite these investigations, no clear consensus sequence has been identified for AMPs.
One of the significant drawbacks to the development of AMPs as therapeutics is poor bioavailability. Generally, the majority of AMPs are sensitive to proteolytic degradation, an especially large hurdle for oral administration of these molecules [28, 29]. Because of this, there has been an increasing interest in the incorporation of non-natural (not found in nature) or non-proteinogenic (found in nature, not normally incorporated into natural proteins) amino acids into AMPs to improve physiochemical properties and reduce the sensitivity to degradation [30,31,32,33]. This also expands the variety of side-chains accessible in the chemical synthesis of AMPs as there are hundreds of non-natural amino acids available and compatible with solid-phase peptide synthesis chemistry. However, there is far less known about the impact these amino acids have on peptide structure and properties compared to the naturally occurring amino acids, thus complicating the design and prediction of activity for novel AMPs.
The AMP C18G was originally developed based on the 13-residue C-terminal fragment of the human platelet factor IV protein [34]. C18G exhibited favorable physical properties and enhanced broad-spectrum activity compared to the 13-residue precursor [34]. The peptide structure was shown to be α-helical, and based on the sequence, C18G would adopt a facially amphiphilic structure when in the helical conformation. Microbiological studies showed that C18G can act as a potent activator of the bacterial two-component sensing system, PhoPQ, normally involved in Mg2+ sensing but also linked to detection of antimicrobial peptides [35, 36]. Structure-activity studies showed that net cationic charge was not sufficient to maintain activity, as C18G variants that incorporated cationic residues with short side-chain arms (such as di-amino propionic acid) were dramatically less effective, while variants with His as the cationic group displayed pH-dependent activity [30, 37]. Similarly, the hydrophobic amino acids were dependent on side chain structure and composition, but activity tracked with overall hydrophobicity, the more hydrophobic the side chain, the greater the antimicrobial activity [33]. These findings paralleled numerous studies in amphiphilic polymers that demonstrated similar trends for both hydrophobic and cationic groups.
This work presents an investigation of the peptide C18G and the impact of non-natural hydrophobic amino acids on activity. The C18G sequence contains six Leu residues as the only hydrophobic amino acids in the peptide. Previous work showed that replacing these Leu residues with other natural hydrophobic amino acids resulted in a spectrum of effects on activity, both antimicrobial and biophysical. The incorporation of a series of non-natural amino acids isomeric to the naturally occurring counterparts was incorporated into C18G to parallel the investigation of the natural amino acids. The results show that side chain length, and likely side chain flexibility, is an important driving force in the antimicrobial and membrane destabilizing activities in AMPs.
Materials and Methods
Materials
C18G and related sequences: ALWKKLLKKLLKSAKKLG, L➔l (C18G-Nle), L➔v (C18G-Nva), L➔f (C18G-Cha), L➔b (C18G-Abu) and L➔A (C18G-Ala) were synthesized using standard FMOC-solid phase chemistry. Natural amino acids were purchased from Bachem (Torrance, CA USA) while non-natural amino acids were from ChemPep Inc. (Wellington, FL USA), and rink-amide resin was from ChemImpex (Wood Dale, IL USA). Peptides were synthesized using 20% piperidine in dimethylformamide (DMF) as a deprotecting agent, 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU) and N,N-diisopropylethylamine (DIEA) activation cocktail and was carried out on a rink-amide resin. Peptides were cleaved from the resin using a cocktail containing 92.5:2.5:2.5:2.5 trifluoroacetic acid (TFA)/H2O/triisopropyl silane (TIPS)/ethanedithiol. Peptides were separated from the resin via filtration and subsequent precipitation by dropwise addition into ice-cold diethyl ether, followed by centrifugation. All peptides were purified by reversed phase HPLC (RP-HPLC) using a linear gradient of water and acetonitrile supplemented with 0.1% TFA on a Jupiter 300 C4 column (Phenomenex). Peptide identity was confirmed by ESI-MS. Eluted HPLC fractions containing peptide were pooled and lyophilized. Lipids: (16:0–18:1) 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphotidylcholine (POPC) and (16:0–18:1) 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphotidylglycerol (POPG), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) were purchased from Avanti Polar Lipids (Alabaster, AL USA), used without further purification and stored as stocks in chloroform at − 20 °C. Buffers used in all assays were PBS (150 mM NaCl, 50 mM sodium phosphate; pH 7.0), 10× diluted PBS (for CD measurements), or Z-Buffer (0.1 M Na2HPO4/NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.05 M β-mercapthoethanol, pH 7.0). All reagents were from VWR (Radnor PA, USA) unless otherwise noted.
Lipid Vesicle Preparation
Appropriate volumes of lipids were taken from stock solutions in chloroform, mixed in glass test tubes, dried under a gentle stream of N2 for approximately 15 min, and further dried under vacuum for at least 60 min. Small unilamellar vesicles (SUVs) used in binding experiments were created by adding 1.5 mL of PBS buffer to the test tube containing the dry lipid film and vortexing to ensure complete resuspension of the film. This sample was placed in a bath sonicator for 17–20 min to create SUVs. For CD and quenching experiments, SUVs were formed using the ethanol dilution method [38]. Briefly, lipid films were dissolved in 10 μL of ethanol to which the appropriate volume PBS were added while vortexing to a total volume of 800 μL accounting for the future addition of peptide.
Antimicrobial Activity
Minimum inhibitory concentration (MIC) was determined using previously described method of serial dilutions based on the guidelines from the Clinical Laboratory Standards Institute [39]. A fixed volume of 10 μL peptide dissolved in a 75:25 H2O/EtOH were added to a 96-well microplate and serially diluted in water. Subsequently, 90 μL of bacterial culture in Mueller-Hinton (MH) broth was added to each well for a final bacterial cell density of approximately 5 × 105 CFU/mL and incubated at 37 °C overnight. Growth was determined after 18 h by measuring optical density at 600 nm (OD600) in the wells. Wells with no peptide were used as a positive growth control reference. The minimal bactericidal concentration (MBC) was determined by plating 1 mL of culture from each well of the MIC plate onto a fresh Luria-Bertani (LB) agar plate. The culture was allowed to grow on the plate for 18 h at 37 °C. The lowest concentration that showed no growth of colonies on the plate was determined to be the MBC. Strains tested were Acetinobacter baumannii ATCC:19606, Pseudomonas aeruginosa ATCC:10145, Staphylococcus aureus ATCC:27660, and the Escherichia coli D31 used was the chromosomal penicillin V-resistant isolate in the 1968 study by Burman et al. [40].
Outer Membrane Permeabilization Assay (β-Lactamase Assay)
A single colony of E. coli was inoculated in sterile LB medium supplemented with 100 μg/mL ampicillin (LB+Amp). The culture was incubated at 37 °C overnight with shaking and subsequently diluted 1:250 in fresh LB+Amp. Culture was then incubated at 37 °C with shaking until it reached an OD600 between 0.2 and 0.5. The culture was centrifuged for 15 min at ~ 1000 RCF in a bench-top clinical centrifuge. The pellet was then resuspended in an equal volume of PBS. Peptides or polymyxin B (positive control) were 2-fold serially diluted as described above. Subsequently, 80 μL of resuspended bacterial cells were added to each well of the plate. Immediately prior to measurement, 10 μL of nitrocefin (EMD Millipore, Billerica, MA) was placed into each of the wells to a final concentration 50 μg/mL. Absorbance was measured at 486 nm in 5-min intervals over 90 min. All experiments were performed in triplicate.
Inner Membrane Permeabilization (β-Galactosidase Assay)
A single colony of E. coli was inoculated in sterile LB medium supplemented with 100 μg/mL ampicillin. The culture was incubated at 37 °C overnight with shaking and subsequently diluted 1:250 in fresh LB+Amp and additionally supplemented with 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Culture was then incubated at 37 °C with shaking until it reached an OD600 between 0.2 and 0.5. Appropriate volumes of the culture were added to wells of a 96-well plate, which contained serially diluted aliquots of peptides or control compound cetyltrimethylammonium bromide (CTAB). Immediately prior to measurement, a 10 μL aliquot of ONPG (ortho-nitrophenyl-β-galactoside, from a 4 mg/mL stock in Z-buffer) was added to each well for a final sample volume of 100 μL. The total volume of each well was maintained at 100 μL by balancing with additional Z-buffer if necessary. Absorbance at 420 nm was measured in 5-min intervals over 90 min. All experiments were performed in triplicate.
Fluorescence Spectroscopy
Samples containing 2 μM peptide dissolved in PBS were added to a semi-micro quartz cuvette. All measurements were performed using a Fluoromax-4 (JY Horiba Scientific, Piscataway, NJ, USA) instrument. Fluorescence emission spectra (λex = 280 nm) were recorded prior to addition of lipid vesicles. Samples were titrated with aliquots of lipid vesicles, and fluorescence was re-measured after each addition. Spectra were corrected for background by subtraction and corrected for dilutions. Barycenters were calculated as previously described [38]. Red edge excitation shift (REES) experiments were performed similarly, except that λex was varied.
Acrylamide Quenching
Samples were prepared similarly as previously described above except lipid vesicles were added in 1 aliquot to a final concentration of 200 μM. Peptide concentration was maintained at 2 μM. Instrument settings were λex = 295 nm and λem = 340 nm. The excitation wavelength of 295 nm was used to minimize inner filter effects and correction for inner filter effects was performed as previously described [38]. Emission was measured prior to addition of acrylamide (F0). Acrylamide was added in 10 μL aliquots from a 4 M solution. Fluorescence was measured after each addition. Fluorescence intensity was corrected for dilution, inner filter effects, and background fluorescence.
Trichloroethanol Quenching
Peptide concentration was maintained at 2 μM. Instrument settings were λex = 280 nm and λem = 340 nm. Emission was measured prior to addition of trichloroethanol (TCE) (F0). TCE was added in 4 μL aliquots from a 10 M stock. Fluorescence was measured after each addition. Fluorescence intensity was corrected for dilution and background fluorescence. Barycenters were calculated as previously described [38].
Model Vesicle Leakage
Leakage from model vesicles was determined by using the fluorophore calcein. Twenty millimolars of lipid film was prepared as previously described (the “Lipid Vesicle Preparation” section). A 75 mM stock of calcein was prepared in HEPES buffer, pH 7.0. The 20 mM lipid film was then dissolved into a 700 μL aliquot of the calcein solution. The solution was then subjected to seven rounds of freeze-thawing by alternating a liquid N2 bath and a 42 °C water bath, finishing with a final thaw. The sample was then sonicated using a VCX 130 Probe sonicator (Sonics & Materials, Inc.) for 60 s at 45% intensity using 1-s pulses. To separate the unincorporated calcein vesicles from those incorporated with the fluorophore, a size exclusion column was utilized using G25 Sephadex. The column was equilibrated in HBS (50 mM HEPES, 150 mM NaCl, pH 7.0) under gravity flow for ~ 1 h. Immediately following the loading of the vesicle solution, ~ 40 1 mL fractions were collected from the column. Fractions that contained the vesicles with the incorporated calcein would elute early and exhibit a cloudy orange color, while those without the calcein would be clear and yellow in color. Two of the most concentrated fractions are diluted 1:1 with HBS buffer, pH 7.0. Vesicles were used on the same day as they were prepared. Ten microliters of peptide solution was dispensed into a black flat-bottom 96-well plate, followed by 20 μL of the diluted vesicle solution, and 70 μL of PBS buffer (50 mM NaHPO4, 150 mM NaCl, pH 7.0). Peptides were prepared in a serially diluted manner with a final concentration range of 15 μM to 0.234 μM. The last row contained only 0.01% acetic acid to act as a negative control. Calcein leakage was measured by intensity of fluorescence emission at 520 nm with an excitation at 495 nm using a 510 nm cutoff filter in a Spectramax M5 multimode plate reader (Molecular Devices, San Jose, CA USA). After the initial reading, 20 μL of the detergent Triton X-100 was added to the last row as a positive control. The plate was shielded from light and allowed to shake at 400 rpm for 1 h. Fluorescence emission was recorded once more following the hour of shaking. The wells containing Triton X-100 were used as the 100% leakage normalization value. Data reported is the average of three replications.
Circular Dichroism Spectroscopy
Circular dichroism (CD) was measured using a JASCO-J810 Spectropolarimeter (Jasco Inc., Easton, MD USA) in continuous scanning mode collecting data over the range 190–260 nm. Samples were prepared similar to above except that 1:10 diluted PBS was used to minimize background in the low UV wavelength region. Peptide concentration was 3 μM for all samples. Spectra were recorded of peptide in buffer, peptide in 1:1 buffer/trifluoroethanol, and peptide with lipid vesicles at 150 μM total lipid. Blank spectra lacking peptide were subtracted for each sample. Sample spectra represent the averages of 64–128 scans.
Hemolysis Assay
Hemolysis of human red blood cells (RBCs) was used to quantify membrane destabilization by leakage of hemoglobin. Blood was collected in 10 mL EDTA-coated vacutainer tubes (provided by the Rowan Department of Health and Exercise Science from a fresh draw ~ 1 h prior to use, IBC protocol 2019-11). A 7 mL aliquot of whole blood was mixed with an equal volume of sterile PBS. The mixture was then sedimented via centrifugation for 7 min. The supernatant was aspirated and the pellet containing RBCs was resuspended to the original volume of 14 mL with sterile PBS. The sedimentation and resuspension was repeated for a total three times. The final cell pellet was resuspended to a final volume of 14 ml, and 90 μL of this cell suspension was added to all wells of a conical-bottom 96-well plate. Prior to addition of RBCs, 10 μL of serially diluted peptide or the detergent, Triton-X 100 (as a positive control), were added to the wells. The covered plate was placed into a shaking incubator for 60 min at 37 °C while shaking (150 rpm). The remaining cells were pelleted using the lowest setting of the centrifuge at 4 °C for 10 min. Next, 6 μL of the supernatant was added to 94 μL of fresh PBS in a flat-bottom plate. Analysis of hemolysis was performed by measuring absorbance at 415 nm. Percent hemolysis was calculated based on the absorbance of each well compared to those wells with no additive and those with Triton-X 100. All data reported is the average of 3 replicates.
Results
Peptide Sequences
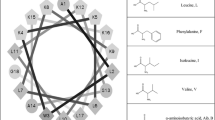
Peptides were synthesized and subsequently purified via RP-HPLC. Major fractions were isolated and analyzed via ESI-MS to determine the molecular weight of the purified fractions (Table 1). The peptide sequences are based on the C18G sequence originally described by Darveau et al. [34]. This peptide is highly cationic (+ 8 charge) and will adopt a facially amphipathic structure when in an α-helical conformation (Fig. 1a). The parent C18G and the C18G-Ala peptides are both predicted to adopt α-helical conformation in solution by the iTasser algorithm (model shown in Fig. 1b) [41]. Notably, the C18G sequence exclusively contained Leu as the hydrophobic amino acids which orient to the non-polar face of the helix. Peptide variants were synthesized in which these Leu residues were replaced with other hydrophobic amino acids norleucine (Nle, l), norvaline (Nva, v), cyclohexyl-alanine (Cha, f), or aminobutyric acid (Abu, b) (Fig. 1c).
Helical models of C18G and hydrophobic amino acid structures. a Helical wheel diagram of C18G is shown. Gray shaded residues are hydrophobic while white residues are polar or cationic. b Ribbon diagram of the predicted helical structure of C18G-Ala. c Chemical structures of the hydrophobic amino acids used in this study. Helical wheel projection was made using the R software. 3D model was constructed using the iTasser software (40) and visualized in PyMol
Antimicrobial Activity
The peptide antimicrobial activity was investigated using the broth microdilution method to determine the minimal inhibitory concentration (MIC). After MIC determination, aliquots from each sample were plated on fresh LB-agar plates to determine the MBC, as MIC may report inhibition of growth and/or bactericidal activity. The MICs and MBCs were determined against four bacterial strains, S. aureus (Gram-positive), E. coli, A. baumannii, and P. aeruginosa (Gram-negative). Notably, the MBC was equal-to or 1-dilution higher than the MIC in all cases, indicating that the peptides likely act in a bactericidal manner (Table 2).
Solution Aggregation Properties
As part of the mechanism of action, AMP transition from an aqueous soluble conformation to a membrane bound conformation upon interaction with bacteria. Any changes in amino acid composition could impact the properties of the peptide in solution, potentially impacting aggregation or oligomerization in solution, which would subsequently impact the thermodynamics of the membrane-binding interaction. One approach to assessing solution state aggregation is by trichloroethanol (TCE) quenching. TCE is a relatively non-polar small molecule that has been shown to be effective at quenching fluorescence from Trp residues buried in the interior of proteins or protein aggregates [42, 43]. TCE quenching, presented as Stern-Volmer plots, are shown in Fig. 2a. The greater the slope of the line fit to the quenching data (Ksv), the greater the extent of quenching and thus burial/occlusion of the Trp side chain. The data show all peptides, with the exception of C18G-Cha, were effectively unaffected by TCE, indicating little to no aggregation that affected the Trp.
Peptide aggregation assays. a Trichloroethanol (TCE) quenching. b Red edge excitation shifts (REES). In all cases samples contained 2 μM peptide. In both panels, filled circles represent C18G-Nle, open squares represent C18G-Nva, filled diamonds represent C18G-Cha, filled triangles represent C18G-Abu, and open circles represent C18G-Ala. In a, lines are to guide the eye while in b, lines represent linear fits to the data (R2 > 0.9). All data are averages of 3–5 samples, and error bars represent the standard deviation. In both assays, data shown is after subtraction of a background sample and TCE quenching data is also corrected for dilution by the quencher
Another approach to investigate aggregation is red-edge excitation shift (REES) which reports on the local mobility of the Trp residue, which in turn can be affected by oligomerization of the peptide [42, 43]. Briefly, REES is dependent on the reorientation of solvent around the fluorophore (Trp) during the excited state lifetime, which is directly affected if the fluorophore is part of a larger order assembly or oligomer compared to free in solution [44, 45]. This approach varies the excitation wavelength used to induce fluorescence and monitors the changes in the emission spectra. An upward curvature of the REES data, resulting in total shifts ~ 5 nm is indicative of oligomerization/aggregation in solution causing restricted motion of the Trp side chain. REES results are shown in Fig. 2b. Similar to the TCE quenching, C18G-Cha is the only peptide that exhibited a marked increase in REES.
Binding and Interactions with Lipid Bilayers
The initial step in most AMPs activity against bacteria is interaction with the bacterial cell surface. The binding of the modified C18G peptides to model lipid membranes was investigated by fluorescence spectroscopy exploiting the environmental sensitivity of the Trp residue as a reporter. The fluorescence emission spectrum of Trp is inherently sensitive to the local environment, resulting in a red-shifted fluorescence spectrum when the Trp is in the aqueous milieu and a more blue-shifted emission spectrum when the Trp is buried in a non-polar environment, such as the hydrophobic core of the lipid bilayer. The binding is saturated, presumably at or near 100% peptide bound, when the barycenter plateaus as a function of lipid added. Peptide binding was investigated using vesicles with three different lipid headgroup compositions: 100% PC (net neutral, zwitterionic); 3:1 PC:PG (net anionic, combination of zwitterionic and anionic), and 3:1 PE:PG (net anionic, combination of zwitterionic and anionic). Lipids with PE headgroups are commonly found in bacterial membranes while PC lipids are the primary component of mammalian lipid membranes [46]. The results are shown in Fig. 3, with representative spectra shown in Supplementary Fig. 1. Only C18G-Nle showed significant binding to the zwitterionic composition, but both C18G-Nle and C18G-Nva exhibited enhanced binding when anionic lipids were incorporated. C18G-Cha exhibited a low level of binding in all compositions, while C18G-Abu and C18G-Ala exhibited minimal binding to any composition.
Peptide binding to small unilamellar vesicles. Peptide binding was assayed using Trp fluorescence emission by titrating SUVs into a sample containing 2 μM peptide. The spectral barycenter was calculated after each addition of lipid vesicles and the Δbarycenter is the difference between the initial barycenter and that after each addition. Binding was assayed using a 100% PC, b 3:1 PC:PG, or c 3:1 PE:PG vesicles. In all panels, filled circles represent C18G-Nle, open squares represent C18G-Nva, filled diamonds represent C18G-Cha, filled triangles represent C18G-Abu, and open circles represent C18G-Ala. All data are averages of 2–3 samples, and the error bars represent the standard deviation. Data shown is after subtraction of a background sample and correction for dilution by the addition of SUVs
Binding to the bilayer is followed by membrane disruption in the activity of AMPs. This requires the burial of hydrophobic side chains into the non-polar core of the membrane, requiring a conformational change on the membrane surface. Acrylamide quenching was used to investigate the occlusion of the Trp after binding to model bilayers. Similar in principle to the TCE experiments, acrylamide is an effective quencher of Trp residues which are exposed to the aqueous milieu, and quenching is significantly reduced if the Trp is in the non-polar core of the bilayer [38]. Acrylamide quenching experiments in solution and in the presence of lipid vesicles are shown in Fig. 4, with Ksv values shown in Supplemental Table 1. In all cases, quenching in solution exhibits the highest Ksv of all conditions tested. In general, there were significant decreases in Ksv when the peptide was bound to vesicles containing anionic lipids, indicating the Trp became more shielded from the aqueous environment when bound. Notably, the C18G-Ala peptide exhibited significant shielding in POPC bilayers, despite only small changes in barycenter in the binding experiments.
Acrylamide quenching and Ksv determination. Peptide fluorescence was measured in the absence of acrylamide and after each subsequent aliquot. In all cases, samples contained 2 μM peptide. Quenching was measured with a C18G-Nle, b C18G-Nva, c C18G-Cha, d CHA-Abu, and e C18G-Ala. In all panels, filled circles represent control (no lipid), open squares represent 100% PC, filled diamonds represent 1:3 PC:PG, and open triangles represent 1:3 PE:PG. All data are averages of 3–6 samples, and error bars represent the standard deviation. Data shown is after subtraction of a background sample and correction for both dilution and inner filter effects
Investigating the membrane disruption mechanism of AMPs in a model bilayer system allows for direct parallels to the binding and quenching experiments. Vesicle leakage experiments rely on the entrapment of a membrane-impermeable fluorophore in the vesicle interior, in this case calcein. Calcein undergoes self-quenching at high concentrations, and therefore exhibits low fluorescence emission when trapped in the vesicle. Upon bilayer disruption, the calcein can leak out of the vesicle which relieves the self-quenching, resulting in a significant increase in fluorescence emission. The results are shown in Fig. 5. If permeabilization occurred, the peptides exhibited a dose-dependent permeabilization of the vesicles, regardless of bilayer composition. The C18G-Nle peptide was the only molecule to induce significant disruption of the zwitterionic PC vesicles. All peptides induced significantly less leakage in vesicles with anionic lipids. This indicates that the final bilayer orientation of the peptides is significantly different in the anionic lipid vesicles resulting in differential activity.
Lipid vesicle permeabilization. Peptide-induced permeabilization was measured by calcein leakage from lipid vesicles. Assays were performed using a 100% PC, b 3:1 PC:PG, or c 3:1 PE:PG vesicles. In all panels, filled circles represent C18G-Nle, open squares represent C18G-Nva, filled diamonds represent C18G-Cha, filled triangles represent C18G-Abu, and open circles represent C18G-Ala. Error bars represent the standard deviation. Percent leakage was calculated by comparison to an internal control treated with the detergent Triton X-100 set as 100% permeabilization, and an untreated sample as the baseline leakage control
Secondary Structure Analysis
The original development of C18G stemmed from isolation of a 13-residue helical fragment of human platelet factor IV. Subsequent studies have shown the C18G peptide is disordered in solution and α-helical when bound to bilayers or bilayer mimetics. This helical propensity was evident regardless of substitutions to the hydrophobic or cationic residues, including several non-proteinogenic amino acids [33, 47]. Circular dichroism spectroscopy was used to investigate the secondary structure of the peptides with non-proteinogenic hydrophobic substitutions (Fig. 6). The peptides exhibited a disordered conformation in PBS while adopting α-helical conformations when bound to anionic lipid vesicles. The overall extent of helical content does appear to vary between peptides. The C18G-Ala and C18G-Abu peptides maintained a significant component of unstructured conformation when interacting with PC vesicles, consistent with the different behavior of these peptides compared to the others. Additional experiments of the peptides in the helix-promoting solvent, TFE, confirm the ability to adopt helical conformations. These results mimic the helical conformations predicted for C18G and C18G-Ala by the iTasser algorithm (Fig. 1). It should be noted that the CD results should be taken qualitatively regarding helical conformation and not quantitatively in terms of helical content.
Circular dichroism (CD) spectroscopy. CD spectra were collected of samples containing 5 μM peptide in 0.1× PBS, or supplemented with TFE or SUVs with peptides a C18G-Nle, b C18G-Nva, c C18G-Cha, d C18G-Abu, and e C18G-Ala. In all panels, solid line represents PBS alone, dotted line represents 100% PC, dash-dot line represents 3:1 PC:PG, dotted square line represents 3:1 PE:PG, and dash-dash line represents 1:1 TFE:PBS. All spectra are the average of 64 scans and have been background corrected by subtraction of spectra lacking peptide
Bacterial Membrane Permeabilization
While biophysical studies in model membranes are informative, the bacterial cell surface is a much more chemically complex environment. Bacterial membranes include significant proportions of proteins and polysaccharides, as well as a greater variety of lipid headgroups and lipid acyl chain identities. This presents a much more complex environment in which the peptides would act under therapeutic conditions. As such, bacterial membrane permeabilization assays were carried out using membrane-impermeable chromogenic substrates of bacterial enzymes. Briefly, if the bacterial membrane is in-tact, the substrate cannot cross the membrane and be converted into a chromophore by the complementary enzyme. However, if the peptide (or other agents) destabilize the bilayer, the substrates can more easily cross the membrane, access the enzyme, and be converted into a product which is monitored spectrophotometrically. The permeability of the E. coli outer membrane is probed using the periplasmic enzyme β-lactamase and the chromogenic substrate, nitrocefin, while permeability of the E. coli inner membrane is probed using the cytoplasmic enzyme β-galactosidase and the chromogenic substrate ortho-nitrophenyl-β-galactoside (ONPG).
The results of the bacterial permeabilization experiments are shown in Fig. 7. The data shown represents leakage at 30 min after the addition of peptide; however full 90-min time courses are shown in the Supplemental Figs. 2 and 3. Both C18G-Nle and C18G-Cha caused significant, dose-dependent permeabilization of the E. coli outer membrane, paralleling the results in lipid vesicles. Similarly, the C18G-Nva peptide exhibited a similar dose dependence, but requiring higher concentrations of peptide for similar levels of permeabilization. The C18G-Abu and C18G-Ala peptides appeared to be inactive against the outer membrane at this time point, however, did show activity at longer time points (Supplemental Fig. 2). Conversely, only the C18G-Nle peptide exhibited any activity against the E. coli inner membrane at the 30-min time point (Fig. 7b), or at longer time points (Supplemental Fig. 3).
Membrane permeabilization of E. coli or red blood cells. a Outer membrane permeability assayed by nitrocefin hydrolysis by β-lactamase in the bacterial periplasm measured at 486 nm. b Inner membrane permeability assayed by ONPG hydrolysis by β-galactosidase in the bacterial cytoplasm measured at 420 nm. c Human red blood cell hemolysis assayed by hemoglobin release. In all panels, filled circles represent C18G-Nle, open squares represent C18G-Nva, filled diamonds represent C18G-Cha, filled triangles represent C18G-Abu, and open circles represent C18G-Ala. All data are averages of 3 samples with error bars representing standard deviation. In all panels, data are subtracted for background absorbance. In c, percent hemolysis was calculated by use of an internal control by adding Triton X-100 to the wells after measurement to achieve full hemolysis, normalized to 100%
Hemolysis Assays
The membrane disrupting activity of AMPs has also been linked to hemolytic and cytotoxic activity. Human red blood cell (RBC) hemolysis assays were carried out as a first approximation of potential cytotoxic activity for the C18G peptides. This also allows identification of potential lead compounds, that is, those which exhibit high activity but low hemolysis at a given concentration. Thus, the assays were performed at the same concentrations as the MIC/MBC experiments. The assay relies on the leakage of hemoglobin from RBCs upon exposure to peptides. The hemolysis data is shown in Fig. 7c. The data are normalized to internal controls using a detergent that completely dissolves the RBC membrane as 100% hemolysis. Interestingly, the C18G-Cha was the only peptide that induced significant hemolysis, up to 65% at the highest concentration tested. C18G-Nle induced up to 24%, but only above background levels at the two highest concentrations tested, and the remaining peptides induced no detectable hemolysis. Notably, the parent peptide, C18G, has been shown to induce < 10% hemolysis over the same concentration range [30].
Discussion
The broader interest in AMPs is driven by multiple factors including the pressing public health needs for development of new antibacterials, the evolutionary persistence of AMPs in higher organisms indicating the inability of bacteria to develop effective resistance mechanisms, and facile chemical synthesis facilitating study. Despite this, there has been no consensus sequence found for AMPs, nor has there been a data-driven, predictive model for design or detection of AMPs. Thus, using the power of solid-phase peptide synthesis coupled with non-natural amino acids, structure-activity relationships can be elucidated and inform the development of novel molecules.
The initial analysis of the data presented here show several important trends. In solution, the peptides show clear and consistent trends in behavior. First, the CD experiments demonstrate that all of the peptides in this study are unstructured in solution. This is consistent with other studies on AMPs which show similar properties. The aggregation studies, TCE quenching and REES, were consistent in that the majority of peptides showed very little evidence of aggregation/oligomerization in solution. However, the only peptide which showed some aggregation propensity was the C18G-Cha sequence, evidenced by the increased Ksv in TCE quenching and the upward curvature in the REES plot. This is not surprising considering the bulky, hydrophobic Cha side chain.
With respect to peptide-lipid interactions, there is a significant difference in behavior for several peptides between zwitterionic and anionic lipid vesicles. As expected, the C18G-Nle and C18G-Nva peptides displayed a clear preference for binding anionic vesicles due to the electrostatic attraction between the cationic peptide and the anionic bilayer surface, and the presence of hydrophobic groups in the peptide sequence. The C18G-Nle peptide, while able to bind the zwitterionic PC vesicles, exhibited an enhanced barycenter shift upon binding anionic vesicles, indicating a different final orientation. The C18G-Cha peptide bound similarly to all vesicles, indicating that electrostatics are not playing a major role and thus the binding is likely driven by hydrophobic forces in this case. This is reasonable considering the bulky, hydrophobic Cha side chain. Also notable was the minimal apparent binding of the C18G-Abu and C18G-Ala peptides, indicating that despite the electrostatic interactions, the peptides did not interact strongly with the anionic bilayers. It should be noted that precise Kd calculations were not carried out as the aggregation state in solution, which appears to differ between peptides (see above), was not quantified. However, the acrylamide quenching experiments indicate the Trp in both peptides was shielded upon interaction with bilayers. Therefore, it is likely that the C18G-Abu and C18G-Ala peptides are binding to bilayers but the Trp is not deeply partitioning in the bilayer core, resulting in a significantly smaller barycenter shift. This is consistent with the physically shorter hydrophobic side chains of C18G-Abu and C18G-Ala, requiring the peptide to adopt more shallowly inserted conformation. Similar effects were observed for C18G variants in which the cationic side chains were shortened, as well as in other peptides and polymers [30, 48]. The smaller shift when bound could also be a result of different rotational orientation of the helix on the membrane surface. These peptides have smaller hydrophobic driving force to bury side chains in the non-polar core, thus making aqueous exposure of these groups less energetically costly. Taken together, it is apparent that both hydrophobicity and electrostatics play a role in C18G binding to bilayers, and that the final conformation is impacted by the side-chain identity of the hydrophobic amino acids. Regarding the peptide structure, all peptides showed some degree of α-helical conformation when bound to anionic vesicles. However, this helical conformation was absent for the C18G-ABU and C18G-Ala peptides bound to PC bilayers. This supports the premise that these peptides adopt a different bilayer-bound conformation in PC, which likely contributes to the changes in barycenter shift and acrylamide quenching.
The hydrophobic balance of AMPs and peptidomimetic polymers has been an area of intensive study by many groups [9, 49,50,51,52]. Overall, a threshold hydrophobic character is required for these molecules to exert antimicrobial activity, presumably providing the ability to partition into and disrupt the bacterial membrane [53]. However, specific detail on this threshold appears tied to both the hydrophobicity and the structure of the hydrophobic groups (see below). Similar results were observed in peptides with non-natural amino acids and in peptidomimetic polymers [9, 31, 32, 49, 54,55,56].
Regarding the mechanism of action of the peptides, many studies have shown membrane disruption is the, or minimally a component of the, mechanism of action of many AMPs. In this work, the three best performing peptides judged by MIC data were also the three that exhibited the greatest extent of E. coli outer membrane permeabilization. This activity is similar to the behavior of C18G peptides in which hydrophobics were altered using only naturally occurring residues, as well as C18G variants in which the cationic residues were varied [30, 33]. Interestingly, the permeabilization of E. coli membranes and RBCs did not parallel to the lipid vesicle leakage assays. In model vesicles, the peptides disrupted zwitterionic bilayers most efficiently. Ramamoorthy and coworkers observed a similar pattern for the peptide MSI-78 (pexiganan) which has similar size and net charge to C18G [57]. One possibility for the discrepancy is the size and curvature of the bilayers. The SUVs created by sonication in the vesicle leakage assay typically average ~ 50-100 nm in diameter as compared to the much larger E. coli cell which averages ~1 × 3 μm [58]. The smaller SUVs have significantly enhanced bilayer curvature compared to the larger E. coli cell. It has been reported that a number of proteins and peptides can sense membrane curvature and have functions which are modulated by membrane curvature [59, 60]. Similarly, AMPs have been shown to both induce and be impacted by bilayer curvature [61, 62]. Matsuzaki and Epand demonstrated a similar result with Magainin-2 where the peptide loses the ability to permeabilize bilayers upon inclusion of PE lipids which induce membrane curvature [63].
The most intriguing results from this study are regarding the C18G-Nva peptide. This sequence showed good antimicrobial activity, on par with the most active C18G-Nle peptide in this study and the parent sequence C18G containing Leu as the hydrophobic residue. The non-natural norvaline side chain enhanced the activity of the peptide compared the variant with Val residues and was the only such isosteric substitution that enhanced activity compared to the naturally occurring counterparts [33]. Additionally, the C18G-Nva peptide exhibited almost no hemolytic activity over the range tested. The major difference between the two is the β-branching in the valine side chain as opposed to the linear chain in norvaline. This results in a slightly longer side chain which can bury into the bilayer but may also impact how lipids pack around the side chain. A linear Nva side chain (and Nle) can likely pack more tightly with lipid acyl chains compared to the branched natural variants, although it is unclear why this may enhance activity for C18G-Nva but not C18G-Nle, nor why the C18G-Nva exhibits significantly less hemolysis. Additional investigations on the impact of these peptides on bilayer properties may shed light on the role and difference between the peptides. The combination of good antibacterial activity and low hemolytic activity make this sequence a promising candidate for further development.
References
Ventola CL (2015) The antibiotic resistance crisis: part 1: causes and threats. P T 40(4):277–283
Spellberg B, Gilbert DN (2014) The future of antibiotics and resistance: a tribute to a career of leadership by John Bartlett. Clin Infect Dis 59(Suppl 2):S71–S75. https://doi.org/10.1093/cid/ciu392
Rossiter SE, Fletcher MH, Wuest WM (2017) Natural products as platforms to overcome antibiotic resistance. Chem Rev 117(19):12415–12474. https://doi.org/10.1021/acs.chemrev.7b00283
Jakubczyk D, Dussart F (2020) Selected fungal natural products with antimicrobial properties. Molecules 25(4). https://doi.org/10.3390/molecules25040911
Kuroda K, Caputo GA (2013) Antimicrobial polymers as synthetic mimics of host-defense peptides. Wiley Interdiscip Rev Nanomed Nanobiotechnol 5(1):49–66. https://doi.org/10.1002/wnan.1199
Kuroki A, Kengmo Tchoupa A, Hartlieb M, Peltier R, Locock KES, Unnikrishnan M, Perrier S (2019) Targeting intracellular, multi-drug resistant Staphylococcus aureus with guanidinium polymers by elucidating the structure-activity relationship. Biomaterials 217:119249. https://doi.org/10.1016/j.biomaterials.2019.119249
Zhou Z, Ergene C, Lee JY, Shirley DJ, Carone BR, Caputo GA, Palermo EF (2019) Sequence and dispersity are determinants of photodynamic antibacterial activity exerted by peptidomimetic oligo(thiophene)s. ACS Appl Mater Interfaces 11(2):1896–1906. https://doi.org/10.1021/acsami.8b19098
Choi S, Isaacs A, Clements D, Liu D, Kim H, Scott RW, Winkler JD, DeGrado WF (2009) De novo design and in vivo activity of conformationally restrained antimicrobial arylamide foldamers. Proc Natl Acad Sci U S A 106(17):6968–6973. https://doi.org/10.1073/pnas.0811818106
Locock KES, Michl TD, Stevens N, Hayball JD, Vasilev K, Postma A, Griesser HJ, Meagher L, Haeussler M (2014) Antimicrobial polymethacrylates synthesized as mimics of tryptophan-rich cationic peptides. ACS Macro Lett 3(4):319–323. https://doi.org/10.1021/mz5001527
Takahashi H, Nadres ET, Kuroda K (2017) Cationic amphiphilic polymers with antimicrobial activity for oral care applications: eradication of S. mutans biofilm. Biomacromolecules 18(1):257–265. https://doi.org/10.1021/acs.biomac.6b01598
Goderecci SS, Kaiser E, Yanakas M, Norris Z, Scaturro J, Oszust R, Medina CD, Waechter F, Heon M, Krchnavek RR, Yu L, Lofland SE, Demarest RM, Caputo GA, Hettinger JD (2017) Silver oxide coatings with high silver-ion elution rates and characterization of bactericidal activity. Molecules 22(9). https://doi.org/10.3390/molecules22091487
Hajizadeh H, Peighambardoust SJ, Peighambardoust SH, Peressini D (2020) Physical, mechanical, and antibacterial characteristics of bio-nanocomposite films loaded with Ag-modified SiO2 and TiO2 nanoparticles. J Food Sci 85:1193–1202. https://doi.org/10.1111/1750-3841.15079
Vincent M, Duval RE, Hartemann P, Engels-Deutsch M (2018) Contact killing and antimicrobial properties of copper. J Appl Microbiol 124(5):1032–1046. https://doi.org/10.1111/jam.13681
O'Brien KT, Noto JG, Nichols-O'Neill L, Perez LJ (2015) Potent irreversible inhibitors of LasR quorum sensing in Pseudomonas aeruginosa. ACS Med Chem Lett 6(2):162–167. https://doi.org/10.1021/ml500459f
Sahariah P, Masson M (2017) Antimicrobial chitosan and chitosan derivatives: a review of the structure-activity relationship. Biomacromolecules 18(11):3846–3868. https://doi.org/10.1021/acs.biomac.7b01058
Cook K, Tarnawsky K, Swinton AJ, Yang DD, Senetra AS, Caputo GA, Carone BR, Vaden TD (2019) Correlating lipid membrane permeabilities of imidazolium ionic liquids with their cytotoxicities on yeast, bacterial, and mammalian cells. Biomolecules 9(6). https://doi.org/10.3390/biom9060251
Hanna SL, Huang JL, Swinton AJ, Caputo GA, Vaden TD (2017) Synergistic effects of polymyxin and ionic liquids on lipid vesicle membrane stability and aggregation. Biophys Chem 227:1–7. https://doi.org/10.1016/j.bpc.2017.05.002
Hice SA, Varona M, Brost A, Dai F, Anderson JL, Brehm-Stecher BF (2020) Magnetic ionic liquids: interactions with bacterial cells, behavior in aqueous suspension, and broader applications. Anal Bioanal Chem 412(8):1741–1755. https://doi.org/10.1007/s00216-020-02457-3
Sharma K, Aaghaz S, Shenmar K, Jain R (2018) Short antimicrobial peptides. Recent Pat Antiinfect Drug Discov 13(1):12–52. https://doi.org/10.2174/1574891X13666180628105928
Mookherjee N, Anderson MA, Haagsman HP, Davidson DJ (2020) Antimicrobial host defence peptides: functions and clinical potential. Nat Rev Drug Discov 19:311–332. https://doi.org/10.1038/s41573-019-0058-8
Ponnappan N, Budagavi DP, Yadav BK, Chugh A (2015) Membrane-active peptides from marine organisms—antimicrobials, cell-penetrating peptides and peptide toxins: applications and prospects. Probiotics Antimicrob Proteins 7(1):75–89. https://doi.org/10.1007/s12602-014-9182-2
Ridgway Z, Picciano AL, Gosavi PM, Moroz YS, Angevine CE, Chavis AE, Reiner JE, Korendovych IV, Caputo GA (2015) Functional characterization of a melittin analog containing a non-natural tryptophan analog. Biopolymers 104(4):384–394. https://doi.org/10.1002/bip.22624
Kang HK, Kim C, Seo CH, Park Y (2017) The therapeutic applications of antimicrobial peptides (AMPs): a patent review. J Microbiol 55(1):1–12. https://doi.org/10.1007/s12275-017-6452-1
Zasloff M (1987) Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A 84(15):5449–5453. https://doi.org/10.1073/pnas.84.15.5449
Ahmed TAE, Hammami R (2019) Recent insights into structure-function relationships of antimicrobial peptides. J Food Biochem 43(1):e12546. https://doi.org/10.1111/jfbc.12546
Wang G (2015) Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol Biol 1268:43–66. https://doi.org/10.1007/978-1-4939-2285-7_3
Huang Y, Huang J, Chen Y (2010) Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein Cell 1(2):143–152. https://doi.org/10.1007/s13238-010-0004-3
Eckert R (2011) Road to clinical efficacy: challenges and novel strategies for antimicrobial peptide development. Future Microbiol 6(6):635–651. https://doi.org/10.2217/fmb.11.27
Costa JR, Silva NC, Sarmento B, Pintado M (2017) Delivery systems for antimicrobial peptides and proteins: towards optimization of bioavailability and targeting. Curr Pharm Biotechnol 18(2):108–120
Kohn EM, Shirley DJ, Arotsky L, Picciano AM, Ridgway Z, Urban MW, Carone BR, Caputo GA (2018) Role of cationic side chains in the antimicrobial activity of C18G. Molecules 23(2). https://doi.org/10.3390/molecules23020329
Narancic T, Almahboub SA, O'Connor KE (2019) Unnatural amino acids: production and biotechnological potential. World J Microbiol Biotechnol 35(4):67. https://doi.org/10.1007/s11274-019-2642-9
Oliva R, Chino M, Pane K, Pistorio V, De Santis A, Pizzo E, D'Errico G, Pavone V, Lombardi A, Del Vecchio P, Notomista E, Nastri F, Petraccone L (2018) Exploring the role of unnatural amino acids in antimicrobial peptides. Sci Rep 8(1):8888. https://doi.org/10.1038/s41598-018-27231-5
Saint Jean KD, Henderson KD, Chrom CL, Abiuso LE, Renn LM, Caputo GA (2018) Effects of hydrophobic amino acid substitutions on antimicrobial peptide behavior. Probiotics Antimicrob Proteins 10(3):408–419. https://doi.org/10.1007/s12602-017-9345-z
Darveau RP, Blake J, Seachord CL, Cosand WL, Cunningham MD, Cassiano-Clough L, Maloney G (1992) Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. J Clin Invest 90(2):447–455. https://doi.org/10.1172/JCI115880
Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI (2003) Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol 50(1):219–230. https://doi.org/10.1046/j.1365-2958.2003.03675.x
Park SY, Groisman EA (2014) Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol Microbiol 91(1):135–144. https://doi.org/10.1111/mmi.12449
Hitchner MA, Santiago-Ortiz LE, Necelis MR, Shirley DJ, Palmer TJ, Tarnawsky KE, Vaden TD, Caputo GA (2019) Activity and characterization of a pH-sensitive antimicrobial peptide. Biochim Biophys Acta Biomembr 1861(10):182984. https://doi.org/10.1016/j.bbamem.2019.05.006
Caputo GA, London E (2019) Analyzing transmembrane protein and hydrophobic helix topography by dual fluorescence quenching. Methods Mol Biol 2003:351–368. https://doi.org/10.1007/978-1-4939-9512-7_15
Institute C-CaLS (2015) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Approved standard - tenth edition. CLSI document M07-A10
Burman LG, Nordström K, Boman HG (1968) Resistance of Escherichia coli to penicillins V. physiological comparison of two isogenic strains, one with chromosomally and one with episomally mediated ampicillin resistance. J Bacteriol 96(2):438
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER suite: protein structure and function prediction. Nat Methods 12(1):7–8. https://doi.org/10.1038/nmeth.3213
Raghuraman H, Chattopadhyay A (2006) Effect of ionic strength on folding and aggregation of the hemolytic peptide melittin in solution. Biopolymers 83(2):111–121. https://doi.org/10.1002/bip.20536
Senetra AS, Necelis MR, Caputo GA (2020) Investigation of the structure-activity relationship in ponericin L1 from Neoponera goeldii. Pept Sci 112(3):e24162. https://doi.org/10.1002/pep2.24162
Chattopadhyay A, Haldar S (2014) Dynamic insight into protein structure utilizing red edge excitation shift. Acc Chem Res 47(1):12–19. https://doi.org/10.1021/ar400006z
Guha S, Rawat SS, Chattopadhyay A, Bhattacharyya B (1996) Tubulin conformation and dynamics: a red edge excitation shift study. Biochemistry 35(41):13426–13433. https://doi.org/10.1021/bi961251g
van Meer G, de Kroon AI (2011) Lipid map of the mammalian cell. J Cell Sci 124(Pt 1):5–8. https://doi.org/10.1242/jcs.071233
Kohn EM, Lee JY, Calabro A, Vaden TD, Caputo GA (2018) Heme dissociation from myoglobin in the presence of the zwitterionic detergent N,N-dimethyl-N-dodecylglycine betaine: effects of ionic liquids. Biomolecules 8(4). https://doi.org/10.3390/biom8040126
Palermo EF, Sovadinova I, Kuroda K (2009) Structural determinants of antimicrobial activity and biocompatibility in membrane-disrupting methacrylamide random copolymers. Biomacromolecules 10(11):3098–3107. https://doi.org/10.1021/bm900784x
Palermo EF, Kuroda K (2010) Structural determinants of antimicrobial activity in polymers which mimic host defense peptides. Appl Microbiol Biotechnol 87(5):1605–1615. https://doi.org/10.1007/s00253-010-2687-z
Thaker HD, Sgolastra F, Clements D, Scott RW, Tew GN (2011) Synthetic mimics of antimicrobial peptides from triaryl scaffolds. J Med Chem 54(7):2241–2254. https://doi.org/10.1021/jm101410t
Juretic D, Vukicevic D, Tossi A (2017) Tools for designing amphipathic helical antimicrobial peptides. Methods Mol Biol 1548:23–34. https://doi.org/10.1007/978-1-4939-6737-7_2
Jiang Z, Mant CT, Vasil M, Hodges RS (2018) Role of positively charged residues on the polar and non-polar faces of amphipathic alpha-helical antimicrobial peptides on specificity and selectivity for Gram-negative pathogens. Chem Biol Drug Des 91(1):75–92. https://doi.org/10.1111/cbdd.13058
Stark M, Liu LP, Deber CM (2002) Cationic hydrophobic peptides with antimicrobial activity. Antimicrob Agents Chemother 46(11):3585–3590. https://doi.org/10.1128/aac.46.11.3585-3590.2002
Avery CW, Palermo EF, McLaughlin A, Kuroda K, Chen Z (2011) Investigations of the interactions between synthetic antimicrobial polymers and substrate-supported lipid bilayers using sum frequency generation vibrational spectroscopy. Anal Chem 83(4):1342–1349. https://doi.org/10.1021/ac1025804
Torres MDT, Sothiselvam S, Lu TK, de la Fuente-Nunez C (2019) Peptide design principles for antimicrobial applications. J Mol Biol 431(18):3547–3567. https://doi.org/10.1016/j.jmb.2018.12.015
Zelezetsky I, Tossi A (2006) Alpha-helical antimicrobial peptides--using a sequence template to guide structure-activity relationship studies. Biochim Biophys Acta 1758(9):1436–1449. https://doi.org/10.1016/j.bbamem.2006.03.021
Lee DK, Brender JR, Sciacca MF, Krishnamoorthy J, Yu C, Ramamoorthy A (2013) Lipid composition-dependent membrane fragmentation and pore-forming mechanisms of membrane disruption by pexiganan (MSI-78). Biochemistry 52(19):3254–3263. https://doi.org/10.1021/bi400087n
Reshes G, Vanounou S, Fishov I, Feingold M (2008) Cell shape dynamics in Escherichia coli. Biophys J 94(1):251–264. https://doi.org/10.1529/biophysj.107.104398
Drin G, Antonny B (2010) Amphipathic helices and membrane curvature. FEBS Lett 584(9):1840–1847. https://doi.org/10.1016/j.febslet.2009.10.022
Fossati M, Goud B, Borgese N, Manneville JB (2014) An investigation of the effect of membrane curvature on transmembrane-domain dependent protein sorting in lipid bilayers. Cell Logist 4(2):e29087. https://doi.org/10.4161/cl.29087
Chen R, Mark AE (2011) The effect of membrane curvature on the conformation of antimicrobial peptides: implications for binding and the mechanism of action. Eur Biophys J 40(4):545–553. https://doi.org/10.1007/s00249-011-0677-4
Schmidt NW, Wong GC (2013) Antimicrobial peptides and induced membrane curvature: geometry, coordination chemistry, and molecular engineering. Curr Opin Solid State Mater Sci 17(4):151–163. https://doi.org/10.1016/j.cossms.2013.09.004
Matsuzaki K, Sugishita K, Ishibe N, Ueha M, Nakata S, Miyajima K, Epand RM (1998) Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry 37(34):11856–11863. https://doi.org/10.1021/bi980539y
Acknowledgments
The authors would like to thank Dr. Dylan Klein for assistance with hemolysis assays and Mr. Calvin Caputo for assistance in synthesis of peptides.
Funding
This work was funded in part by NIH 1R15GM094330 to G.A.C.
Author information
Authors and Affiliations
Contributions
Caputo participated in research design; Hitchner, Necelis, and Shirley conducted the experiments and performed data analysis; Caputo, Necelis, and Hitchner contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1403 kb)
Rights and permissions
About this article
Cite this article
Hitchner, M.A., Necelis, M.R., Shirley, D. et al. Effect of Non-natural Hydrophobic Amino Acids on the Efficacy and Properties of the Antimicrobial Peptide C18G. Probiotics & Antimicro. Prot. 13, 527–541 (2021). https://doi.org/10.1007/s12602-020-09701-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-020-09701-3