Abstract
This study aimed to elucidate the targets and mechanisms of anti-staphylococcal effects from bioactive metabolites produced by lactic acid bacteria. We aimed to better understand the safety and efficacy of these bioactive metabolites in in vivo systems, typically at topical sites. The cell-free supernatant and protein-rich fraction from Lactobacillus plantarum USM8613 inhibited staphyloxanthin biosynthesis, reduced (p < 0.05) the cell number of Staphylococcus aureus by 106 CFU/mL and reduced biofilm thickness by 55% in S. aureus-infected porcine skins. Genome-wide analysis and gene expression analysis illustrated the production of several plantaricins, especially the plantaricins EF and JK that enhanced the anti-staphylococcal effects of L. plantarum USM8613. In vivo data using rats showed that the protein-rich fraction from L. plantarum USM8613 exerted wound healing properties via direct inhibition of S. aureus and promoted innate immunity, in which the expression of β-defensin was significantly (p < 0.05) upregulated by 3.8-fold. The protein fraction from L. plantarum USM8613 also significantly enhanced (p < 0.05) the production of cytokines and chemokines through various stages of wound recovery. Using ∆atl S. aureus, the protein-rich fraction from L. plantarum USM8613 exerted inhibitory activity via targeting the atl gene in S. aureus. Taken altogether, our present study illustrates the potential of L. plantarum USM8613 in aiding wound healing, suppressing of S. aureus infection at wound sites and promoting host innate immunity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the past 100 years, changes in society and technology have led to lifestyle changes and improvement of many basic life needs globally. Simultaneously, these changes have resulted in new diseases and modification of existing dermatoses. The discovery and extensive use of antibiotics has generated various antibiotic-resistant variants, giving rise to new health risks [1]. Staphylococcus aureus is a transient opportunistic skin pathogen of humans and other animals. S. aureus is well equipped with various virulence factors that cause mild to severe infections, ranging from cutaneous to systemic infections. In addition to virulence factors, S. aureus also contains several surface components such as Protein A and extracellular adhesion proteins which facilitate S. aureus to evade the inhibitory actions of antibiotics and recognition by the host immune system [2, 3]. The emergence of antibiotic-resistant strains has further reduced treatment and therapeutic options [4]. Hence, there is a need for natural alternative compounds against S. aureus.
Lactic acid bacteria (LAB) are Gram-positive, non-sporulating, microaerophilic bacteria that produce lactic acid as the main end product of carbohydrate fermentation. LAB have a long history of use in the food industry due to their ability to exert various beneficial effects [5, 6]. In addition to its additive effect on food content, the intake of LAB also confers health benefits to the host via an improved gut ecosystem, reduced serum cholesterol and enhanced host immune system. Among them, members of the genera Lactobacillus are the most commonly and commercially used. Despite the long-term use of these beneficial lactobacilli in the food industry, it was not until recently that the use of lactobacilli was extended to improve dermal health. Several studies have suggested the use of these beneficial LAB to maintain cutaneous homeostasis and improve regulation of the skin immune system [7, 8].
Although LAB have been widely documented to produce antimicrobial substances, antimicrobial peptides such as bacteriocins remain the most extensively studied compounds. Bacteriocins are generally cationic and constitute a heterogenous group of peptides that could be classified into five main classes based on their molecular size, chemical structure, antimicrobial spectrum, mode of action and target cell receptors [9]. The bacteriocins produced by Lactobaclllus plantarum are termed plantaricins. The plantaricin locus contains five operons: the plnABCD operon encodes for the signal-transducing pathway, the plnEFI and plnJKLR operons encode for bacteriocins and its immunity proteins, plnGHSTUVW operon encodes for an ABC transport system that processes and secretes peptides containing double-glycine N-terminal leaders, and lastly, the plnMNOP operon encodes genes with unknown functions in bacteriocin synthesis [10]. In the past, antimicrobial peptides produced by Lactobacillus lactis such as nisin, which has been extensively used for food preservation in over 50 countries, have been applied as biopreservatives in the food industry [11]. Antimicrobial peptides are now used for applications such as incorporation into nanofibre scaffolds for medical applications beyond the food industry [12], demonstrating its therapeutic potential for pharmaceutical demands.
Meanwhile, limited data are available on topical applications of LAB that specifically target S. aureus in an in vivo model, towards both S. aureus cells and/or the infected host. Lactobacillus plantarum USM8613 has demonstrated promising anti-staphylococcal activity via inhibitory metabolites that are proteinaceous in nature (data not shown). However, the efficacy, safety and immune-modulating effects of L. plantarum USM8613 in the presence of opportunistic S. aureus remain largely unknown. Thus, we aimed to elucidate these properties using ex vivo and in vivo wound models.
Materials and Methods
Bacterial Cultures
L. plantarum USM8613 was isolated from a meat product (pork minced meat) purchased at a local market (Penang, Malaysia), following the methods from Najjari et al. [13]. It was activated successively three times in sterile de Man, Rogosa and Sharpe (MRS) broth (HiMedia, Mumbai, India) at 37 °C before experimental use. A clinical isolate of S. aureus (identified via 16s rDNA) was obtained from the Penang General Hospital (Penang, Malaysia). The atl null (∆atl) and its wild-type (WT) S. aureus strains were obtained from Hiroshima University, Japan. S. aureus was activated in trypticase soy broth (TSB; HiMedia) at 37 °C for 24 h prior to use.
Cell-Free Supernatant and Crude Protein Fractionation
L. plantarum USM8613 (grown overnight for 20 h at 37 °C) was diluted (OD600nm = 1.0) with MRS broth (HiMedia) prior to centrifugation at 3500×g for 15 min at 4 °C to obtain cell-free supernatant (CFS). The CFS was neutralised to pH 7.0 using 1.0 M NaOH and then filter-sterilised through 0.22 μm mixed cellulose filter membrane prior to use (Merck Millipore, Darmstadt, Germany). Crude protein was extracted using an ammonium sulphate (at 80% saturation) precipitation method with stirring at 4 °C for 18 h. The mixture was centrifuged at 8000×g for 10 min at 4 °C and the pellet was suspended with 1.0 M ammonium acetate buffer, pH 6.5 in a ratio of 1:1 (v/w) [14]. Then, the solution was filtered and the protein content was determined via the Bradford assay [15]. The crude protein obtained is then designated as a protein-rich fraction. Unfermented MRS broth fractionated in a similar manner was used as a control. Anti-staphylococcal activity against S. aureus was tested using the 96-well microtitre plate assay following the method from Romero-Tabarez et al. [16].
Staphyloxanthin Biosynthesis Inhibition Assay
Inhibition of staphyloxanthin biosynthesis was qualitatively determined according to the method by Liu et al. [17]. Briefly, 15 mL of trypticase soy agar (TSA; HiMedia) supplemented with 10% (v/v) unfermented MRS broth (HiMedia) was poured into a Petri dish and left to be solidified. Half of the agar was cut and dug out using a sterilised scalpel and tweezer. Then, 7.5 mL of TSA supplemented with 10% (v/v) CFS from L. plantarum USM8613 was added into the excavated area. S. aureus was streaked on both of the half area separately and incubated at 37 °C for 24 h.
Agarose Gel Electrophoresis
The DNA of L. plantarum USM8613 was extracted using a QIAamp DNA Mini Kit (Qiagen, Venlo, Netherlands), following the manufacturer’s protocol. Polymerase chain reaction (PCR) was performed using a Taq PCR Master Mix Kit (Qiagen) and a Biometra Tone Thermal Cycler (Analytik Jena, Jena, Germany). The PCR amplicons were subjected to 2.0% (w/v) agarose gel electrophoresis at 100 V for 60 min. The gel was visualised using the GeneGenius Imaging System (Syngene, Cambridge, UK). The primers used are listed in Table S1.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction
Total RNA of L. plantarum USM8613 (grown overnight for 20 h at 37 °C) was extracted using RNeasy Mini kits (Qiagen) and converted to cDNA using SuperScript™ III Reverse Transcriptase (Life Technologies, CA, USA). Equal amounts of cDNA were used for qRT-PCR using QuantiTech SYBR® Green PCR master mix (Qiagen). The reactions were performed using an ABI 7500 Fast Real-Time PCR system (Applied Biosystems, CA, USA). All primer sequences are listed in Tables S1.
Porcine Skin
The dorsal skin part of a 60-kg domestic farm pig was obtained from a local market (Penang, Malaysia). The body hair on the skin was removed using a scalpel and the skin was cleaned using 70% (v/v) isopropanol (QRec, Malaysia). The skin samples were then cut into 4-cm2 (2 cm × 2 cm) pieces. The 4-cm2 skin samples were subjected to different treatments: (a) inoculated with 200 μL of unfermented MRS broth (HiMedia) and incubated at 37 °C for 24 h; (b) inoculated with S. aureus (106 CFU/mL) in 200 μL of unfermented MRS and incubated at 37 °C for 24 h; and (c) inoculated with S. aureus (106 CFU/mL) in 200 μL of protein-rich fraction of L. plantarum USM8613 prior to incubation at 37 °C for 24 h.
Confocal Scanning Laser Microscopic Analysis
The porcine skin samples were fixed in 10% (v/v) neutral buffered formalin for 24 h in separated glass Petri dishes. Following fixation, the skin samples were washed and dehydrated through an ascending series of ethanol, specifically 30%, 50%, 70%, 90% and 100% (v/v), and finally kept at − 20 °C. The dehydrated skin samples were then re-wetted via soaking in sterilised distilled water prior to the staining process. The rehydrated skin samples were then stained with 1% (v/v) acridine orange (AO) dye (Sigma-Aldrich, Steinheim, Germany) for 15 min, followed by washing with PBS (10 mM, pH 7.4) for 30 min. The skin samples were further stained with 1% (v/v) 2-(4-amidinophenyl)-1H-indole-6- carboxamide (DAPI; Sigma-Aldrich) for 30 min, and washed again with PBS (10 mM, pH 7.4) for 30 min. The skin samples were then examined using a confocal scanning laser microscope (CSLM; Eclipse-Ti, NY, USA). The thickness of the biofilm was quantified using the COMSTAT programme written as a script in the MATLAB 5.2 software suite [18].
Microbial Enumeration
The total microbial count of the porcine skins was enumerated via the spread plate method. Briefly, the porcine skin was immersed in 1 mL PBS (10 mM, pH 7.4) and homogenised using the Seward Stomacher 400C (Cole-Palmer, IL, USA) at 160 rpm for 30 s. The homogenised solution was obtained and a tenfold serial dilution was performed. One hundred microlitres of each sample dilution was spread onto mannitol salt agar (MSA; HiMedia). Three replicates were carried out for each dilution, and the agar plates were incubated at 37 °C for 24 h. The colonies were counted and the results were tabulated.
Animals
Thirty-six male Wistar rats weighting 150–200 g each were obtained from the Animal Research Unit and Service Centre (ARASC), Universiti Sains Malaysia (USM), and were housed individually in polypropylene cages under controlled light conditions with a 12-h light-dark cycle at 22 ± 2 °C and 50 ± 5% humidity. Water and food were provided to animals ad libitum. The 36 rats were divided into treatment and control groups with n = 18 rats in each group. The experimental protocol for treating animals was approved and in accordance with the guidelines recommended by the USM Committee on Animal Care; reference number: USM/Animal Ethics Approval/2012/(81)(433).
Formulation of Skin Ointment
A 10% (v/w) formulated ointment was prepared by mixing 50 μL of the protein-rich fraction from L. plantarum USM8613 with 500 mg of yellow soft paraffin [19]. Control ointment was prepared by mixing the protein-rich fraction precipitated from unfermented MRS broth with yellow soft paraffin (Unicorn Petroleum Industries, Mumbai, India).
Wounding and Infection
The animals were anaesthetised using ketamine/xylazine (75 mg/kg; 10 mg/kg). Using a sterile scalpel, a full thickness wound of size 1.5 cm × 1.5 cm was created by removing the skin on the shaved dorsal part of the animals, at a depth down to the underlying fascia of the panniculus carnosus muscle. Wound sites were inoculated with 50 μL (106 CFU/mL) of virulent S. aureus. On the following day, the wounds of L. plantarum USM8613-treated rats were topically treated with 10% formulated protein-rich fraction of L. plantarum UMS8613 ointment, while control group rats were topically treated with 10% formulated control ointment. The wounds were then covered with plain gauze and reinforced with cohesive bandage. The ointment was reapplied twice per day. A total of three rats from each group were sacrificed at intervals of 4 days after treatment (days 1, 4, 8, 12 and 16). Blood samples were collected and the entire wound tissues were cut out and stored at − 80 °C until further analysis. The expression of β-defensin at the wound sites was determined via the qRT-PCR method. Wound sites were evaluated at days 1, 4, 8, 12 and 16. The decrease in wound diameter during the healing process was measured with a calliper, with the percentage of wound closure (wound contraction) calculated as [19]:
Histological Analysis
Fresh tissue samples were collected and fixed in 10% (v/v) neutral formalin buffer for 24 h at 4 °C. Conventional techniques of paraffin wax sectioning and haematoxylin-eosin staining were used for histological studies. The formalin-fixed tissues were dehydrated via series of ethanol [30%, 50%, 70%, 90% and 100% (v/v)] and cleared in xylene [100% (v/v); QRec] and embedded in paraffin wax (Sigma-Aldrich). Sections of 5–8-μm thickness were cut using a microtome (Leica Biosystems, Nussloch, Germany) and stained using aqueous haematoxylin and eosin (H&E). The prepared slides were viewed under a light microscope (Nikon Instruments, NY, USA).
Serum Cytokine Analyses
Blood samples were collected in vacutainer tubes (Becton Dickinson, NJ, USA) and centrifuged at 3500×g for 10 min at 25 °C to obtain serum. The levels of interleukin-4 (IL-4), interleukin-6 (IL-6), transforming growth factor-β (TGF-β), interferon-γ (IFN-γ) and tumour necrosis factor-α (TNF-α) in the blood serum were determined using a commercial ELISA kit (R&D System Inc., MN, USA) according to the manufacturers’ protocol.
MMP Gelatine Zymography
The presence of matrix metalloproteinases (MMPs) on the granulation tissue was analysed by gelatine zymography [19].
Transmission Electron Microscopic Analysis
The activated ∆atl and WT S. aureus cells were diluted to OD600nm = 0.3 (106 CFU/mL) and treated with 5 mL of 200 AU/mL protein-rich fraction in a 1:1 ratio at 37 °C for 5 h until the mid-exponential growth phase. The cells were collected via centrifugation (2200×g at 25 °C for 15 min) and washed twice with 0.1 M phosphate buffer (pH 7.2). The cell pellets obtained were pre-fixed with McDowell-Trump fixative for 24 h at 4 °C. The fixed samples were washed twice with 0.1 M phosphate buffer, followed by post-fixation in 1% osmium tetroxide (Sigma-Aldrich) for 1 h at 25 °C. Cell pellets were subsequently encapsulated in 2% (w/v) batch agar and cut into 1 mm × 5 mm rectangular shapes and dehydrated through a series of ethanol washes [50%, 75%. 95%, and 100% (v/v)] and a 100% (v/v) acetone wash prior to being embedded in Spurr resin. Five pieces of each sample were embedded in gelatine capsules filled with Spurr resin and polymerised at 60 °C for 24 h. Ultra-thin sections (70 to 90 nm) were collected onto uncoated 300-mesh nickel grids and stained using 2% (v/v) uranyl acetate and lead citrate. Samples were viewed and captured using an EF-TEM Libra 120 (Carl Zeiss, Oberkochen, Germany).
Quantitative Reverse Transcriptase-Polymerase Chain Reaction
Total RNA of tissue samples, WT S. aureus and ∆atl S. aureus (grown overnight for 20 h at 37 °C) were extracted using RNeasy Mini kits (Qiagen) and converted to cDNA using SuperScript™ III Reverse Transcriptase (Life Technologies, USA). Equal amounts of cDNA were used for qRT-PCR using QuantiTech SYBR® Green PCR master mix (Qiagen). The reactions were performed using an ABI 7500 Fast Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA). All primer sequences are listed in Tables S2, S3 and S4.
Statistical Analyses
All data were presented as the mean ± standard deviation from triplicate runs (n = 3), unless stated otherwise, and analysed using SPSS© version 20.0. One-way ANOVA was used to determine the significant differences between means at a significance level of ρ < 0.05. Tukey’s test was used to perform multiple comparisons between means. The independent t test was used to determine the significant differences between two means at a given time point with a significance level of ρ < 0.05.
Results
Anti-staphylococcal Activity of CFS and Plantaricin Genes
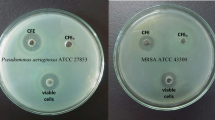
S. aureus that grew on TSA supplemented with 10% (v/v) unfermented MRS broth appeared as golden-carotenoid pigmentation colonies but lost the golden-carotenoid pigment and grew as pale-white colonies in TSA supplemented with 10% of CFS from L. plantarum USM8613 (Fig. 1A). The ability of CFS to inhibit and remove biofilm formed by S. aureus was demonstrated using S. aureus-infected porcine skins. The number of viable S. aureus cells was significantly lower (ρ < 0.05) in the CFS treatment group (104 CFU/mL) compared to the unfermented MRS treatment group (1010 CFU/mL) (Fig. 1B). In addition, the biofilm formed was also significantly (ρ < 0.05) thicker in the unfermented MRS control group (118.92 ± 2.87 μm) in comparison to the CFS treatment group (53.67 ± 5.56 μm) (Fig. 1C).
(A) Staphyloxanthin production of S. aureus illustrated on TSA supplemented with 10% (v/v) of MRS and cell-free supernatant of L. plantarum USM8613 (CFS). (B) Number of viable S. aureus cell count on porcine skin samples inoculated with 200 μL of unfermented MRS, S. aureus (106 CFU/mL) in 200 μL of unfermented MRS or, S. aureus (106 CFU/mL) in 200 μL of CFS prior to incubation at 37 °C for 24 h. (C) Thickness of biofilm formed on S. aureus-infected porcine skins. Results are expressed as mean ± standard deviation of means (n = 3). a, bMeans between different sample groups were significantly different (ρ < 0.05)
Gel electrophoresis revealed the presence of structural pln genes such as plnA, plnC, plnEF, plnJ and plnK, and non-structural pln genes such as plnB, plnD, plnG, plnI, plnN and plnS in L. plantarum USM8613. Meanwhile, the non-structural plnNC8 and structural plnW genes were both not detected (Fig. 2A). The positive response regulator PlnC was upregulated (ρ < 0.05) while the negative response regulator PlnD was suppressed (based on expression level = 1.00; ρ < 0.05; Fig. 2B). Gene expression of plnB and plnEF was neither upregulated nor suppressed, while the plnJ and plnK genes were significantly upregulated (ρ < 0.05) and plnI and plnS were significantly downregulated (ρ < 0.05; Fig. 2B).
(A) Gel electrophoresis of PCR amplicons of plantaricin genes (pln) in L. plantarum USM8613. (B) Gene expression levels of plantaricin genes in L. plantarum USM8613. Results are expressed as mean ± standard deviation of means (n = 3). *Means between different sample groups were significantly different (ρ < 0.05)
Anti-staphylococcal Activity in Porcine Skin
The viable cell count of S. aureus in porcine skin treated with the protein-rich fraction of CFS from L. plantarum USM8613 was lower (ρ < 0.05) as compared to the porcine skin treated with the protein-rich fraction of unfermented MRS control at all time points studied (Fig. 3A). The protein-rich fraction also reduced the thickness of biofilm formed as compared to the control group at all time points studied (ρ < 0.05; Fig. 3B). Confocal images of the porcine skin treated with the MRS control revealed a thick layer of biomass on the surface, covering the porcine basal skin layer, as compared to porcine skin treated with the protein-rich fraction at all time points studied (Fig. 3C). In addition, colonies of S. aureus (represented as red coloured spots and indicated with white arrows) were observed in the protein-rich fraction-treated group, but not in the MRS control group (Fig. 3C).
(A) Viable cell count and (B) biofilm thickness of S. aureus on porcine skin samples upon treatment with the protein-rich fraction from unfermented MRS or CFS of L. plantarum USM8613. (C) Confocal micrographs of porcine skin cells (blue) stained with DAPI were differentiated from S. aureus cells (red) and its product (green) which was stained using AO dye. Magnification: 100x. Results are expressed as mean ± standard deviation of means (n = 18). a, bMeans within the same sample group and A, Bmeans between different treatment groups at the same time point were significantly different (ρ < 0.05)
Wound Healing, Inhibitory and Inflammatory Properties
After 1 day, the population of S. aureus maintained at 106 CFU/g in control rats, but reduced to 105 CFU/g in rats treated with the protein-rich fraction of CFS from L. plantarum USM8613. Subsequent reductions were observed over time for both treatment groups. By day 12, the population of S. aureus was reduced to 104 CFU/g in the control rats while the L. plantarum USM8613-treated rats showed a reduction of 50% as compared to the initial inoculum to reach 103 CFU/g (Fig. 4A). The L. plantarum USM8613-treated rats also showed an enhanced wound contraction percentage (54%) compared to their control counterparts (43%; ρ < 0.05) by day 4, although all rats showed increased wound contractions over time (Fig. 4B). Rats in the control group only achieved a wound contraction percentage of above 50% by day 8 (79%).
(A) Population of S. aureus cells at wound sites (1.5 cm × 1.5 cm) of infected rats treated with 500 mg of yellow soft paraffin containing 50 μL of the protein-rich fraction from CFS of L. plantarum USM8613 or empty paraffin (control), applied twice daily over 16 days. (B) Wound contraction percentage of S. aureus-infected rats. Results are expressed as mean ± standard deviation of means (n = 18). *Means between treatment groups within the same sampling day are significantly different (ρ < 0.05); a, b, c, dMeans between sampling day within the same group were significantly different (ρ < 0.05)
The production of β-defensin at wound sites of the L. plantarum USM8613-treated rats were upregulated (ρ < 0.05) in the initial stages of infection and healing, with a marked peak of 3.83-fold higher expression by day 4 as compared to the control group (Table 1). The expression of β-defensin of the L. plantarum USM8613-treated rats decreased over time (days 4–12) and was significantly lower compared to the control group at day 16 (ρ < 0.05).
H&E staining of skin biopsies showed that the protein-rich fraction from CFS of L. plantarum USM8613 accelerated keratinocyte migration over the wound edge towards the centre area over time (days 4–16; Fig. 5a–e) and achieved better wound closure and complete re-epithelisation by day 12 (Fig. 5d). Rats in the control group displayed slower wound closure over time (Fig. 5f–j), where complete re-epithelisation was only observed by day 16 (Fig. 5j). In addition, the presence of inflammatory cells remained visible at wound sites of the control rats at day 16 (Fig. 5j) as compared to day 12 of L. plantarum USM8613-treated rats (Fig. 5c).
Haematoxylin and eosin (H&E)-stained sections of the granulation tissue at wound sites from rats treated with 500 mg of yellow soft paraffin containing 50 μL of the protein-rich fraction from CFS of L. plantarum USM8613 or empty paraffin (control), applied twice daily over 16 days. L. plantarum USM8613-treated rats at (a) day 1, (b) day 4, (c) day 8, (d) day 12 and (e) day 16, and control rats at (f) day 1, (g) day 4, (h) day 8, (i) day 12 and (j) day 16. Magnification: × 400; Scab (SC), re-epithelialisation (Ep) and inflammatory cells (Ic)
In the early days of wound recovery (days 1–4), rats treated with the protein-rich fraction from CFS of L. plantarum USM8613 showed higher concentrations of Il-4, IL-6 and TGF-β at wound sites as compared to the control group (Fig. 6a, c and e), but showed lower concentrations of TNF-α than the control group during the final stages of wound recovery (day 16; Fig. 6b). The concentrations of IFN-γ at wound sites were increased after day 1 and decreased over time in both groups, but the L. plantarum USM8613-treated group decreased more by day 8 as compared to the control group, which only decreased by day 12 (Fig. 6d). Meanwhile, the levels of MMPs in wound sites of L. plantarum USM8613-treated rats were gradually reduced over time but this was not observed in the control rats (Fig. 7).
Production of various cytokines, (A) interleukin (IL)-4, (B) tumour necrosis factor-α (TNF-α), (C) IL-6, (D) interferon-γ (IFN-γ) and (E) transforming growth factor-β (TGF-β) at wound sites of infected rats treated with 500 mg of yellow soft paraffin containing 50 μL of the protein-rich fraction from CFS of L. plantarum USM8613 or empty paraffin (control), applied twice daily over 16 days. Results are expressed as mean ± standard deviation of means (n = 18). *Means between treatment groups with the same sampling day were significantly different (ρ < 0.05); a, b, cMeans between sampling days within the same group were significantly different (ρ < 0.05)
Matrix metalloproteinases (MMPs) zymogram at wound sites of infected rats treated with 500 mg of yellow soft paraffin containing 50 μL of the protein-rich fraction from CFS of L. plantarum USM8613 (A) or empty paraffin (control; B), applied twice daily over 16 days. (Red arrows) MMP-9, 92 kDa; (yellow arrows) MMP-8, 75 kDa
Autolysis Mechanisms
The gene expression of global regulators such as sigB, agrA and ROT in ∆atl S. aureus were unaffected upon treatment with the protein-rich fraction from CFS of L. plantarum USM8613 (Fig. 8a), while TRAP expression was significantly upregulated (ρ < 0.05). The same phenomena were observed in the pathogenicity factors of ∆atl S. aureus, in which expression of genes hlb, clfB, icaA and lytM were unaffected by the protein-rich fraction from L. plantarum USM8613 (Fig. 8b), while only the expression of the pathogenicity factor spa gene, which encodes for protein A, was suppressed (ρ < 0.05) upon treatment with the protein-rich fraction from CFS of L. plantarum USM8613 (Fig. 8b). Under TEM, the control Δatl S. aureus was distinguishable by its rough outer surface, in contrast with the smooth outer surface appearance of the WT S. aureus (Fig. 8c). L. plantarum USM8613 treated-WT S. aureus also failed to maintain an intact cell wall, where leakage of the intracellular content was observed (Fig. 8c).
Gene expression of (A) global regulators and (B) pathogenicity factors in Δatl and WT S. aureus upon treatment with the protein-rich fraction from CFS of L. plantarum USM8613. C) TEM images of Δatl and WT S. aureus cells that were untreated (top images) and upon treatment with the protein-rich fraction from CFS of L. plantarum USM8613 until the mid-exponential phase (5 h; bottom images). Magnification: 25,000X. Results are expressed as mean ± standard deviation of means (n = 3). a, bMeans between Δatl and WT S. aureus within the same gene were significantly different (ρ < 0.05). *Means between treated-S. aureus and the untreated control (expression level = 1.00) were significantly different (ρ < 0.05)
Most of the autolysis-related genes studied were expressed normally and not affected by the protein-rich fraction in both Δatl and WT S. aureus. The expression of the mgrA gene was the only one that was upregulated, while the expression of atl was not detected in Δatl S. aureus upon protein-rich fraction treatment (ρ < 0.05). Meanwhile, expression of norA, cidA and sigB in WT S. aureus was upregulated and that of sarA and agrA was downregulated (ρ < 0.05) upon treatment with the protein-rich fraction of CFS from L. plantarum USM8613 (Fig. 9).
Expression of autolysis-related genes in Δatl and WT S. aureus upon treatment with the protein-rich fraction from CFS of L. plantarum USM8613. Results are expressed as mean ± standard deviation of means (n = 3). abMeans between Δatl and WT S. aureus within the same gene were significantly different (ρ < 0.05). * Means between treated S. aureus and the untreated control (expression level = 1.00) were significantly different (ρ < 0.05)
Discussion
Biofilm formation is a virulence factor in the pathogenesis of staphylococcal infections, contributing to over 80% of bacterial infections, and is one of the strategies used by S. aureus to survive in various environments [20]. L. plantarum USM8613 inhibited staphyloxanthin biosynthesis and reduced biofilm formation by S. aureus. Staphyloxanthin serves as a reactive oxygen radical scavenger that contributes to the survival of S. aureus during stress and host response. Since staphyloxanthin is membrane-bound, it primarily protects the membrane lipid against reactive oxygen radical threat and also protects the protein and DNA from free radicals [21]. By inhibiting staphyloxanthin biosynthesis, L. plantarum USM8613 could compromise the survival of S. aureus under stress conditions.
Another proposed mechanisms involved the inhibition of quorum sensing, namely, the agr system and expression of the ica operon. The ica operon encodes for the production of polysaccharide intercellular adhesion, which is crucial for biofilm formation [22]. The upregulation of plnJ and plnK genes indicates the ability of L. plantarum USM8613 to produce the two-peptide plantaricin JK, which exerts activity via complementary actions of two different peptides and facilitates initial contact with the negatively charged membrane of S. aureus via electrostatic interaction [23]. Plantaricin JK permeabilises the inner membrane, leading to influx and efflux of various molecules across the transmembrane barrier, and altering electrical potential and pH gradients across the membrane, leading to cell death [24].
In addition to inhibition of the growth and pathogenicity of S. aureus, L. plantarum USM8613 also improved wound healing properties. Wound healing is a complex biological process that consists of four phases: the formation of blood clots, inflammation, proliferation and maturation. Blood clots are formed to seal and protect against further fluid loss and potential infections, while inflammation at wound sites in the early stages of infection triggers the wound healing process and blocks the invasion of pathogens [25]. The protein-rich fraction from L. plantarum USM8613 stimulated the production of β-defensin at wound sites. β-defensin, which is a group of small (3–5 kDa), cationic, cysteine-rich β-sheet peptides, plays an important role in cutaneous host immune defences. It confers a broad spectrum of antibacterial properties by disrupting membrane integrity leading to pathogenic cell lysis [26].
In addition to β-defensin, L. plantarum USM8613-treated rats also showed peak production of TNF-α, IL-4 and IL-6 during the early stages of wound healing, followed by a gradual decrease of TNF-α towards the end of the study. As a pro-inflammatory cytokine, TNF-α initiates the pro-inflammatory cascade and induces IL-1β synthesis to start the inflammatory phase. TNF-α gradually decreases in the latter stages of wound healing to prevent overexpression, which causes a delay in the wound healing process [27]. Meanwhile, IL-4 aids in the recruitment of mast cells and other inflammatory cells to wound sites [25], and IL-6 is expressed in the early stages of wound healing to influence the accumulation of neutrophils [27]. TH1 cells, NK cells and macrophages often produce IFN-γ, an immunomodulator that activates cytotoxic activity to kill foreign invaders [25]. In both groups, the production of IFN-γ was elevated in the initial inflammatory phase to aid against S. aureus infection. The levels of IFN-γ gradually decreased over time upon the clearance of S. aureus infections in the later stages, as overproduction of IFN-γ causes negative feedback and interrupts the wound healing process [28].
During the proliferation phase, the number of macrophages decreases at wound sites, while migration and proliferation of keratinocytes, fibroblasts and capillary endothelial cells results in re-epithelialisation and tissue granulation in close context with neovascularisation [28]. Several growth factors such as TGF-β and MMPs are secreted to stimulate proliferation, synthesis of extracellular matrix proteins and angiogenesis. TGF-β is a key regulator for the proliferation of keratinocytes at wound edges for re-epithelialisation by increasing collagen synthesis [25]. TGF-β transformed wound clots produced by activated fibroblasts into myofibroblasts, generating a strong contractile force for wound closure [25]. The levels of TGF-β were higher in L. plantarum USM8613-treated rats in the early stages of wound recovery than in the control rats, indicating a faster wound healing effect. During the maturation phases, epidermal migration ceases and a new stratified epidermis with underlying basal lamina is re-established from the outer margins of the wound inwards. A reduction in MMP-9 levels during the late stages of wound recovery as observed in L. plantarum USM8613-treated rats is crucial, as prolonged and excessive production of MMP-9 and TNF-α would result in negative feedback, leading to a loss of keratinocytes and disordered wound healing [29]. The overall antimicrobial and wound healing-promoting activity of the protein fraction from L. plantarum USM8613 in S. aureus-infected rats is summarised in Fig. S1.
During cell division of S. aureus, the atl gene product appears as a ring structure on the septal region of the cell surface for cell separation. A proper balance between cell wall synthesis and degradation by autolysin during bacteria growth is needed to prevent autolysis of the cell due to overproduction of autolysin [30]. Treatment with a lytic concentration of penicillin reportedly induced autolysis of S. aureus cells while penicillin-induced autolysis was eliminated upon treatment with antisera against the atl gene product [31], illustrating the physiological involvement of the atl gene in autolysis. Hence, the role of the atl gene during autolysis, as induced by the protein-rich fraction from L. plantarum USM8613, was investigated using an atl null mutant strain of S. aureus.
Only the TRAP gene showed upregulated expression in ∆atl S. aureus. The same phenomena were observed in the pathogenicity factors of ∆atl S. aureus, in which gene expression of hlb, clf, icaA, and lytM were unaffected by the protein-rich fraction from L. plantarum USM8613. The results obtained indicated a directional relationship between the regulatory and downstream effector genes, in which the unaffected gene expression of sigB and agrA in ∆atl S. aureus enables their effector pathogenicity factors to express normally. Meanwhile, expression of the pathogenicity factor spa gene, which encodes for protein A, was suppressed after treatment with the protein-rich fraction from L. plantarum USM8613, resulting in reduced pathogenesis in ∆atl S. aureus.
After treatment with the protein-rich fraction from L. plantarum USM8613, most of the autolysis-related genes (except mgarA) in both Δatl and WT S. aureus were expressed normally and not affected by the protein-rich fraction from L. plantarum USM8613. The mgrA gene played an important role in suppressing the expression of autolysins to maintain the balance between cell wall synthesis and lysis for cell survival [32]. The norA and norB efflux pump system, which is regulated by mgrA, is another defence mechanism of S. aureus that enables cell adaptation to various environmental stresses [33]. In the current study, expression of both norA and norB of ∆atl S. aureus was unaffected, while expression of norA in WT S. aureus was upregulated upon treatment with the protein-rich fraction from L. plantarum USM8613. This indicated that upregulation of the mgrA gene was insufficient to upregulate expression of the norA and norB genes in ∆atl S. aureus upon treatment with the protein-rich fraction of L. plantarum USM8613.
Under the stresses of the stationary phase, environmental and host signals, sar proteins alter the expression and transcription of the agr system, virulence determinants and autolysis in S. aureus cells [34]. Downregulation of sarA expression in WT S. aureus indicated the ability of the protein-rich fraction from L. plantarum USM8613 to affect major regulators and effector genes in WT S. aureus towards autolysis, whereas gene expression of sarA in ∆atl S. aureus was unaffected by the protein-rich fraction from L. plantarum USM8613, enabling all of the autolysis-related genes to express normally. The atl gene also directly affected the high expression levels of the sigB gene in WT S. aureus. In addition to the major autolysin atl gene, the cidABC operon has reportedly also enhanced murein hydrolase activity and eventually cell death [35]. The expression of the cidABC operon is dependent on the stress regulator, sigB [36]. This indicates that the high expression level of the cidA gene in WT S. aureus was attributed to upregulation of the sigB gene upon treatment with the protein-rich fraction from L. plantarum USM8613, while such a phenomenon was absent in ∆atl S. aureus. The overall effect of the protein-rich fraction from L. plantarum USM8613 on the autolysis pathways of S. aureus is summarised in Fig. 10.
Overall effect of the protein-rich fraction from L. plantarum USM8613 on the autolysis pathways of S. aureus. Upon treatment, the global regulator, sarA was suppressed, leading to activation of the autolysis process and cell death in S. aureus. Induction of stress (sigB gene) by the protein-rich fraction led to the expression of the cidA gene that eventually caused autolysis in S. aureus. Expression of the major autolysin atl gene was upregulated by 8.5-fold upon protein-rich fraction treatment, contributing to major cell death in S. aureus via autolysis. Expression of the global regulator mgrA gene and the efflux system norA gene was upregulated by 1.2- and 2.4-fold, respectively, to counteract the protein-rich fraction-induced autolysis in S. aureus, but it was insufficient to overcome cell death through autolysis as the atl gene was upregulated by 8.5 fold
Conclusions
The protein-rich fraction from L. plantarum USM8613 exerted wound healing properties via inhibition of opportunistic S. aureus and promotion of innate immunity via stimulated expression of β-defensin at wound sites. The protein fraction from L. plantarum USM8613 also enhanced the production of cytokines and chemokines (TNF-α, IFN-γ, IL-4, IL-6, TGF-β and MMPs) through various stages of wound recovery starting from inflammation to re-epithelialisation, resulting in faster wound recovery. The protein-rich fraction from L. plantarum USM8613 also reduced the survival and resistance of S. aureus via autolysis pathways. The results from this study suggest the possible use of L. plantarum as a natural alternative for treatment of S. aureus dermal infections and wound healing amid concerns of antibiotic resistance.
References
Amini R, Sahib AA, Wong CB, Jahansiri F, Chung C, Hematian A, Sekawi Z, Jalilian FA (2013) Identification and screening of methicillin-resistant Staphylococcus aureus from fingernails among university students in Malaysia. Asia Life Sci 22:323–340
Chavakis T, Hussain M, Kanse SM, Peter G, Bretzel RG, Flock JI, Herrmann M, Preissner KT (2002) Staphylcoccus aureus extracellular adherence protein (Eap) serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat Med 8:687–693
Lee LY, Miyamoto YJ, Mclntyre BW, Höök M, McCrea KW, McDevitt D, Brown EL (2002) The Staphylococcus aureus Map protein is an immunomodulator that interferes with T cell mediated responses. J Clin Invest 110:1461–1471
Foster TJ (2005) Immune evasion by staphylococci. Nat Rev Microbiol 3:948–958
Jay J (2000) Fermentation and fermented dairy products. In: Jay J (ed) Modern food microbiology. Aspen Food Science Text Series. Springer US, New York, pp 113–130
Holzapfel WH, Haberer P, Geisen R, Björkroth J, Schillinger U (2001) Taxonomy and important features of probiotic microorganisms in food and nutrition. Am J Clin Nutr 73:365s–373s
Kalliomäki M, Salminen S, Poussa T, Isolauri E (2007) Probiotic during the first 7 years of life: a cumulative risk reduction of eczema in a randomised, placebo-controlled trial. J Allergy Clin Immunol 119:1019–1021
Kalliomäki M, Salminen S, Poussa T, Arvilommi H, Isolauri E (2003) Probiotics and prevention of atopic disease: 4-year follow-up of a randomized placebo-controlled trial. Lancet 361:1869–1871
Gillor O, Etzion A, Riley MA (2008) The dual role of bacteriocins as anti-and probiotics. Appl Microbiol Biotechnol 81:591–606
Havarstein LS, Diep DB, Nes IF (1995) A family of bacteriocin transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol 16:229–240
de Arauz LJ, Jozala AF, Mazzola PG, Penna TCV (2009) Nisin biotechnological production and application: a review. Trends Food Sci Technol 20:146–154
Amariei G, Kokol V, Boltes K, Leton P, Rosal R (2018) Incorporation of antimicrobial peptides on electrospun nanofibres for biomedical applications. RSC Adv 8:28013–28023
Najjari A, Ouzari H, Boudabous A, Zagorec M (2008) Method for reliable isolation of Lactobacillus sakei strains originating from Tunisian seafood and meat products. Int J Food Microbiol 121:342–351
Ivanova I, Kabadjova P, Pantev A, Danova S, Dousset X (2000) Dectection, purification and partial characterization of a novel bacteriocin substance produced by Lactococcus lactis subsp. lactis B14 isolated from boza-Bulgarian traditional cereal beverage. Photonics Nanostruct Fundam Appl 41:47–53
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilising the principle of protein-dye binding. Anal Biochem 72:248–254
Romero-Tabarez M, Jansen R, Sylla M, Lunsdorf H, Häußler S, Santosa DA, Timmis KN, Molinari G (2006) 7-O-malonyl macrolactin A, a new macrolactin antibiotic from Bacillus subtilis active against methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, and a small-colony variant of Burkholderia cepacia. Antimicrob Agents Chemother 50:1701–1709
Liu CI, Liu GY, Song Y, Yin F, Hensler ME, Jeng WY, Nizet V, Wang AH, Oldfield E (2008) A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 319(5868):1391–1394
Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S (2000) Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407
Sasidharan S, Logeswaran S, Latha LY (2012) Wound healing activity of Elaeis guineensis leaf extract ointment. Int J Mol Sci 13:336–347
Van Loosdrecht MCM, Lyklema J, Norde W, Zehnder AJ (1990) Influence of interfaces on microbial activity. Microbiol Rev 54:75–87
Clauditz A, Resch A, Wieland KP, Peschel A, Götz F (2006) Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun 74:4950–4953
Lauderdale KJ, Boles BR, Cheung AL, Horswill AR (2009) Interconnection between sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun 77(4):1623–1635
Nissen-Meyer J, Rogne P, Oppegard C, Haugen HS, Kristiansen PE (2009) Structure-function relationships of the non-lantionine-containing peptide (classII) bacteriocins produced by Gram-positive bacteria. Curr Pharm Biotechnol 10:19–37
Anderssen EL, Diep DB, Nes IF, Eijsink VG, Nissen-Meyer J (1998) Antagonistic activity of Lactobacillus plantarum C11: new two-peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Appl Environ Microbiol 64:2269–2272
Akdis M, Burgler S, Crameri R, Eiwegger T, Fujita H, Gomez E, Klunker S, Meyer N, O’Mahony L, Palomares O, Rhyner C, Quaked N, Schaffartzik A, Van de Veen W, Zeller S, Zimmemann M, Akdis CA (2011) Interleukins, from 1 to 37, and interferon- Ɣ: receptors, functions, and roles in diseases. J Allergy Clin Immunol 127:701–721
Froy O, Hananel A, Chapnik N, Madar Z (2005) Differential expression of rat β-defensins. IUMBM Life 57(1):41–43
Steenfos HH, Hunt TKL, Scheuenstuhl H, Goodson WHIII (1989) Selective effects of tumour necrosis factor-alpha on wound healing in rats. Surgery 106:171–176
Ishida Y, Kondo T, Takayasu T, Iwakura Y, Mukaida N (2004) The essential involvement of cross-talk between IFN- Ɣ and TGF- β in the skin wound healing process. J Immunol 172:1848–1855
Gutiérrez-Fernández A, Inada M, Balbín M, Fueyo A, Pitiot AS, Astudillo A, Hirose K, Hirata M, Shapiro SD, Noël A, Werb Z, Krane SM, López-Otín C, Puente XS (2007) Increased inflammation delays wound healing in mice deficient in collagenase-2 (MMP-8). FASEB J 21:2580–2591
Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Gotz F (2006) Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett 259:260–268
Sugai M, Yamada S, Nakashima S, Komatsuzawa H, Matsumoto A, Oshida T, Suginaka H (1997) Localised peroration of the cell wall by a major autolysin: atl gene products and the onset of penicillin-induced lysis of Staphylococcus aureus. J Bacteriol 179:2958–2962
Trotonda MP, Xiong YQ, Memmi G, Bayer AS, Cheung AL (2009) The role of mgrA and sarA in autolysis and resistance to cell-wall-active antibiotics in methicillin-resistant Staphylococcus aureus. J Infect Dis 199:209–218
Truong-Bulduc QC, Hooper DC (2010) Phosphorylation of mgrA and its effect on expression of the norA and norB efflux pumps of Staphylococcus aureus. J Bacteriol 192:2525–2534
Cheung GYC, Wang R, Khan BA, Sturdevant DE, Otto M (2011) Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun 79:1927–1935
Rice KC, Patton JB, Yang S-J, Dumoulin A, Bischoff M, Bayles KW (2004) Transcription of the Staphylococcus aureus cidABC and lrg murein hydrolase regulators is affected by sigma factor B. J Bacteriol 186:3029–3037
Yang SJ, Rice KC, Brown RJ, Patton TG, Liuo LE, Park YH, Bayles KW (2005) A Lys-type regulator, CidR, is required for induction of the Staphylococcus aureuscidABC operon. J Bacteriol 187:5893–3900
Acknowledgements
The Δatl S. aureus strain was kindly provided by Prof. Dr. Hiroshi Ohno from the RIKEN Center for Integrative Medical Sciences (Japan).
Funding
This research was funded by the USM-RIKEN International Centre for Ageing Science (URICAS) grant (1001/PEKIND/870030).
Author information
Authors and Affiliations
Contributions
Ohno H and Liong MT designed the experiments. JS Ong, Yong CC, Khoo BY, Sasidharan S and Choi SB performed the analyses. Taylor TD, JS Ong, Yong CC, Ohno H and Liong MT wrote the manuscript.
Corresponding authors
Ethics declarations
The experimental protocol for treating animals was approved and in accordance with the guidelines recommended by the USM Committee on Animal Care; reference number: USM/Animal Ethics Approval/2012/(81)(433).
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 613 kb)
Rights and permissions
About this article
Cite this article
Ong, J.S., Taylor, T.D., Yong, C.C. et al. Lactobacillus plantarum USM8613 Aids in Wound Healing and Suppresses Staphylococcus aureus Infection at Wound Sites. Probiotics & Antimicro. Prot. 12, 125–137 (2020). https://doi.org/10.1007/s12602-018-9505-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-018-9505-9