Abstract
Three hundred and sixty presumptive lactic acid bacteria (LAB) isolated from pregnant sows, newborn, suckling, and weaned piglets were preliminarily screened for anti-Salmonella activity. Fifty-eight isolates consisting of Lactobacillus reuteri (n = 32), Lactobacillus salivarius (n = 10), Lactobacillus mucosae (n = 8), Lactobacillus johnsonii (n = 5), and Lactobacillus crispatus (n = 3) were selected and further characterized for probiotic properties including production of antimicrobial substances, acid and bile tolerance, and cell adherence to Caco-2 cells. Eight isolates including Lact. johnsonii LJ202 and Lact. reuteri LR108 were identified as potential probiotics. LJ202 was selected for further use in co-culture studies of two-bacterial and multiple-bacterial species to examine its inhibitory activity against Salmonella enterica serovar Enteritidis DMST7106 (SE7106). Co-culture of LJ202 and SE7106 showed that LJ202 could completely inhibit the growth of SE7106 in 10 h of co-culture. In co-culture of multiple-bacterial species, culturable fecal bacteria from pig feces were used as representative of multiple-bacterial species. The study was performed to examine whether interactions among multiple-bacterial species would influence antagonistic activity of LJ202 against SE7106 and fecal coliform bacteria. Co-culture of SE7106 with different combinations of fecal bacteria and probiotic (LJ202 and LR108) or non-probiotic (Lact. mucosae LM303) strains revealed that the growth of SE7106 was completely inhibited either in the presence or in the absence of probiotic strains. Intriguingly, LJ202 exhibited notable inhibitory activity against fecal coliform bacteria while LR108 did not. Taken together, the results of co-culture studies suggested that LJ202 is a good probiotic candidate for further study its inhibitory effects against pathogen infections in pigs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salmonellosis, an infection with bacteria called Salmonella, is a leading cause of foodborne illness affecting humans and animals. Although animals, like pigs, infected with Salmonella do not normally show clinical sign of symptoms, the carcasses and meats are main reservoirs of the pathogen [1]. Salmonellosis outbreaks have been annually reported in the USA and European Union. Antibiotics have been used for prevention and treatment of pathogenic diseases. However, the extensive use of antibiotics in livestock has linked to the emergence of antibiotic-resistant bacteria found in humans and animals [2]. As a result, the European Union has banned the use of several antibiotics in farm animals (Regulation 1831/2003/EC) [3]. Since then, the use of probiotics has received great attention as an alternative to antibiotics.
Probiotics are live microorganisms which, when administered in adequate amounts, confer a health benefit to the host [4]. Probiotic Lactobacillus has been used for promoting and improvement of animal health [5,6,7,8]. The use of probiotics to combat Salmonella infection has been widely studied in poultry [9,10,11]. However, little attention has been paid to find probiotics with protective activity against Salmonella infection in pigs. The following are few examples of the studies of pig probiotics with protective activity against Salmonella infection. Lactobacillus acidophilus LAP5 isolated from swine was shown to inhibit the growth of Salmonella Typhimurium in vitro and reduce invasion of Salmonella Choleraesuis to human Caco-2 cell line [12, 13]. Casey et al. showed that feeding pigs with mix strains of probiotic Lactobacillus and Pediococcus bacteria reduced incidence, severity, and duration of diarrhea in pigs challenged with Salmonella Typhimurium [14].
In this study, it aimed to search for probiotic Lactobacillus with protective effects against Salmonella infection in pigs. To do this, Lactobacillus isolated from pigs of four ages, pregnant sows, newborn, suckling, and weaned piglets, were preliminarily screened for anti-Salmonella activity. The strains exhibiting anti-Salmonella activity were selected and further characterized for probiotic properties such as acid and bile salt tolerance, adhesion to Caco-2 cells, and production of antimicrobial substances. Eight potential probiotic strains were obtained. A potential probiotic strain, Lactobacillus johnsonii LJ202, was chosen for co-culture studies of two-bacterial species and multiple-bacterial species to gain insight into its inhibitory effects on the growth of Salmonella Enteritidis DMST7106 and fecal coliform bacteria.
Materials and Methods
Isolation of Lactic Acid Bacteria
Fecal samples were collected from three pigs of each age group comprising pregnant sows (90 days of pregnancy),newborn piglets (3 days of age), suckling piglets (10 days of age), and weaned piglets (30 days of age, weaning at 28 days of age).Twenty-five grams of each fecal sample were mixed with 225 ml of 0.1% (w/v) peptone-buffered water. The fecal samples were beaten in a stomacher (IUL instruments, Barcelona, Spain) at 2000 rpm with 3 cycles of 1 min beating and 1 min pause. The fecal slurry was filtered through gauze. A hundred microliters of the filtrate was diluted in a decimal series, and 100 μl of appropriate dilution were spread on de Man Rogosa, Sharpe (MRS) agar (BD, MD, USA) supplemented with 0.5% CaCO3.The plates were incubated at 37 °C for 24–48 h in anaerobic jars using the Gas Pak system (AnaeroPack®_Anaero, MCG, Japan).The colonies surrounding with clear zone were selected from each fecal sample and tested for catalase activity. The isolates which did not exhibit catalase activity were selected for further analysis.
Anti-Salmonella Activity
The presumptive lactic acid bacteria (LAB) were preliminarily screened for the ability to suppress the growth of Salmonella spp. using agar spot test [15]. Briefly, three microliters of LAB cultures were individually spotted on bottom agar containing 1.5% MRS. Then, the molten top agar containing 0.7% tryptic soy agar (TSA) was mixed with 10 μl of indicator culture (109 CFU/ml) and poured on top of the bottom agar. The indicator strains used in this experiment were Salmonella Choleraesuis DMST5880, Salmonella Choleraesuis DMST8014, Salmonella Enteritidis DMST7106, and Salmonella Typhimurium DMST562. The plates were incubated at 37 °C for 18–24 h. The inhibition zones were recorded.
16S rRNA Sequence Analysis
Bacterial DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega, MD, USA). The primers 27F (5′–AGAGTTTGATCMTGGCTCAG̶–3′) and 1492R (5′–TACGGYTACCTTGTTACGACTT3–′) were used for PCR amplification of 16S ribosomal RNA (rRNA) region. The PCR products were subjected to nucleotide sequencing (Macrogen, Seoul, Korea). BLAST nucleotide algorithm was used to search for similar sequences in the NCBI database.
Antimicrobial Activity of Cell-Free Culture Supernatant
Well diffusion was performed to investigate whether the CFS of Lactobacillus spp. contained antimicrobial substances which could inhibit the growth of Salmonella spp. [16]. Briefly, 1.5% TSA was melted, mixed with an indicator culture, and poured into 90 mm Petri dish plates. Then 7-mm wells were made, and 80 μl of neutralized and non-neutralized CFS of a Lactobacillus culture were added into each well. For the control, 80 μl of MRS pH 6.8 were added into wells. The indicator strains used in this experiment were Salmonella Choleraesuis DMST5880, Salmonella Choleraesuis DMST8014, Salmonella Enteritidis DMST7106, and Salmonella Typhimurium DMST562. Inhibition zones were recorded. Three replicates were performed for each sample.
Acid and Bile Salt Tolerance
The Lactobacillus isolates were anaerobically grown in MRS for 18 h at 37 °C. The cells were collected by centrifugation, washed once with PBS, and suspended in MRS. The initial cell number was determined by plate count. One hundred microliters of cell suspension were inoculated into 10 ml of MRS adjusted to pH 2.5 with 1 M HCl. The cell suspensions were incubated at 37 °C for 4 h. After incubation, 1 ml of the cell suspensions was used for determination of viable cells by plate count. The remaining was centrifuged, washed, and suspended in 9 ml of MRS containing 0.5% oxgall (Sigma-Aldrich; MD, USA). The cell suspensions were incubated at 37 °C for 24 h, and viable cell counts were determined. Three replicates were performed for each strain. The ability of isolates to tolerate to acid or bile salts was indicated as percentage of survival which was calculated as follows.
% survival (after incubation in acid) = (log10 N 1/log10 N 0) × 100
% survival (after incubation in bile) = (log10 N 2/log10 N 1) × 100
Where log10 N 0 is the initial cell count (number of bacterial cells before incubation in acid), and log10 N 1 and log10 N 2 are viable cells after incubation in acid and bile salts, respectively.
Adhesion Assay
Cell adhesion assay was followed the method described by Chauvière et al. [17]. Briefly, Caco-2 cells (ATCCHTB-37) were grown in Dulbecco’s modified Eagle’s minimal essential medium (DMEM, 25 mM glucose, 4 mM l-glutamine, 1 mM sodium pyruvate) (Gibco, Life Technologies, NY, USA), supplemented with 10% (v/v) fetal bovine serum (Gibco, Life Technologies, NY, USA). The cells were seeded at 104 cells/well in a 24-well plate (Corning Incorporated, NY, USA) and incubated until a confluent monolayer formed. Prior to performing the assay, Caco-2 monolayers were washed twice with phosphate-buffered saline (PBS). Overnight culture of LAB isolates were centrifuged, washed twice with PBS, and suspended in DMEM. The initial bacterial cell counts were determined by plate count. Five hundred microliters of the cell suspensions were added into three replicate wells. The plates were incubated at 37 °C. After 90 min of incubation, the bacterial cell suspension was removed and the Caco-2 cells were washed three times with PBS to remove unbound bacteria. The bound bacteria were removed from Caco-2 cells by addition of 500 μl of 0.1% Triton X-100, and the number of adherent cells was determined by plate count method. Three replicates were performed for each strain.
Hemolytic Activity
LAB cultures were streaked in triplicates on sheep blood agar (Department of Medical Science, National Institute of Health, Nonthaburi, Thailand). The plates were incubated at 37 °C for 24–48 h and examined for hemolytic patterns. Three types of hemolysis can occur and are classified as β-hemolysis (clear zones around colonies), α-hemolysis (green-hued zones around colonies), and γ-hemolysis (no zones around colonies).
Antagonistic Activity Test
The potential probiotics were examined for their antagonistic activity against bacteria isolated from pigs, such as Streptococcus gallolyticus, Enterococcus faecium, Enterococcus faecalis, Enterococcus villorum, and Lactobacillus spp., using agar spot test [15]. In brief, 3 μl of overnight cultures of probiotic strains were spotted on the bottom agar containing 1.5% MRS. Then, the molten top agar containing 0.7% TSA was mixed with 10 μl of an indicator culture (109 CFU/ml) and poured on top of the bottom agar. Reciprocally, Streptococcus, Enterococcus, and Lactobacillus isolated from pigs or other probiotic strains were spotted on appropriate bottom agar. Then, overnight culture of a probiotic strain was used to make bacterial lawn. Antagonistic activity was recorded by measuring the radii of inhibition zones. Two replicates were performed for each strain.
Co-Culture of Potential Probiotic and Salmonella
Overnight culture of probiotic Lact. johnsonii LJ202 (106 CFU/ml) and Salmonella Enteritidis DMST7106 (103 CFU/ml) were inoculated into 10 ml of co-culture medium containing an equal volume of double strength MRS and double strength tryptic soy broth (TSB), (MRS/TSB). The medium was incubated at 37 °C, without shaking. The co-culture medium containing only LJ202 or SE7106 was used as controls. Viable cells of LJ202 and SE7106 were monitored at 0, 2, 4, 6, 8, 10, 12, 14, and 24 h by plate count on MRS and xylose lysine deoxycholate (XLD) agar plates (BD, MD, USA), respectively. The pH values of the culture medium were determined at 0, 2, 4, 6, 8, 10, 12, 14, and 24 h. Three replicates were performed for each sample.
Co-Culture of Probiotic with Salmonella and Fecal Bacteria
Ten grams of pig feces were collected from a healthy suckling piglet (age 25 days). The fecal sample was determined as Salmonella free. The fecal sample was mixed with 90 ml of 0.1% buffered peptone water, and beaten using a stomacher (IUL instruments, Barcelona, Spain) at 200 rpm per minute for 2 min. The fecal slurry was filtered through gauze, and the filtrate was collected. One milliliter of the supernatant was inoculated into 9 ml of co-culture medium (MRS/TSB) and incubated at 37 °C for 18 h. The culturable fecal culture was used as a representative of mixed-bacterial species from pigs. Lact. johnsonii LJ202, Lactobacillus reuteri LR108, and Lactobacillus mucosae LM303 were individually grown in MRS/TSB at 37 °C for 18 h. SE7106 was grown in MRS/TSB at 37 °C for 18 h. To evaluate the inhibitory activity of probiotic and non-probiotic strains on the growth of fecal coliform bacteria and SE7106, four types of bacterial cultures were made: (i) fecal culture (1% (v/v) inoculum) was inoculated into 10 ml of MRS/TSB and served as the control; (ii) fecal culture (1% (v/v) inoculum) and SE7106 (106 CFU/ml) were inoculated into 10 ml of MRS/TSB; (iii) overnight culture of LJ202, LR108, and LM303 were individually inoculated into 10 ml of MRS/TSB at 106 CFU/ml, followed by 1% (v/v) of fecal culture; and (iv) overnight culture of LJ202, LR108, and LM303 were individually inoculated into 10 ml (106 CFU/ml) of MRS/TSB followed by fecal culture (1% (v/v) inoculum) and SE7106 at 106 CFU/ml. The culture samples were incubated at 37 °C for 24 h without shaking. Cell numbers of presumptive lactic acid bacteria (LAB), Salmonella, and fecal coliforms were monitored at 0 and 24 h by plate counts on MRS containing 0.5% CaCO3, XLD, and MacConkey plates (BD, MD, USA), respectively. Three replicates were performed for each sample. The mean cell counts of presumptive LAB and fecal coliforms of each culture sample were compared with those of fecal culture alone using paired t test.
Results
Preliminary Screening for Lactic Acid Bacteria with Anti-Salmonella Activity and Strain Identification
A total of three hundred and sixty isolates which showed clear zone on MRS containing 0.5% CaCO3 and exhibited no catalase activity were collected from fecal samples of pregnant sows, newborn, suckling, and weaned piglets. The isolates were preliminarily tested for inhibitory activity against Salmonella Choleraesuis DMST5880, Salmonella Choleraesuis DMST8014, Salmonella Enteritidis DMST7106, and Salmonella Typhimurium DMST562 by agar spot test. Sixty isolates which inhibited the growth of Salmonella spp. were selected.
The sixty isolates were subjected to16S rRNA sequencing. The analysis revealed that the sixty isolates comprised of five Lactobacillus species and two Enterococcus species which were Lact. reuteri (n = 32, 53.3%), Lactobacillus salivarius (n = 10, 17.6%), Lact. mucosae (n = 8, 13.3%), Lact. johnsonii (n = 5, 8.3%), Lactobacillus crispatus (n = 3, 5%), Ent. faecalis (n = 1, 1.6%), and Ent. faecium (n = 1, 1.6%). Lact. reuteri was the most abundant species among the isolates. All Lactobacillus isolates were further evaluated for their probiotic properties.
Evaluation of Probiotic Properties
CFSs of the fifty-eight Lactobacillus isolates were investigated for their abilities to produce antimicrobial substances by agar well diffusion assay. The assay showed that the non-neutralized CFSs of 17 isolates of Lact. reuteri, 10 isolates of Lact. salivarius, 2 isolates of Lact. johnsonii, and 3 isolates of Lact. mucosae could suppress the growth of one of the four Salmonella serovars used in this study (data not shown; the strains which their CFS showed inhibitory activity against Salmonella spp. were marked with asterisks (Fig. 1)). However, antimicrobial activity of the CFSs was drastically reduced when the CFSs were neutralized to pH 6.8.
The ability of Lactobacillus isolates to tolerate to acid and bile salts. a, b Survival rate of the Lactobacillus isolates after sequential incubation in acidic medium (pH 2.5) for 4 h (white bars) and 0.5% oxgall for 24 h (grey bars). LR Lactobacillus reuteri, LJ Lactobacillus johnsonii, LS Lactobacillus salivarius, LM Lactobacillus mucosae, LC Lactobacillus crispatus. The numbers 1xx, 2xx, 3xx, and 4xx behind the Lactobacillus species names referred to pregnant sows, newborn, suckling, and weaned piglets, respectively. Asterisks marked the isolates whose cell-free supernatant inhibited the growth of Salmonella spp., as determined by well diffusion assay
The abilities of Lactobacillus isolates to tolerate to acid and bile salts were further examined. The result showed that most of the isolates were able to survive at low pH (pH 2.5) after incubation for 4 h in acid medium (Fig. 1), except for the three isolates of Lact. crispatus which were highly sensitive to acidic environment. After exposure to acid, the cells were subsequently incubated with 0.5% oxgall for 24 h. Incubation with bile salts caused substantial cell death in most of Lact. salivarius and Lact. mucosae isolates (survival rate <60%). In contrast, most of Lact. reuteri and Lact. johnsonii isolates showed high tolerance to bile salts (survival rate >70%).
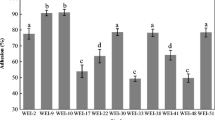
The Lactobacillus isolates which tolerated to acid and bile salts and had the abilities to produce antimicrobial substances were selected for cell adhesion assay. The isolates showed variable adhesion abilities. Most of Lact. reuteri, Lact. johnsonii, and Lact. salivarius isolates had higher adherence capability (8 log10–11 log10 CFU/ml) than Lact. mucosae and Lact. crispatus isolates(6 log10–9 log10 CFU/ml) (Fig. 2).
The ability of Lactobacillus isolates to adhere to Caco-2 cells. a Adherence capabilities of Lactobacillus reuteri isolates. b Adhesion abilities of Lactobacillus salivarius, Lactobacillus mucosae, Lactobacillus johnsonii, and Lactobacillus crispatus. The initial bacterial cell counts used for cell adhesion assay are depicted as white bars. The black bars represent the number of bacterial cells adhered to Caco-2 cells
Selection of Probiotic Candidates
Three criteria were used for selection of potential probiotics: (i) the isolates must have survival rate higher than 80% after exposure to acid and bile salts, (ii) they could inhibit the growth of Salmonella spp.(the four serovars used), and (iii) the number of bacterial cells adhering to Caco-2 cells should be higher than 7 log10 CFU/ml. Based on the selection criteria, six isolates of Lact. reuteri (LR105, LR108, LR111, LR310, LR311, LR401), one isolate of Lact. johnsonii (LJ202), and one isolate of Lact. salivarius (LS404) were selected.
Potential Probiotic Possessed No Hemolytic Activity
For safety use in pigs, the probiotic strains were tested for hemolytic activity on sheep blood agar. The test showed that none of the probiotic strains exhibited β-hemolysis. Most of the isolates exhibited γ-hemolysis (LR105, LR108, LR111, LR310, LR311, LR401) while a few isolates displayed α-hemolysis (LJ202, LS404). Therefore, all probiotic strains were considered to be safe for use in pigs.
Antagonistic Activity Among Probiotics and Some Bacteria Isolated from Pigs
The potential probiotics were tested for antagonistic activity among them and against some of the bacteria isolated from pigs. The result revealed that probiotic Lact. reuteri, Lact. johnsonii, and Lact. salivarius strains had strong antagonistic activities against Strep. gallolyticus, Ent. faecium, Ent. faecalis, and Ent. villorum and a weak antagonistic effect on some Lactobacillus isolates, but not Lact. mucosae strains (Fig. 3). None of Streptococcus and Enterococcus used as indicator bacteria had antagonistic activity against the probiotic strains. However, it was found that most of probiotic Lact. reuteri strains and Lact. johnsonii LJ202 as well as some Lact. salivarius isolates could inhibit the growth of Lact. salivarius LS404 (data not shown).
Antagonistic effects of potential probiotic strains against Streptococcus, Enterococcus, and Lactobacillus isolated from pig feces. The figure shows a part of the data obtained from antagonistic activity test. Antagonistic activity of probiotic strains against the bacteria isolated from pig feces was determined by measuring the radii of inhibition zones. White and grey bars show the radii of inhibition zones produced by LR108 and LJ202 against the indicator strains
Lact. johnsonii LJ202 Inhibited the Growth of Salmonella Enteritidis DMST7106 in the Co-Culture Study
Since all of the probiotic candidates showed the ability to inhibit Salmonella spp. in agar spot test and well diffusion assay, it was of interest to gain insight into how the probiotic strains inhibit the growth of Salmonella spp. Probiotic Lact. johnsonii LJ202 whose CFS showed notable inhibitory activity against Salmonella spp. was chosen for co-culture study with SE7106. Prior to co-culture study, LJ202 and SE7106 were preliminarily tested for their ability to grow on the co-culture medium, MRS/TSB. The result showed that both bacteria grew very well in MRS/TSB (Fig. 4a). Co-culture of LJ202 and SE7106 revealed that the number of SE7106 increased from 3 log10 to 5 log10 CFU/ml in 2 h. After that, the SE7106 count slightly decreased to 4 log10 CFU/ml at 4 h of co-culture and remained almost constant until 8 h (Fig. 4a). In contrast to SE7106, the number of LJ202in the co-culture showed a steady increase until 8 h. At 10 h of co-culture, the number of LJ202 decreased by 2.5 log10 CFU/ml while the number of SE7106 sharply decreased to undetectable level (˂10 CFU/ml) through the end of the experiment. After 10 h of co-culture, the number of LJ202 slowly increased and reached the maximum cell count at 24 h of co-culture. The maximum counts of LJ202 in mono- and co-culture were similar. Monitoring pH of the mono- and co-culture media of LJ202 revealed that the pH gradually decreased from 6.8 to approximately 4.0 at 8 h of co-culture and remained at pH 4.0 through the end of the experiment (Fig. 4b).
Antagonistic Activity of LJ202 Against Fecal Coliform Bacteria and SE7106 in a Mixed Bacterial Community
In the gastrointestinal (GI) tract of pigs, there are many species of bacteria that interactions with one another occur. Interactions among bacterial species may modulate the diversity, behaviors, and activities of the individual species existing in the complex communities [18]. Therefore, it was of interest to investigate whether interactions among multiple-bacterial species might influence antimicrobial activity of LJ202 against SE7106. To do this, co-culture of probiotic strains with multiple-bacterial species and SE7106 was performed. The culturable fecal bacteria were used as representative of multiple-bacterial species from pigs. Probiotic LJ202 and Lact. reuteri LR108were used to compare their inhibitory activities. The non-probiotic Lact. mucosae LM303, which possessed antimicrobial activity against Salmonella Typhimurium DMST562 but not Salmonella Enteritidis DMST7106, was included in the experiment. In the co-culture study, total presumptive LAB, Salmonella, and fecal coliform counts were monitored at 0 and 24 h of incubation. At 0 h, the number of presumptive LAB in culture samples containing only fecal bacteria (F) or fecal bacteria spiked with SE7106 (F + SE) was 6.3 log10 CFU/ml. Inoculation of LJ202 or LR108 into fecal culture(F + LJ or F + LR) caused a slight increase of total counts of presumptive LAB (Table 1). After 24 h of co-culture, total numbers of presumptive LAB in F and F + SE increased by 7.6 log10 CFU/ml. Co-culture of probiotic or non-probiotic strains with fecal bacteria resulted in the reduction in cell numbers of presumptive LAB in F + LR, F + LJ, and F + LM compared with those in F and F + SE (about 1.3–3 log10 CFU/ml reduction). Addition of SE7106 into fecal culture containing LR108 or LJ202 (F + SE + LR and F + SE + LJ) did not cause significant change in the number of presumptive LAB. But, in F + SE + LM, the number of presumptive LAB decreased by 2.3 log10 CFU/ml compared with that of F + LM. Monitoring pH of the culture media after 24 h of co-culturing revealed that the pH of culture media of F and F + SE was 4.14. In F + LJ and F + SE + LJ, the pH of the culture media was lowered to 4.02 which was the lowest pH among the culture samples. The pH values of the culture media of F + LR, F + SE + LR, F + LM, and F + SE + LM were in a range of 4.21–4.29 which was higher than those of F and F + SE.
Determination of Salmonella numbers at 0 h revealed that Salmonella counts in most of the culture samples were below the detection limit (detection limit ≥10 CFU/ml), except for those spiked with SE7106 (F + SE, F + SE + LR, F + SE + LJ, F + SE + LM) where Salmonella counts were about 6 log10 CFU/ml. After 24 h of co-culture, the viable cells of Salmonella were not detected in all culture samples. Determination of fecal coliforms in the culture samples revealed that the number of fecal coliforms at 0 h was in a range of 5.6–5.7 log CFU/ml. After 24 h of co-culture, the decrease in fecal coliformcounts was observed in all culture samples. Significant reduction of fecal coliform counts was observed in F + LJ and F + SE + LJ where the viable cells were not detected (˂10 CFU/ml). In F + LR, the number of fecal coliforms was comparable to that of F + SE; however, fecal coliform count significantly increased in F + SE + LR. In F + LM, the number of fecal coliforms was less than those of F, F + SE, and F + LR. But, addition of SE7106 (F + SE + LM) into the co-culture sample caused a significant increase in fecal coliform count.
Discussion
With the three selection criteria, eight potential probiotic strains which possessed the abilities to produce antimicrobial substances to inhibit the growth of Salmonella, to resist to acid and bile salts, and to adhere to Caco-2 cells were selected. The eight probiotic strains comprised of six strains of Lact. reuteri (LR105, LR108, LR111, LR310, LR311, LR401) and one strain each of Lact. johnsonii (LJ202) and Lact. salivarius (LS404). The three Lactobacillus species are known as inhabitants of pig GI tracts [19]. This would facilitate the probiotic strains to establish in GI niches and provide beneficial effects to pigs. Beneficial effects of probiotic Lact. reuteri, Lact. johnsonii, and Lact. salivarius on pigs have been reported. Daily feeding of probiotic Lact. reuteri I5007 was shown to protect newborn piglets from bacterial infections, particularly through the expression of tight junction protein [20]. Administration of Lact. johnsonii XS4 to sows during the end of pregnancy and during lactation increased litter weight at birth and weaning, and enhanced survival rate of newborn piglets [6]. Oral administration of Lactobacillus salivarius B1 to neonatal piglets showed modulatory effects on piglets by increasing the amount of intra-epithelial lymphocyte cells and IgA-producing cells in the intestinal tracts [21].
As we were interested in finding Lactobacillus with protective effect against Salmonella infection, we thus conducted further experiment to gain insight into how the potential probiotics affect the growth of Salmonella. By employing an in vitro co-culture model, it was found that the growth of SE7106 was retarded at the early hours of co-culturing with LJ202. At 4 h of co-culture, reduction of Salmonella counts occurred simultaneously with the decrease in pH of the culture medium. It has been shown that low pH environment affected the growth of Salmonella spp. [22, 23]. In our study, the effect of pH lowering seemed not strong enough to completely inhibit the growth of SE7106 even pH of the co-culture medium was lowered to 4.0. At 10 h of co-culture, decrease in LJ202 numbers and loss of viable cells of SE7106 was observed, suggesting that competition for limiting nutrients might occur at this hour. After 10 h, only LJ202 could resume its growth, indicating that LJ202 had the ability to compete for the remaining nutrients but SE7106 did not. Thereby, the growth of SE7106 was then completely inhibited. Competition for nutrients was shown to affect the growth of pathogen in a co-culture study. By using transcriptome and biochemical analyses, Nouaille et al. revealed that competition for consumption of glucose between Lactococcus lactis and Staphylococcus aureus occurred when the nutrients were limited and growth rates of both bacteria were concomitantly retarded [24]. Competition for nutrients is considered as one of the mechanisms by which a bacterium used for survival in the gut [25].
The co-culture of two-bacterial species clearly showed that the probiotic LJ202 exhibited inhibitory effects on the growth of SE7106. In the subsequent co-culture study, it was interesting to investigate whether the presence of multiple-bacterial species where interactions among bacteria occurred may influence the antagonistic activity of LJ202. The use of culturable fecal bacteria as representative of multiple-bacterial species from pigs allowed us to investigate the inhibitory effect of LJ202 on fecal coliforms, apart from Salmonella. Also, the number of presumptive LAB was monitored since the previous antagonistic activity test showed that LJ202 displayed inhibitory activity against some lactic acid bacteria. Determination of Salmonella counts revealed that the number of Salmonella in all culture samples were below the detection limit (˂10 CFU/ml) after 24 h of co-culture. The absence of Salmonella even in F + SE sample indicated that the presence of only culturable fecal bacteria could inhibit growth of SE7106. The ability of fecal bacteria to inhibit SE7106 can be explained as follows. (i) After 24 h of co-culture, the fecal culture contained presumptive LAB as high as 13 log10 CFU/ml. These LAB produced acids which lowered pH of the culture media to 4.14. The effect of pH lowering is considered as one factor that affected the growth of SE7106. (ii) Competition for nutrients, particularly among multiple-bacterial species, must be vigorous; thereby, SE7106 may lose the ability to compete for the limiting nutrients. (iii) Antimicrobial substances (like bacteriocins) produced by presumptive LAB may involve in growth inhibition of SE7106 [26].
Although inhibitory effect of LJ202 and LR108 on the growth of SE7106 was not clearly seen in the co-culture of multiple-bacterial species, it was clearly shown that LJ202 had inhibitory effect on the growth of fecal coliforms. Fecal coliforms in pig feces are known as potential pathogens (such as Escherichia coli) which can cause diarrhea in pigs [27]. In many studies, feeding pigs and chickens with probiotics could reduce coliform counts and reduced the incidence of diarrhea [28,29,30,31,32,33]. The presence of LJ202 in the co-culture with fecal bacteria alone(F + LJ) or fecal bacteria with SE7106 (F + SE + LJ) completely inhibited the growth of fecal coliforms. The result suggested that LJ202 exhibited antagonistic activity against fecal coliforms, and the activity was not interfered with the interactions among multiple-bacterial species in the co-culture. Inhibitory effect of LJ202 against fecal coliforms would be due to acids which lowered the pH of co-culture medium to 4.02. However, a study of Haberbecket al. showed that 75% of the 188 different E. coli strains used in their study had high capability to survive under low pH conditions (a pH range of 3.8–4.8) [34]. Therefore, pH lowering might not be the sole factor that inhibited the growth of fecal coliforms. It is possible that other antimicrobial substances like bacteriocin(s) produced by LJ202 might participate in inhibiting the growth of fecal coliforms [6, 35]. In the complex community, LR108 showed no inhibitory activity against fecal coliforms while LM303 displayed the activity only if SE7106 was absent. Inhibitory activity of LM303 might not be due to acids or pH lowering, since the pH values of co-culture media of F + LM and F + SE + LM were similar. It is possible that other antimicrobial substances, like bactericoins, may play a role in inhibiting the growth of SE7106. LM303 may be able to produce bacteriocins like Lact. mucosae strain Marseille. Draft genome sequencing of Lact. mucosae strain Marseille revealed that the bacterium has 12 different genes which possibly encode bacteriocins ranging from 38 to 67 amino acids [36]. Loss of inhibitory activity of LM303 against fecal coliforms in F + SE + LM was surprised. One possible explanation is that the presence of SE7106 and fecal coliforms may initiate synergistic interactions between the bacteria. The interactions may lead to stabilize or increase population of fecal coliform bacteria [37, 38].
Monitoring the number of presumptive LAB in the co-culture study revealed that LR108, LJ202, and LM303 caused significant reduction of presumptive LAB counts when they were co-cultured with fecal bacteria (F + LR, F + LJ, F + LM), particularly LJ202. In a more complex system where SE7106 was added, the number of presumptive LAB further decreased in F + SE + LM while those in F + SE + LR or F + SE + LJ did not change. From the antagonistic activity test, it was found that LR108, LJ202, and LM303 showed the capability to inhibit the growth of Streptococcus and Enterococcus but had little effect on Lactobacillus. Therefore, it assumed that the decrease in presumptive LAB counts was the result of the decrease in the number of Streptococcus and Enterococcus rather than Lactobacillus. Some species of Streptococcus and Enterococcus are opportunistic pathogens, feeding pigs with probiotics which exhibit inhibitory activity against Streptococcus and Enterococcus would benefit to pig health [39].
In conclusion, probiotic characterization and co-culture studies provided evidence to suggest that LJ202 is a good probiotic which exhibits inhibitory activity against Salmonella spp. and fecal coliform bacteria. Therefore, LJ202 is a suitable candidate for further study its protective effects against pathogen infections in pigs.
References
Kreuzer S, Janczyk P, Assmus J, Schmidt MF, Brockmann GA, Nockler K (2012) No beneficial effects evident for Enterococcus faecium NCIMB 10415 in weaned pigs infected with Salmonella enterica serovar Typhimurium DT104. Appl Environ Microbiol 78:4816–4825
Mathew AG, Cissell R, Liamthong S (2007) Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog Dis 4:115–133
Regulation 1831/2003/EC. Regulation (EC) No 1831/2003 of the European Parliament and of the Council of 22 September 2003 on additives for use in animal nutrition. Official Journal of the European Union 268
FAO/WHO (2001) Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. FAO/WHO, Rome
Veizaj-Delia E, Piu T, Lekaj P, Tafaj M (2010) Using combined probiotic to improve growth performance of weaned piglets on extensive farm conditions. Livest Sci 134:249–251
Wang J, Ji HF, Hou CL, Wang SX, Zhang DY, Liu H, Shan DC, Wang YM (2014) Effects of Lactobacillus johnsonii X54 supplementation on reproductive performance, gut environment, and blood biochemical and immunological index in lactating sows. Livest Sci 164:96–101
Lähteinen T, Lindholm A, Rinttilä T, Junnikkala S, Kant R, Pietilä TE, Levonen K, von Ossowski I, Solano-Aguilar G, Jakava-Viljanen M, Palva A (2014) Effect of Lactobacillus brevis ATCC 8287 as a feeding supplement on the performance and immune function of piglets. Vet Immunol Immunopathol 158:14–25
Lähteinen T, Rinttilä T, Koort JMK, Kant R, Levonen K, Jakava-Viljanen M, Björkroth J, Palva A (2015) Effects of a multispecies Lactobacillus formation as a feeding supplement on the performance and immune function of piglets. Livest Sci 180:164–171
Higgins JP, Higgins SE, Wolfenden AD, Henderson SN, Torres-Rodiguez A, Vicente JL, Hargis BM, Tellez G (2010) Effect of lactic acid bacteria probiotic culture treatment timing on Salmonella Enteritidis in neonatal broilers. Poult Sci 89:243–247
Chen CY, Tsen HY, Lin CL, Yu B, Chen CS (2012) Oral administration of a combination of select lactic acid bacteria strains to reduce the Salmonella invasion and inflammation of broiler chicks. Poult Sci 91:2139–2147
Feng J, Wang L, Zhou L, Yang X, Zhao X (2016) Using in vitro immunomodulatory properties of lactic acid bacteria for selection of probiotics against Salmonella infection in broiler chicks. PLoS One 11(1):e0147630. doi:10.1371/journal. pone.0147630
Tsai CC, Hsih HY, Chiu HH, Lai YY, Liu JH, Yu B, Tsen HY (2005) Antagonistic activity against Salmonella infection in vitro and in vivo for two Lactobacillus strains from swine and poultry. Int J Food Microbiol 102:185–194
Lin CK, Tsai HC, Lin PP, Tsen HY, Tsai CC (2008) Lactobacillus acidophilus LAP5 able to inhibit the Salmonella Choleraesuis invasion to the human Caco-2 epithelial cell. Anaerobe 14:251–255
Casey PG, Gardiner GE, Casey G, Bradshaw B, Lawlor PG, Lynch PB, Leonard FC, Stanton C, Ross RP, Fitzgerald GF, Hill C (2007) A five-strain probiotic combination reduced pathogen shedding and alleviates disease signs in pigs challenged with Salmonella enterica serovar Typhimurium. Appl Environ Microbiol 73:1858–1863
Barbosa TM, Serra CR, La Ragione RM, Woodward MJ, Henriques AO (2005) Screening for Bacillus isolates in the broiler gastrointestinal tract. Appl Environ Microbiol 71:968–978
Schillinger U, Lücke FK (1989) Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol 55:1901–1906
Chauvière G, Coconnier MH, Kernéis S, Fourniat J, Servin AL (1992) Adhesion of human Lactobacillus acidophilus strain LB to human enterocyte-like Caco-2 cells. J Gen Microbiol 138:1689–1696
WigneswaranV, AmadorCI, JelsbakL, SternbergC, JelsbakL (2016) Utilization and control of ecological interactions in polymicrobial infections and community-based microbial cell factories [version 1; referees:3 approved]. F1000 Research. doi:10.12688/f1000research.7878.1
Leser TD, Amenuvor JZ, Jensen TK, Lindecrona RH, Boye M, Møller K (2002) Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl Environ Microbiol 68:673–690
Yang F, Wang A, Zeng X, Hou C, Liu H, Qiao S (2015) Lactobacillus reuteri I5007 modulates tight junction protein expression in IPEC-J2 cells with LPS stimulation and in newborn piglets under normal conditions. BMC Microbiol 15:32. doi:10.1186/s12866-015-0372-1
Zhang J, Deng J, Wang Z, Che C, Li YF, Yang Q (2011) Modulatory effects of Lactobacillus salivarius on intestinal mucosal immunity of piglets. Curr Microbiol 62:1623–1631
Rubin HE, Nerad T, Vaughan F (1982) Lactate acid inhibition of Salmonella Typhimurium in yogurt. J Dairy Sci 65:197–203
Fayol-Messaoudi D, Berger CN, Coconnier-Polter MH, Liévin-LeMoal V, Servin AL (2005) pH-, lactic acid-, and non-lactic acid-dependent activities of probiotic Lactobacillus against Salmonella enterica serovar Typhimurium. Appl Environ Microbiol 71:6008–6013
Nouaille S, Even S, Charlier C, Le Loir Y, Cocaign-Bousquet M, Loubière P (2009) Transcriptome response of Lactococcus lactis in mixed culture with Staphylococcus aureus. Appl Environ Microbiol 75:4473–4482
Savage DC (1977) Microbiology of the gastrointestinal tract. Annu Rev Microbiol 31:107–133
De Vuyst L, Leroy F (2007) Bacteriocins from lactic acid bacteria: production, purification, and food applications. J Mol Microbiol Biotechnol 13:194–199
Melin L, Mattsson S, Katouli M, Wallgren P (2004) Development of post-weaning diarrhoea in piglets. Relation to presence of Escherichia coli strains and rotavirus. J Vet Med B Infect Dis Vet Public Health 51:12–22
Shu Q, Qu F, Gill HS (2001) Probiotic treatment using Bifidobacterium lactis HN019 reduces weanling diarrhoea associated with rotavirus and Escherichia coli infection in a piglet model. J Pediatr Gastroenterol Nutr 33:171–177
Huang CH, Qiao SY, Li DF, Piao XS, Ren JP (2004) Effects of lactobacilli on the performance, diarrhea incidence, VFA concentration and gastrointestinal microbial flora of weaning pigs. Asian Australas J Anim Sci 17:401–409
Mallo JJ, Rioperez J, Honrubia P (2010) The addition of Enterococcus faecium to diets improves piglet’s intestinal microbiota and performance. Livest Sci 133:176–178
Liu H, Ji HF, Zhang DY, Wang SX, Wang J, Shan DC, Wang YM (2015) Effects of Lactobacillus brevis preparation on growth performance, fecal microflora and serum profile in weaned pigs. Livest Sci 178:251–254
Peng Q, Zeng XF, Zhu JL, Wang S, Liu XT, Hou CL, Thacker PA, Qiao SY (2016) Effects of dietary Lactobacillus plantarum B1 on growth performance, intestinal microbiota, and short chain fatty acid profiles in broiler chickens. Poult Sci 95:893–900
Wang S, Peng Q, Jia HM, Zeng XF, Zhu JL, Hou CL, Liu XT, Yang FJ, Qiao SY (2017) Prevention of Escherichia coliinfection in broiler chickens with Lactobacillus plantarum B1. Poult Sci. doi:10.3382/ps/pex061
Haberbeck LU, Oliveira RC, Vivijis B, Wenseleers T, Aertsen A, Michiels C, Geeraerd AH (2015) Viability in growth/no growth boundaries of 188 different Escherichia colistrains reveals that approximately 75% have a higher growth probability under low pH conditions than E. coliO157:H7 strain ATCC 43888. Food Microbiol 45:222–230
Abee T, Klaenhammer TR, Letellier L (1994) Kinetic studies of the action of lactacin F, a bacteriocin produced by Lactobacillus johnsonii that forms poration complexs in the cytoplasmic membrane. Appl Environ Microbiol 60:1006–1013
Drissi F, Merhej V, Blanc-Tailleur C, Raoult D (2015) Draft genome sequence of the Lactobacillus mucosae Marseille. Genome Announc 3(4):e00841–e00815. doi:10.1128/genomeA.00841-15
Ramsey MM, Rumbaugh KP, Whiteley M (2011) Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog 7(3):e1002012. doi:10.1371/journaLact.ppat.1002012
Chubiz LM, Granger BR, Segrè D, Harcombe WR (2015) Species interactions differ in their genetic robustness. Front Microbiol 6:271. doi:10.3389/fmicb.2015.00271
Chanter N (1997) Streptococci and enterococci as animal pathogens. J Appl Microbiol Symp Suppl 83:100S–109S
Acknowledgements
This study was funded by the National Center for Genetic Engineering and Biotechnology (BIOTEC), Thailand (grant number P-14-50177). We thank Ms. Sukitaya Veeranondha for her assistance in the culture of Caco-2 cells.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Abhisingha, M., Dumnil, J. & Pitaksutheepong, C. Selection of Potential Probiotic Lactobacillus with Inhibitory Activity Against Salmonella and Fecal Coliform Bacteria. Probiotics & Antimicro. Prot. 10, 218–227 (2018). https://doi.org/10.1007/s12602-017-9304-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-017-9304-8