Abstract
A specific strain of naturally occurring oral lactobacilli was isolated and identified based on morphological, biochemical, and 16S rRNA gene sequencing. The phylogenetic affiliation of the isolate confirmed that the NK02 strain had close association with the Lactobacillus salivarius. An effective mouthwash was developed for treatment of periodontitis and suppression of the indicator bacterium Aggregatibacter actinomycetemcomitans which is an obvious pathogen of periodontal disease. The mouthwash containing L. salivarius NK02 was tested at a dose level of 108 (colony forming units (CFU) ml−1), monitoring over a period of 4 weeks. The study was a randomized double-blind placebo control trial, and the patients were treated in two groups of control and test by using scaling and root planing (SRP) + placebo and scaling and root planing (SRP) + probiotic, respectively. It appeared that the probiotic mouthwash was able to inhibit the bacterial growth on both saliva and sub-gingival crevice and exhibited antibacterial activity against A. actinomycetemcomitans. The results also showed that SRP+ probiotic treatment led to a significant decrease of gingival index (GI) and bleeding on probing (BOP) compared with that of SRP + placebo for the probiotic group. The rate of decrease in pocket depth was displayed in the group with SRP + probiotic treatment equal to 1/2 mm, and probing pocket depth (PPD) value was decreased in the probiotic bacteria treatment group that can explain the decrease in inflammation in gingiva. Our findings suggest that probiotic mouthwash is healthy for daily use as an alternative for maintaining dental and periodontal health.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The use of probiotics in the medical field is growing as more evidence accumulates supporting their benefits. Given that probiotic bacteria impact human health, applying probiotics to prevent or treat a wide range of diseases has appeared to be a promising new therapy [1,2,3]. However, the act of choosing microorganisms for use as probiotic treatments should follow careful consideration procedure. Application of probiotics for oral health care has received much greater attention during the past few years; however, most of the efforts are currently focusing on adopting gastrointestinal probiotic bacterium [4]. Periodontal diseases are one of the most common diseases in human societies. Unbalanced oral microbial flora is an important risk for human body against these diseases. Various pathogens are involved in causing periodontal disease. These pathogens mostly live in biofilm in human mouth and aggravate their pathogenic activity there [5]. In fact, periodontitis is mostly related to disproportionate microflora resulting in overgrowth of periodontal pathogens such as Porphyromonas gingivalis, Prevotella intermedia, and A. actinomycetemcomitans, [6]. Among these bacteria, A. actinomycetemcomitans (previously Actinobacillus actinomycetemcomitans) is a true pathogen for the occurrence of periodontal diseases. This bacterium is one of the most important factors affecting the initiation and progression of these diseases due to its high level of virulence factors [7]. The treatment of periodontal disease at very early stages depends upon omitting pathogens from the oral environment in different ways such as scaling and root planing (SRP), antibiotic therapy, and surgical treatment.
Although successful treatment of periodontitis will result in tooth surfaces free of plaque in a long-term perspective, it will not be able to maintain an environment which is free of plaque. Antibiotics are used as complementary therapies for treating periodontal disease that accompany some disadvantageous effects, including changes in the bacterial flora of body, destruction of all strains of the bacterial flora, antibiotic resistance, etc., while probiotic bacteria are complementary therapeutic means that do not have disadvantages of antibiotics. In fact, considering the effect of probiotic bacteria on pathogen bacteria in the body as well as other useful properties that they have makes them a good alternative for treatment of periodontal diseases [8, 9]. The ability of probiotics to stick to saliva-coated surfaces varies among different species. It has been reported that Lactobacillus rhamnosus and L. paracasei strains have durable binding activity. Haukioja et al. [10] have shown that probiotic lactobacilli (L. rhamnosus GG, L. casei) may affect oral ecology by preventing adherence of other bacteria by altering the protein composition of the salivary pellicle. Eventually, once a species is firmly attached to the oral biofilm structures, it could be anticipated to disturb the development of the pathogenic character of the species based on antimicrobial activity, which indeed is another evaluation principle for probiotic bacteria. It was found that destruction and inflammation at periodontal sites are closely associated with reduced level of certain lactic acid-dependent bacteria such as streptococci and aerobic coryneforms [11]. Sookhee et al. [12] isolated 3790 strains of lactic acid bacteria from 130 individuals and found that L. paracasei and L. rhamnosus had a high capacity to control important oral pathogens including Streptococcus mutants and P. gingivalis. Here, we are the first to mention the isolation and identification of an indigenous probiotic strain of Lactobacillus salivarius with significant capability to produce antibacterial compounds that may be appropriate for application in managing periodontal diseases. This work was accomplished by the following:
-
1.
Isolation and testing of a selected probiotic strain from oral cavity
-
2.
Reviewing and testing the effects of probiotics against pathogens
-
3.
Mouthwash preparation for transferring probiotic bacteria to oral cavity
-
4.
Evaluating effect of probiotics against periodontitis
Materials and Methods
Isolation and Identification of Lactobacilli Isolates and Aggregatibacter actinomycetemcomitans
For isolation of probiotic Lactobacillus strains from mouth saliva, 25 patients with moderate to severe chronic periodontitis (according to radiographic and clinical findings confirmed by two periodontists) and 25 healthy volunteers were selected.
Inclusion criteria for periodontal patients were as follows:
-
1.
Presence of bone loss due to periodontal infection
-
2.
Presence of periodontal pockets more than 5 mm
-
3.
Having complete health situation
The patients were not included in the following cases:
-
1.
Patients suffering from systemic diseases (intolerance to lactose and immune deficiency diseases, etc.)
-
2.
Administration of immunosuppressive medicines by the patients
-
3.
Usage of antibiotics by the patients in the last 3 months
-
4.
Smoking
-
5.
Pregnant patients or breastfeeding mothers
-
6.
Using other probiotic products in the last 3 months
-
7.
Patients undergone periodontal treatment in the last 3 months
After providing a verbal and written consent, sampling was done from unstimulated saliva of selected patients (after at least 2 h of not eating anything). Individual saliva samples were collected in sterile-capped plastic vials and transferred to the laboratory immediately on an ice pack. One milliliter of collected saliva samples was dissolved in 9 ml sterile normal saline to obtain 10-fold serial dilution series. An appropriate diluted solution (0.1 ml) was spread onto the MRS agar plates (Merck, Germany) for isolation of lactobacilli. All plates were incubated at 30 °C for 48–72 h under anaerobic conditions. After subsequent culture and sub-culture to obtain distinct colonies, the non-spore-forming cells with Gram-positive short rod-shaped and catalase-negative characteristics are generally considered as lactobacilli. These colonies were sub-cultured into the MRS broth, and glycerol stocks (15% v/v) of the isolates were prepared for further studies.
Aggregatibacter actinomycetemcomitans is an important pathogen which is associated with periodontal disease. To isolate this bacterium, sub-gingival samples from patients with chronic periodontitis were collected and inoculated into a highly selective medium for A. actinomycetemcomitans (AASM), and incubated at 37 °C for 48 h in a 5% CO2 incubator [13]. Several colonies (which seemed to be A. actinomycetemcomitans based on catalase production and colony morphology on AASM plates) were sub-cultured to affirm the existence of A. actinomycetemcomitans. Pure cultures of each isolate were recognized on the basis of colony morphology, Gram staining, and catalase activity.
Strain Selection Based on Antibacterial Activity Against A. actinomycetemcomitans
To evaluate the antibacterial activity of the 15 lactobacillus isolates against A. actinomycetemcomitans, each fresh culture of the Lactobacillus isolates were individually streaked (on a straight line) onto BHI (Brain Hearth Infusion) agar plate and incubated at 30 °C for 24 h. A. actinomycetemcomitans was then perpendicularly streaked across the line of the isolates, and plates were incubated anaerobically (as mentioned above) accordingly. The zone of inhibition close to lactobacillus isolates were measured and considered as positive results. From all the lactobacilli isolates examined, an isolate which showed a superior antibacterial activity against A. actinomycetemcomitans was selected for further examinations. Experiments were repeated twice in any case, and each determination was performed in triplicate.
Identification of the Selected Isolate
After obtaining axenic cultures of 15 lactobacilli isolates, an isolate (with highest antibacterial activity against A. actinomycetemcomitans) was selected and identified on the basis of its biochemical characteristics accompanied by 16S ribosomal RNA (rRNA) gene sequencing [14]. The single bands of PCR products demonstrating the size of about 540 bp were purified using a PCR purification kit (Roche Applied Science, Germany). Sequencing of the amplified fragment was performed on an ABI PRISM 377 automated analyzer. 16S rRNA sequence homology with other reference strains was examined using the Basic Local Alignment Search Tool (BLAST) version 2.2.12 provided by the National Center for Biotechnology Information (NCBI). Finally, the multiple sequence alignments were achieved using Clustal W software and a consensus neighbor-joining tree was constructed using the molecular evolutionary genetic analysis (MEGA) software 6.0 [15].
Probiotic Features of Isolated Lactobacilli
Bile Salt Hydrolase Gene Analysis by PCR
PCR was done to screen the presence of the bsh gene responsible for encoding bile salt hydrolase (BSH) by a selected Lactobacillus strain. Bile salt hydrolase is one of the most important properties of probiotic bacteria. PCR primers were used with the following sequences: 5′CGTATCCAAGTGCTCATGGTTTAA3′ (nucleotide position 150,568 to 150,593 of the bsh gene), 5′ATGTGTACTGCCATAACTTATCAATCTT3′ (bsh rev). The primers were designed to produce a PCR product length of 919 bp. DNA amplifications were set as follows: 4 min at 94 °C and 30 cycles of 30 s at 94 °C, 30 s at 64 °C, and 1 min at 72 °C; the final extension step consisted of 10 min at 72 °C.
Tolerance to Acidic pH
To evaluate the tolerance of the selected Lactobacillus strain to acidic pH, the isolate was grown in MRS broth at 37 °C overnight. Then, 0.5-ml aliquots of cell suspension was withdrawn and adjusted to pH 2.0, 5.0, 7.0, and 9.0 with 1 M HCl and 0.5 M NaOH, and incubated at 37 °C for 120 min (final inoculum was around 5 × 105 CFU/ml). Samples were taken every hour, and the viable number of bacterium was calculated by pour plate counts of all samples using 10-fold serial dilutions prepared in 0.1% peptone water. Concurrently, bacterial growth was examined by evaluating absorbance at 600 nm with a spectrophotometer [16]. Survival of isolates was evaluated as the percentage of growth compared to the control. This experiment was repeated twice with three independent replicates per experiment (in the same conditions). Values were regarded as mean ± standard deviation.
Bile Resistance
The ability of the selected Lactobacillus strain to grow in percent of 0.3, 0.5, and 1.0% of bile (w/v) was determined according to the method of Vinderola and Reinheimer [17]. According to this method, Lactobacillus strain was inoculated (2% v/v) into MRS with either 0.3, 0.5, or 1% (w/v) of bile and culture was incubated at 37 °C for 24 h. Thereafter, viable counts of Lactobacillus strain were determined by pour plate counts and the growth rate of the bacterium was measured at A560 nm and compared to a control culture. The results were expressed as the percentage of growth (A560 nm) in the presence of bile salts compared to the control (culture with 0% bile considered as a control sample). This experiment was repeated twice with three independent replicates per experiment (in the same conditions). Values signify mean ± standard deviation.
Antibacterial Activity
Detection of the selected Lactobacillus strain with antibacterial activity and evaluation of its effectiveness against some indicator microorganisms were done by well diffusion assay. For the agar well diffusion assay, an overnight culture of the indicator organisms including Staphylococcus aureus PTCC 1112, Bacillus subtilis PTCC 1715, B. cereus PTCC 1015, E. coli PTCC 1338, Salmonella typhimurium (wild type), Klebsiella pneumonia PTCC 1290, Pseudomonas aeruginosa PTCC 1310, and E. coli O-157 were engaged to spray on Muller Hinton agar plates (approximately 106 cells ml−1 of indicator strains were overlaid onto MHA agar plates) at 37 °C. Then, wells of 5 mm diameter were cut into agar plates (under sterile conditions) and 100 μl of the cell-free culture of the selected Lactobacillus strain (with OD = 1) was transferred into the formed wells in the agar plates, while MRS medium served as control. Inhibitory zone of the Lactobacillus strain was checked after 24 h incubation at 37 °C.
Screening for Bile Salt Hydrolytic Activity
For BSH activity, overnight cultures of the Lactobacillus isolate were spotted on MRS agar plates containing 0.37 g l−1 CaCl2 and 0.5% sodium salt of glycodeoxycholic acid (GDCA) (Sigma-Aldrich). Plates were anaerobically incubated at 37 °C for 72 h. Visible halos around the punctures (a granular precipitate around the white colonies) indicate the positive BSH activity of the strain [18]. Negative control was the inocula of each strain in MRS without supplementation. This experiment was repeated twice with three independent replicates per experiment (in the same conditions). Values denote mean ± standard deviation.
Tolerance to NaCl
NaCl tolerance of the selected Lactobacillus strain was determined by providing test tubes containing MRS broth with different concentrations of NaCl (1–3–5%). After sterilization, each test tube was inoculated with 1% (v/v) fresh overnight culture of the Lactobacillus isolate and incubated at 37 °C for 24 h. After 24 h of incubation, growth was determined by observing turbidity (optical density of the samples measured at a wavelength of 600 nm), compared with MRS without NaCl as control. This experiment was repeated twice with three independent replicates per experiment (in the same conditions). Values denote mean ± standard deviation.
Tolerance to Simulated Gastric Juice
Pellet of overnight culture of the selected Lactobacillus strain was obtained by centrifugation, washed twice with phosphate buffer (0.1 M, pH 7.0), resuspended in SES (sterile electrolyte solution), and immediately added to the same volume of “gastric” solution [0.6% (w/v) pepsin, 1% (w/v) NaCl]. Cell suspensions were immediately placed in a water bath (37 °C) and gradually acidified, under gentle agitation, from pH 5.0 to 2.2 in 90 min [19]. Cell counts on MRS agar were performed at times 0, 30, 60, 70, 80, and 90 min. After 90 min, an aliquot of each cell suspension was taken and pelleted by centrifugation, and the cells were suspended in phosphate buffer (0.1 M, pH 8.0) containing 0.3 and 0.1% (w/v) of bile and pancreatin (Sigma-Aldrich), respectively. The cells were kept for a further 60 min at 34 °C and then counted by pour plate counts. Washed cells were suspended in phosphate buffer and subjected to the same conditions as treated samples were used as controls. Survival rate was calculated as percentage of the CFU ml−1 after 30, 60, 70, 80, and 90 min compared to the CFU ml−1 at time 0. This experiment was repeated twice with three independent replicates per experiment (in the same conditions). Values denote mean ± standard deviation.
Lysozyme Resistance
Overnight culture of the isolate were pelleted by centrifugation, washed twice with phosphate buffer (0.1 M, pH 7.0), and suspended in 2 ml of Ringer solution (Sigma-Aldrich). Then, 10% of the bacterial suspension was inoculated in a sterile electrolyte solution consisting of SES (0.22 g l−1 CaCl2, 6.2 g l−1 NaCl, 2.2 g l−1 KCl, 1.2 g l−1, and NaHCO3) in the presence of 100 mg l−1 of lysozyme (Sigma-Aldrich) [20]. Bacterial suspension in SES without lysozyme was considered as control. Samples were incubated at 37 °C, and colony count of samples was carried out after 30 and 120 min on MRS agar (48 h, 30 °C). Survival rate was calculated as percentage of the CFU ml−1 after 30 and 120 min compared to the CFU ml−1 at time 0. This experiment was repeated twice with three independent replicates per experiment (in the same conditions). Values denote mean ± standard deviation.
Determination of Antibiotic Resistance
The selected Lactobacillus strain was tested for resistance to different antibiotics including amoxicillin, ampicillin, cefixime, azithromycin, tetracycline, gentamicin, chloramphenicol, and streptomycin. The test was performed using the standard disc diffusion method [21]. The results were expressed as sensitive (S) or resistant (R).
Preparing Probiotic Mouthwash
The probiotic mouthwash product contained the selected species of interest with a possible functional role in maintaining a healthy oral environment. The mouthwash product was supplied to study subjects as a dry powder in a 20-ml amber glass bottle with a rubber stopper and crimp. Each bottle contained approx. 108 colony forming units (CFU) ml−1 of the selected Lactobacillus strain. The freeze-dried bacterium was blended with food grade maltodextrin as a bulking agent. The single-dose product containers were stored and sealed at room temperature until use. The subjects were instructed to mix the product with approx. 15 ml of bottled or tap water prior to application. The reconstituted mouthwash was then swished for 30 s before being expectorated.
Mouthwash and placebo were numbered by the manufacturer. Placebo mouthwash and the probiotic one were quite similar in shape, nutrients, and taste; the only difference was the presence or absence of bacterial strain.
Clinical Trial Design
This research was a randomized double-blind placebo control to evaluate the effects of probiotic mouthwash and scaling and root planing (SRP) on clinical and microbiological parameters of moderate to severe periodontitis. A total of 50 patients with chronic periodontitis referred to the Department of Periodontology, and 50 periodontal healthy subjects were selected for this research. An informed consent form was obtained from all subjects. In addition, a written signed consent document was obtained from each human subject. Healthy subjects were included in this study as having no radiographic or clinical signs of periodontal destruction. Each patient was examined by two periodontists to assure the accurate diagnosis. All participants were evaluated by full mouth probing. Diagnosis of moderate to severe periodontitis was made based on the following criteria:
-
1.
PPD ≥4 mm
-
2.
CAL ≥3 mm
-
3.
Bone loss ≥3 mm
Patients with the following criteria were excluded:
In case they had used antibiotics (in the past 3 months), had a background of systemic diseases associated with chronic periodontitis, had received previous periodontal treatment (in the past 1 year), used probiotic products (in the past 3 months), have lactose tolerance, are smoking, and are using drugs. Accordingly, a study followed 40 patients and 50 periodontal healthy subjects aged between 24 and 52 years including 45 female and 45 male. After patients’ arrival to the clinic, periodontal index and microbiological samples were taken. Periodontal indices such as plaque index (PI) and O’Leary index (Full-Mouth Plaque Score (FMPS)) were expressed in percent, and based on observation of plaque on four surfaces of each tooth in a limited proportion to the whole surfaces of every tooth, gingival index (GI) [22], periodontal disease index (Ramfjord), bleeding on probing index (Ainamo and Bay) were used [23]. Bleeding was observed for 10–15 s in the gingival pocket (probing number “1” was given as scale of presence of bleeding and number “0” was considered in case there was no bleeding), and periodontal pocket depth (PPD) measured from the free gingival margin to the base of the periodontal pocket with slight manual force (0.25 N) for six points (mesio-buccal, mid-buccal, disto-buccal, mesio-lingual, mid-lingual, and disto-lingual) from first molar to first molar in each jaw was recorded. These parameters were evaluated by an expert (after calibration) out of the study via Williams probe (Williams calibrated, Hu-Friedy, USA).
Sampling and Clinical Examinations
Participants were randomly allocated by a computer-generated list to receive one of the two treatments (SRP and probiotic mouthwash or SRP and placebo). A balance block randomization was used to prepare the randomization tables in order to avoid unequal balance between the two treatment groups. Allocation was implemented by a person not involved in the study. To maintain randomization and blindness, testing and marking mouthwashes was done by a statistical consultant. After this stage, scaling and root planing was done for all the patients via hand instruments (Gracey curettes, Hu-Friedy) and ultrasonic scalars (Cavitron Select, Dentsply). Scaling and root planing were performed until the root surface felt smooth and clean. After that, oral hygiene instructions like brushing techniques (Modified Stillman twice a day, flossing once a day, and interproximal brushing) were explained by a specialist to the patients. Mouthwashes were given to patients, and they were asked to use it for 28 days. Twenty milliliters of mouthwash was used twice a day after brushing the teeth. After washing by mouthwash, patients had to avoid eating or drinking for 2 h.
After probiotic mouthwash treatment, the samples were collected from whole saliva (non-irritation saliva) and sub-gingival plaque from the deepest gingival pocket. These samples were collected with three paper points that inserted into the deepest part of the pockets. The samples were transferred to the lab in sterile containers containing anaerobic transferring medium (mineral salt-base semi-solid media with reductive agents designed as a holding medium for maintaining viability of microorganisms). One hundred microliters of the proper dilutions of these samples was plated, in triplicate, on AASM agar plates. All plates were incubated for 72 h at 37 °C in a 5% CO2 incubator. Subsequently, the number of colony forming units was calculated. Finally, each patient’s clinical parameters were recorded at baseline and 14 and 28 days.
Statistics
All (continuous) variables were compared by ANCOVA in which the post-intervention values of indices were considered as dependent variables. The pre-intervention values and intervention were considered as covariate and independent variables, respectively. Before conducting the ANCOVA, the normality of data, Levene’s test of homogeneity of variance, and homogeneity of regression (slope) were checked. P values less than 0.05 were considered as statistically significant. Statistical analysis was performed using IBM SPSS Statistics (version 20).
Results
Selection of Best Lactobacilli Strain on the Basis of Antibacterial Activity
Among 15 Lactobacillus strains examined in this study, a strain with the highest antibacterial activity against A. actinomycetemcomitans was selected among all the isolates and identified by 16S rRNA gene sequencing. Morphological and biochemical characteristics of the selected isolate showed that the strain NK02 was a Gram-positive, catalase-negative, non-spore-forming rod. Results from 16S rRNA gene sequencing identified our strain as L. salivarius with 100% similarity to L. salivarius DSPV 022SA. We tentatively labeled our strain as L. salivarius NK02 and deposited it in GenBank with the accession number JX129916.1 (Fig. 1).
Probiotic Features of the Selected Lactobacillus Strain
BSH activity and bile tolerance is one of the most critical features for probiotic bacteria. To search for the presence of bsh genes, associating with probiotic features, the selected strain was examined on the basis of species-specific PCR for bsh gene. The results indicated that the NK02 strain produced the expected size of amplicon for bsh gene (data not shown here).
The overall resistance of the strain to lysozyme was expressed in percentage of survival with the value of 90.87% (after 120 min). The strain showed a capability to grow in the presence of bile with a maximum value of 79.87%. Regarding resistance of the NK02 strain in simulated gastric juice conditions (Table 1), results showed that no significant difference was observed within the first 60 min when pH decreased from 5.0 to 2.5. L. salivarius NK02 did not show a significant decrease in number of cells after 70 min (pH 2.4) and 80 min (pH 2.3). At the end of the test, when the simulated gastric juice reached pH of 2.2 (after 90 min), the selected strain still showed survival higher than 7 logs CFU ml−1.
L. salivarius NK02 demonstrated the ability to hydrolyze sodium salt of glycodeoxycholic acid, as shown by halos and granular precipitates around the colonies after growth in MRS-GDCA. The current results showed that the NK02 strain was able to tolerate 1–5% of NaCl and the highest growth was achieved at 1% of NaCl (not shown here).
The effect of pH on the NK02 strain was tested, and the number of viable cells and survival percentage at each pH were determined. The bacterial survival and viability to pH changes was evaluated, and results showed that the NK02 strain had a high survival rate at acidic pH (not shown). The test strain showed antibacterial effects against different Gram-positive, Gram-negative bacteria (Table 2). The results indicated that the selected isolate was susceptible to most of the antibiotics tested, while resistance to the antibiotics was observed in some rare cases. It was also found that the NK02 strain was susceptible to the major classes of antibiotics used in human clinical therapy. The NK02 strain was susceptible to amoxicillin, ampicillin, cefixime, azithromycin, tetracycline, gentamicin, and chloramphenicol and resistant to streptomycin (data not shown here).
Bacterial and Clinical Parameters
Our results showed that the ANCOVA assumptions are met. The Kolmogorov–Smirnov tests were not significant, and the data are normally distributed. Levene’s test confirmed the hypothesis of equality of the error variances between the two groups (all p values were greater than 0.05 for both the pre- and post-intervention values of all the variables). The homogeneity of regression slope assumption was not violated, where interactions between independent variables (intervention) and covariates (pre-intervention values) were not significant (all p values >0.05). The baseline data and the adjusted means were reported as follows:
Some baseline periodontal indices in the two groups are as follows: In the control group, plaque index, bleeding on probing (BOP), probing pocket depth (PPD), gingival index (GI), and periodontal index were 58.6, 62.7, 2.55, 1.6, and 2.97, respectively, and for the test group they were 61.7, 62.3, 2.67, 1.42, and 1.91, respectively (Fig. 2). After using mouthwashes, periodontal indices were changed. BOP and PPD and gingival index were significantly decreased in the test group compared to the control group. Changes in PDI (periodontal disease index) and PI were not significantly different.
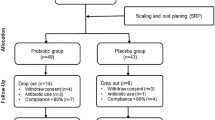
The colony counts of A. actinomycetemcomitans in saliva and sub-gingival crevice was as follows: control 6.77 and 7.08 test 6.62 and 7.15 (Table 3). After using the probiotic mouthwashes for a 28-day period, the reduced bacterial colony count of A. actinomycetemcomitans in the test group was significant compared to that of the control group (Table 4). The whole experimental setup is shown in Fig. 3.
Discussion
New horizons have appeared in the field of periodontal disease treatment thanks to probiotic bacteria especially Lactobacillus species and Bifidobacteria. Presence of probiotics in microflora of human mouth can guarantee more successful treatment of these diseases since it proves these bacteria are consistent with the ecosystem of human mouth.
Bacteria are under a lot of stresses in human mouth, for example, lysozyme enzymes in human saliva first put these bacteria under stress, then in stomach these bacteria are exposed to pH levels of 1.5 to 3, after which they have to tolerate bile [24]. These bacteria spices must be resistant to a dose of 100 mg ml−1 of lysozyme as well. So, one special feature of probiotics is BSH activity, hydrolyzing the bile salts which it makes up at the first place, and it is one of the mechanisms through which the bacteria are protected against bile. This is a vitally important mechanism of probiotic bacteria especially lactobacilli [25, 26]. The current study showed that L. salivarius NK02 is resistant to pH level of 2.5, and also it has an average resistance to bile exposure. In addition, the concentration of bile ranges from of 0.3 to 1%; however, Mathara et al. [27] used a 0.3% dose of this component in their study before, as well (average level of resistance was shown when more than 50% of the bacteria grew in the case compared to the control). Using similar doses of the NK02 strain with the same characteristics in the GI tract, we showed more successful results (1.0 h incubation at pH 2.2 in the presence of 0.3% bile) [28]. In the current study, we investigated the effects of a selected probiotic Lactobacillus strain isolated from the mouth on some of the clinical and microbiologic indices of periodontal disease. The study was a randomized double-blind placebo-controlled trial, and the patients were treated in two groups of control and test by using (SRP + placebo) and (SRP + probiotic), respectively. The periodontal and microbiologic indices were studied in patients during treatment. The results showed that SRP + probiotic treatment led to a significant decrease of GI and BOP compared with that of SRP + placebo for the probiotic group. This result is consistent with Krass et al. [29]. They studied how Lactobacillus reuteri decreases gingivitis and plaque index and came into the conclusion that probiotic L. reuteri is significantly efficient in decreasing gingivitis and plaque index in patients with moderate to severe gingivitis. In addition, Vivekananda et al. [30] showed that probiotic-based mouthwash (probiotic L. reuteri) is significantly efficient in decreasing BOP level, compared to just SRP (35% decrease in BOP in day 42), which proves our findings, as well (48% decrease in BOP in the day 28).
They also found that the use of probiotic mouthwash liquid led to a more meaningful decrease of PPD, compared with that of placebo. The highest rate of decrease in depth of pocket was shown in the group with SRP + probiotic treatment (1/31 mm) which was twice greater than that of SPR alone and probiotic treatment only. In our study, a decreased level of PPD for SRP + probiotic was 1.2 mm which was similar to the results of Vivekananda et al. [30], as well. In addition, Twetman et al. [31] showed that probiotic chewing gums have a positive effect on depletion of gingival inflammation and inflammatory meditator rate in GGF of patients suffering from gingivitis which was in line with our findings.
In our study, PPD decreased in the probiotic bacteria treatment group which can explain the decrease of inflammation in gingiva. Earlier, Shimauchi et al. [32] conducted a randomized double-blind placebo control study in which a meaningful improvement of PPD and PI was shown for the probiotic group (L. salivarius WB21), but a meaningful difference was not shown between placebo and probiotic groups, in comparison to our study in which there was a meaningful difference between the two groups. We used a new species of L. salivarius NK02 which can have a different antimicrobial activity. The studies revealed great differences for antimicrobial activity of the lactobacilli group on periodontal pathogens of P. gingivalis and A. actinomycetemcomitans (unpublished data). Our study showed that A. actinomycetemcomitans was the most prone bacteria to probiotic NK02 strain in vitro. We studied the effects of L. salivarius NK02 on A. actinomycetemcomitans for the first time and then examined the effects of this strain on periodontal indices of our patients under study. Unlike all the studies mentioned before, a meaningful difference in PI and PDI was not shown for the two groups. One of the reasons for the lack of difference in PI is the personal differences of patients in following hygienic instructions and differences in mouth brushing skills among the patients, and in some cases, patients forgot to brush their teeth or wash their mouth with a mouthwash. Other than this, may be the amount of plaque made on the teeth of patients in the probiotic group was the same as in the placebo group, but this plaque is safe with a reduced amount of pathogenic bacteria in comparison to the control group. Actually, it is emphasized that the quantity of plaque was the same but the quality of plaque was different, so as the amount of pathogenicity of plaque was reduced. The current study showed a significant difference between GCF and saliva in A. actinomycetemcomitans pathogen reduction. It was previously shown that one session of SRP treatment can deplete or omit a certain form of sub-gingival periodontal pathogens and the microbial flora need 42 days to reach the baseline state afterwards [33,34,35]. These studies showed the effect of SRP on sub-gingival microbiota depletion of mobile bacteria and anaerobic spirochetes, and enhancement of cocci and immobile bacteria. Two RCT studies in Japan were designated to evaluate the effects of probiotic on periodontal, and the results were consistent with our findings. The results showed that oral use of tablets rich in probiotic L. salivarius resulted in depletion of the number of P. gingivalis in sub-gingival and saliva of healthy patients [36, 37].
Conclusions
All of the effects of probiotics on periodontal health have not been determined completely in the present studies. Early data that have been collected from a variety of studies in this area have shown a number of positive effects of probiotics on periodontal health; however, a higher number of long-term clinical trials are needed to validate that probiotics always have a positive influence on periodontal health. The current study confirms anti-bacterial effects of L. salivarius NK02 against A. actinomycetemcomitans, an important causative agent of periodontitis. Accordingly, it is recommended as an adjunctive treatment for SRP and preventive maintenance phases. To provide stronger evidences, however, more clinical trial studies are recommended in a longer period of time.
References
Castiblanco GA, Yucel-Lindberg T, Roos S, Twetman S (2016) Effect of Lactobacillus reuteri on cell viability and PGE2 production in human gingival fibroblasts. Probiotics Antimicro Prot. doi:10.1007/s12602-016-9246-6
Sharif MR, Haddad-Kashani H, Taghavi-Ardakani A, Kheirkhah D, Tabatabaei F, Sharif A (2016) The effect of a yeast probiotic on acute diarrhea in children. Probiotics Antimicro Prot 8:211–214
Gillian H, Lee M, McGrath C, Yiu CKY (2016) Evaluating the impact of caries prevention and management by caries risk assessment guidelines on clinical practice in a dental teaching hospital. BMC Oral Health 16:58
Meurman JH (2005) Probiotics: do they have a role in oral medicine and dentistry? Eur J Oral Sci 113:188–196
Newman MG, Takei H, Klokkevold PR, Carranza F (2015) Carranza clinical periodontology. 12th edition, Elsevier
Kumar SK, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL (2006) Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol 44:3665–3673
Rylev M, Kilian M (2008) Prevalence and distribution of principal periodontal pathogens worldwide. J Clin Periodontol 35:346–361
Reddy MS, Babu MN (2011) How beneficial is bacterial prophylaxis to periodontal health? J Investig Clin Dent 2:95–101
Henderson B, Ward JM, Ready D (2010) Aggregatibacter (Actinobacillus) actinomycetemcomitans: a triple A* periodontopathogen. Periodontol 54:78–105
Haukioja JH, Stamatova I (2007) Probiotics: contributions to oral health. Oral dis 13:443–451
Koll-Klais P, Mandar R, Leibur E, Mikelsaar M (2005) Oral microbial ecology in chronic periodontitis and periodontal health. Microbial Ecol Health Dis 17:146–155
Sookhee S, Chulasiri M, Prachyabrued W (2001) Lactic acid bacteria from healthy oral cavity of Thai volunteers: inhibition of oral pathogens. J Appl Microbiol 90:172–179
Tsuzukibashi O, Takada K, Saito M, Kimura C, Yoshikawa T, Makimura M, Hirasawa M (2008) A novel selective medium for isolation of Aggregatibacter (Actinobacillus) actinomycetemcomitans. J Periodontal Res 43:544–548
Heilig HGHJ, Zoetendal EG, Vaughan EE, Marteau P, Akkermans ADL, de Vos WM (2002) Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol 68:114–123
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Singhal K, Joshi H, Chaudhary BL (2010) Bile and acid tolerance ability of probiotic Lactobacillus strains. J Global Pharma Tech 2:17–25
Vinderola CG, Reinheimer JA (2000) Enumeration of Lactobacillus casei in the presence of L. acidophilus, bifidobacteria and lactic starter bacteria in fermented dairy products. Int Dairy J 4:271–275
Dashkevicz MP, Feighner SD (1989) Development of a differential medium for bile salt hydrolase-active Lactobacillus spp. Appl Environ Microbiol 55:11–16
Blanquet S, Zeijdner E, Beyssac E, Meunier JP, Denis S, Havenaar R, Alric M (2004) A dynamic artificial gastrointestinal system for studying the behavior of orally administered drug dosage forms under various physiological conditions. Pharm res 21:585–591
Vizoso-Pinto MG, Franz CM, Schillinger U, Holzapfel WH (2006) Lactobacillus spp. with in vitro probiotic properties from human faeces and traditional fermented products. Int J Food Microbiol 109:205–214
Clinical and Laboratory Standard Institute (CLSI) (2010). Performance Standards for Antimicrobial Susceptibility testing; 20th Informational Supplement CLSI document M100-S20-U. Wayne, PA
Loe H, Silness J (1963) Periodontal disease in pregnancy I. Prevalence and severity. Acta Odontol Scand 21:533–551
Ainamo J, Bay I (1975) Problems and proposals for recording gingivitis and plaque. Int Dent J 25:229–235
Corzo G, Gilliland SE (1999) Bile salt hydrolase activity of three strains of Lactobacillus acidophilus. J Dairy Sci 82:472–480
Candela M, Perna F, Carnevali P, Vitali B, Ciati R, Giochetti P, Rizzello F, Campieri MP (2008) Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol 125:286–292
De Smet I, Hoorde LV, Woestyne MV, Christiaens H, Verstraete W (1995) Significance of bile salt hydrolytic activities of lactobacilli. J Appl Bacteriol 29:292–301
Mathara JM, Schillinger U, Kutima PM, Mbugua SK, Guigas C, Franz C, Holzapfel WH (2008) Functional properties of Lactobacillus plantarum strains isolated from Maasai traditional fermented milk products in Kenya. Curr Microbiol 56:315–321
Marteau P, Minekus M, Havenaar R, Huis In’t Veld JH (1997) Survival of lactic acid bacteria in a dynamic model of the stomach and small intestine: validation and the effects of bile. J Dairy Sci 80:1031–1037
Krass P, Carlsson B, Dahl C, Paulsson A, Nilsson A, Sinkiewicz G (2006) Decreased gum bleeding and reduced gingivitis by probiotic Lactobacillus reuteri. Swed Dent J 30:55–60
Vivekananda MR, Vandana KL, Bhat KG (2010) Effect of probiotic Lactobacillus reuteri (Prodentis) in the management of periodontal disease: a preliminary randomized clinical trial. J Oral Microbiol 2:5344. doi:10.3402/jom.v2i0.5344
Twetman S, Derawi B, Keller M, Ekstrand K, Yucel-Lindberg T, Stecksen-Blicks C (2009) Short–term effect of chewing gums containing Lactobacillus reuteri on the level of inflammatory mediators in gingival crevicular fluid. Acta Odontol Scand 67:19–24
Shimauchi H, Mayanagi G, Nakaya S (2008) Improvement of periodontal condition by probiotics with Lactobacillus salivarius WB21: a randomized double blind placebo controlled study. J Clin Periodontol 10:897–905
Amornchat C, Rassameemasmaung S, Sirpairojthikoon W, Swasdison S (2003) Invasion of Porphyromonas gingivalis into human gingival fibroblasts in vitro. J Int Acad Periodontol 5:98–105
Socransky SS, Haffajee AD, Cuqini MA, Smith C, Kent RL Jr (1998) Microbial complexes in subgingival plaque. J Clinc Periodontol 25:134–144
Mousques T, Listgarten MA, Philips RW (1980) Effect of scaling and root planing on the composition of the human subgingival microbial flora. J Periodontol res 15:144–151
Ishikawa H, Aiba Y, Nakanishi M, Oh-hashi Y, Koga Y (2003) Suppression of periodontal pathogenic bacteria in the saliva of humans by the adminstration of Lactobacillus salivarius TI2711. J Jpn Soc Periodontol 45:105–112
Matsouka T, Sugano N, Takigawa S, Takane M, Yoshimura N, Ito K (2006) Effect of oral Lactobacillus salivarius TI 2711 (LS1) administration on periodontal pathogenic bacteria in subgingival plaque. J Jpn Soc Periodontol 48:315–324
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
The study was autonomously reviewed and approved by the ethics committee of Tehran University of Medical Sciences, the review board. Our clinical trial was registered in IRCT and allocated a unique code as follows: IRCT201201258825N1.
Funding
The current work was supported by the National Institute of Genetic Engineering and Biotechnology (NIGEB), affiliated to the Ministry of Science, Research & Technology (IMSRT), Iran.
Competing Interests
The authors declare that they have no competing interests.
Informed Consent
The present study was conducted in full accordance with the World Medical Association Declaration of Helsinki and a written consent form signed by all the subjects or the parents in case the participants were under 18.
Rights and permissions
About this article
Cite this article
Sajedinejad, N., Paknejad, M., Houshmand, B. et al. Lactobacillus salivarius NK02: a Potent Probiotic for Clinical Application in Mouthwash. Probiotics & Antimicro. Prot. 10, 485–495 (2018). https://doi.org/10.1007/s12602-017-9296-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-017-9296-4