Abstract
Many plant virus diseases of major economic importance in sub-Saharan Africa are caused by disease complexes resulting from synergistic interaction of two or more viral agents known to enhance disease severity inflicting heavier crop losses. The most destructive of these are maize lethal necrosis disease (MLND), sweet potato virus disease (SPVD), cassava mosaic disease (CMD), cowpea mosaic disease (CPMD), groundnut rosette disease (GRD) and tobacco bushy top disease (TBTD). MLND, SPVD, CMD and CPMD are caused by synergistic interaction of two independent viruses in mixed infection, whereas GRD and TBTD represent helper-dependent synergism in which the multiple agents involved in the disease complexes which include an umbravirus, a polerovirus and a satellite RNA synergistically interact with each other for their survival and spread. Mixed virus infections can cause disease synergism due to viral suppression of RNA silencing of host defense, an increase in viral replication, enhanced viral movement or any combination of these. Each disease complex has its own characteristics, and a variety of factors affecting its epidemiology must be considered when devising diagnostic tools and management options. All the causal viruses are transmitted by insect vectors such as aphids, whiteflies, thrips, or beetles while some are transmitted by seeds or vegetative propagation. Although the diagnosis of the multiple agents is more complicated than those with single infections, multiplex methods primarily based on serology, PCR and next generation sequencing are available and widely used. This paper briefly addresses the etiology, symptoms, economic importance, synergistic mechanisms, diagnosis, field spread and management practices of these disease complexes and discusses future research needs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
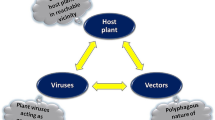
Plant virus diseases are among the major constraints to global agricultural production because they substantially reduce crop yields and damage produce quality. The annual global economic impact of crop virus diseases is estimated at more than US$ 30 billion (Rao & Reddy, 2020). Crops in sub-Saharan Africa (SSA) are particularly vulnerable to virus diseases because in addition to the conducive tropical environment that favor disease development, control measures are often either unavailable or not readily accessible by the mostly small-scale farmers (Thresh, 2003). Virus diseases can result either by infection by a single virus or a mixed infection of two or more viruses and/or sub-viral agents. In many cases, mixed infection by different viral agents results in synergistic interactions which lead to a drastic increase in symptom severity compared to either of the single infections. Synergism is a situation in which mixed infection of a plant with two or more viruses results in an increased multiplication of one or more of the viruses (Syller, 2012). Viral synergism in crop plants has considerable biological, epidemiological, and economic implications in that the increased multiplication of one or more interacting viral partners may have modifying effects on the virus ecology that may include increased symptom severity, increased virus movement in the host plant, expanded host range and enhanced rate of vector transmission (Moreno & López-Moya, 2020), often resulting in higher crop losses.
Synergistic virus-virus interaction can occur either between two or more independent viruses or between a helper and a dependent virus or sub-viral agent which need each other for essential functions, for which the term helper-dependence is used (Syller, 2012). The sub-viral agent involved can be a satellite virus or a satellite RNA that may alter the result of coinfection as in enhanced symptom severity. In such interactions, in addition to an increase in viral load or symptom severity, the virus infection cycle may also be affected. Several diseases caused by a mixed infection of umbravirus-polerovirus complexes in which the polerovirus provides its coat protein to the umbravirus and/or its satellite RNA to make the latter aphid-transmissible exemplify such interactions (Naidu et al., 1999). Many devastating crop diseases in SSA are caused by mixed infection of viruses resulting in complex synergistic interactions in an independent or a helper-dependent manner. In this review, an overview of economically important synergistic virus disease complexes affecting the major crops in SSA is provided. Some of these disease complexes occur exclusively in SSA while others are of considerable economic importance in the region although they may exist elsewhere. In addition, the synergistic mechanisms involved and the different detection, diagnosis, and management strategies that are available for the disease complexes are discussed. The key characteristics of viruses or sub-viral agents involved in the interactions such as particle type, genome type and structure, size and transmission methods are presented in Table 1.
The disease complexes
Maize lethal necrosis disease
Maize (Zea mays L.) is the most important cereal crop in SSA, covering over 40 million ha, largely in smallholder farming systems. Maize lethal necrosis disease (MLND) is currently the most important constraint in maize production in SSA (Prasanna et al., 2020). The disease is characterized by severe symptoms such as leaf necrosis, premature aging and plant death and small cobs with no or few deformed seed which dramatically reduce crop yield. MLND is caused by synergistic interaction of two unrelated viruses, maize chlorotic mottle virus (MCMV, genus Machlomovirus, family Tombusviridae) and one of the maize-infecting members of family Potyviridae (Mahuku et al., 2015). MCMV was first recorded in Peru in 1971 (Castillo & Herbert, 1974). In Africa, MLND has first been reported in 2011 in Kenya where it caused an extensive to complete yield loss (Wangai et al., 2012). Yield losses of 23–100% were reported in affected areas in Kenya (Prasanna et al., 2020). At present, MLND has been identified in Ethiopia, Kenya, South Sudan, Uganda, Rwanda, Mozambique, Tanzania, and Democratic Republic of the Congo (Redinbaugh & Stewart, 2018). Sugarcane mosaic virus (SCMV, genus Potyvirus, family Potyviridae) is the most common potyviral component of MLND in Africa although recent evidence indicates that johnsongrass mosaic virus (JGMV), another potyvirus, can cause MLND in mixed infection with MCMV (Mahuku et al., 2015; Stewart et al., 2017). Since SCMV and to a lesser extent JGMV commonly occur on maize in east African countries for decades causing mosaic symptom (Kulkarni, 1973; Lencho et al., 1997), the new and the most important component of MLND is MCMV.
MCMV and SCMV can individually cause mosaic symptoms in maize in single infections resulting in significant yield losses. The much more severe symptoms of MLND that occur in mixed infection result because MCMV concentration increases many-folds causing drastic increase in symptom severity while that of SCMV remains unchanged (Goldberg & Brakke, 1987; Xia et al., 2016). MCMV is transmitted by thrips (e.g. Frankliniella williamsi) and beetles such as Oulema melanopus and Diabrotica spp (Cabanas et al., 2013; Nault et al., 1978), and by seed at a very low rate of 0.025–0.040% in east African maize varieties (Kimani et al., 2021; Regassa et al., 2021a). Although these rates of MCMV seed transmission appear to be very low, they may be sufficient to provide initial foci to cause disease outbreaks in the presence of high vector populations and to introduce the virus to a new geographical area with seed movement. MCMV is also transmitted by soil via infected plant residue (Mahuku et al., 2015; Regassa et al., 2022). On the other hand, SCMV is transmitted by aphids such as Rhopalosiphum maidis, R. padi and Myzus persicae (Brunt et al., 2000) and seed at even lower rate (Regassa et al., 2021a).
Sweet potato virus disease
Sweet potato (Ipomea batata) is an important food security and subsistence crop in SSA. Virus diseases occur wherever sweet potato is cultivated. The most serious disease of sweet potato in Africa is sweet potato virus disease (SPVD) caused by synergistic interaction of sweet potato feathery mottle virus (SPFMV, genus Potyvirus, family Potyviridae) and sweet potato chlorotic stunt virus (SPCSV, genus Crinivirus, family Clostroviridae) (Karyeija et al., 2000). Mixed infection of SPCSV and Sweet potato virus C, a recently recognized potyvirus species earlier used to be SPFMV strain C also causes SPVD (Kreuze et al., 2021). The disease was first observed in what is now Democratic Republic of Congo in 1939 (Carey et al., 1999). Symptoms include severe stunting, leaf distortion, vein clearing and chlorosis, leading to an 80–90% decrease in tuber yield (Clark et al., 2012). In addition to the direct yield losses in the growing season, SPVD infection can cause degeneration, a progressive yield decline in the next seasons due to infection via vine cuttings (Gibson & Krueze, 2015). Single infection of any of the viruses causes milder symptoms and lesser yield losses and does not cause SPVD. Studies showed that in the interaction leading to SPVD, SPCSV enhances the accumulation of SPFMV by approximately 600-fold with consequent increase in disease severity (Karyeija et al., 2000). SPCSV is transmitted by whitefly Bemisia tabaci while SPFMV is transmitted by aphids such as Myzus persicae and Aphis gossypi (Shaefers & Terry, 1976). SPVD is most prevalent in drier regions of East Africa where whiteflies are also more abundant (Aritua et al., 1998). However, due to vegetative propagation of sweet potato, transmission by vine cuttings plays a more important role in disease spread to the next crop as well as to new geographical areas (Gibson & Krueze, 2015).
SPCSV can also cause synergistic diseases with other sweet potato viruses, including sweet potato mild mottle virus (SPMMV, genus Ipomovirus, family Potyviridae) (Untiveros et al., 2007). In east Africa, synergistic interaction of SPCSV and SPMMV causes a severe symptom of chlorosis, rugosity, leaf strapping and dark green islands resulting in a disease designated sweet potato severe mosaic disease causing about 80% losses (Mukassa et al., 2006). Similarly, the presence of a third different virus in plants affected with SPVD increased the severity of symptoms even further compared with SPVD alone (Mukassa et al., 2006, Untiveros et al., 2007). SPCSV, as a partner in SPVD, is therefore considered the most significant virus contributing to sweet potato yield losses in Africa and elsewhere. Recent studies in South Africa (Mulabisana et al., 2019) however demonstrated that SPFMV and begomoviruses in different combinations of mixed infection can also cause up to total yield losses depending on sweet potato cultivars used, highlighting the economic importance of multiple virus infections even if SPCSV is absent.
Cassava mosaic disease
Cassava (Manihot esculanta Crantz) is an important food security crop for about one billion people. SSA produces more than 60% of global cassava production mostly by small holders. Cassava mosaic disease (CMD) is a devastating virus disease caused by a complex of at least 11 distinct but closely related begomoviruses (genus Begomovirus, family Geminiviridae) worldwide, of which nine occur in Africa (Legg et al., 2015). All these begomoviruses elicit the same mosaic diseases in infected cassava plants which typically include an irregular yellow or yellow–green chlorotic mosaic of leaves, leaf distortion and extreme narrowing near leaf let base, and stunted growth. CMD was first recorded in present-day Tanzania 130 years ago (Warburg, 1894) which makes it the first recorded plant virus disease in Africa. Since then, CMD has been reported from nearly all countries of Africa where cassava is cultivated with an estimated total loss of more than US$1 billion incurred annually (Legg et al., 2019). The begomoviruses are transmitted between plants by the whitefly Bemisia tabaci and spread to the next crop or new areas mainly through infected cuttings, which are the usual mode of cassava propagation (Legg et al., 2015).
While single infection of cassava begomoviruses alone can cause huge losses by itself, evidence that emerged in late 1990’s indicated that in mixed infection, two of the CMD causal viruses, African cassava mosaic virus (ACMV) and a recombinant Ugandan strain of East African cassava mosaic virus (EACMV-UG) interact synergistically, leading to increased viral titers and enhanced symptom severity than both single infections (Harrison et al., 1997). Such synergism due to dual infection was first observed in Uganda, Tanzania, and southern Sudan. A similar synergistic interaction was reported in Cameroon in mixed infection of ACMV and East Africa cassava mosaic Cameroon virus (EACMCV) in which case a many-fold increase in viral DNA of both viruses and enhanced symptom severity was reported (Fondong et al., 2000). This work also provided the first experimental evidence of begomoviral synergism. Such considerable increase in virus accumulation in mixed infection is believed to have epidemiological implication as it will dramatically increase the transmission efficiency of the viruses by whitefly vector. This biological phenomenon is believed to be among the key factors in the genesis and spread of the CMD pandemic in East and Central Africa (Pita et al., 2001). Moreover, the impact of synergistic interaction of the two viruses on cassava yield was demonstrated by field experiment in Uganda in which compared to virus-free cassava control, yield reduction on plants affected by ACMV and EACMV-UG (severe isolate) was reduced by 42% and 68% respectively, while those of mixed infection of the two viruses was reduced by 82% (Owor et al., 2004).
Cowpea mosaic disease
Cowpea (Vigna unguiculata) is the most widely cultivated indigenous legume in SSA, especially by smallholder farmers. SSA accounts for about 70% of total world production. Virus diseases are among the major constraints to cowpea productivity in SSA reducing grain yield by 20–80% (Legg et al., 2019; Thottappilly & Rossel, 1992). The most widespread and economically important viruses are cowpea aphid-borne mosaic virus (CABMV), bean common mosaic virus (black eye cowpea strain) (BCMV-BlCM), formerly black eye cowpea mosaic virus (both in genus Potyvirus, family Potyviridae) and cucumber mosaic virus (CMV, genus Cucumovirus, family Bromoviridae) (Hampton et al., 2003; Legg et al., 2019). These viruses occur wherever the crop is grown and cause significant yield loss in either single or mixed infection among themselves or with any other cowpea viruses (Hema et al., 2014; Legg et al., 2019). CABMV and BCMV produce an indistinguishable mosaic, veinal and interveinal chlorosis, or dark-green vein banding. on susceptible cowpeas while CMV causes some yellow mosaic and distortion of leaves. All the three viruses are seed-transmitted to a varying extent and transmitted by different aphids such as Aphis craccivora and Myzus persicae (Brunt et al., 2000).
Mixed infections of different viruses including those of CABMV, BCMV and CMV have been reported to occur wherever cowpeas are grown causing serious damage to crop productivity (Hema et al., 2014; Legg et al., 2019). This is partly explained by the fact that these three viruses are transmitted by seeds and share aphids vector species for field spread. Co-infection of Moroccan strains of CABMV and CMV cause a synergistic effect on cowpea in Morocco sometimes resulting in premature death of the plant (Fischer & Lockhart, 1976). In another study, Mih et al. (1991) reported from Nigeria that in a susceptible cowpea variety, mixed infection of CABMV and CMV caused a significantly lower yield compared to singly infected or uninoculated control. In the USA, a severe stunting disease named cowpea stunt caused by a strong synergistic interaction between CMV and BCMV (black-eye cowpea mosaic virus strain) results in serious losses in cowpeas (Pio-Ribeiro et al., 1978). The authors further indicated that each virus caused relatively mild disease when inoculated singly and cowpea plants showed significantly reduced stunting. Although cowpea stunt syndrome like that reported in the USA is not reported in SSA, mixed virus infection is very common in cowpea and research efforts are being made to obtain multiple virus-resistant cultivars (Legg et al., 2019). However, information on virus synergism, the effect of co-infection on virus accumulation and symptom severity as well as the mechanisms of the interaction are lacking and should be addressed in future research.
Groundnut rosette disease
Groundnut (Arachis hypogaea L.), also known as peanut, is an important food and cash crop in SSA predominantly grown by crop small scale farmers as a source of dietary protein, oil, and fodder. Groundnut rosette disease (GRD) caused by viruses is the most destructive biotic constraint of groundnut in SSA (Waliyar et al., 2007). It was first reported in 1907 in the present-day Tanzania and occurs only in the African continent and its offshore islands (Naidu et al., 1999). Disease symptoms include very severe stunting, shortened internodes, and reduced leaf size resulting in bushy plant appearance usually with bright yellow curled leaves (chlorotic rosette) or dark or light green leaves (green rosette) (Naidu et al., 1998). The disease is sporadic with an average annual yield loss estimated between five and 30% in non-epidemic years and epidemics often result in 100% yield loss (Olorunju et al., 1991). Numerous epidemics of rosette have been reported in Africa resulting in substantial crop losses (Naidu et al., 1999).
GRD is a classic example of a complex and helper-dependent synergism. It is caused by a synergistic interaction of three viral agents that depend on each other and contribute to the biology and perpetuation of the disease: groundnut rosette assistor virus (GRAV, genus unassigned, family Solemoviridae), groundnut rosette virus (GRV, genus Umbravirus, family Tombusviridae) and its satellite RNA (GRV-SatRNA) (Naidu et al., 1999). In the interaction, GRAV provides GRV and satellite RNA with its coat protein for encapsidation and transmission by aphid vector as GRV does not encode a coat protein. GRV helps the satellite RNA in replication. SatRNA is primarily responsible for disease symptoms (Murant et al., 1988). It also helps in encapsidation of GRV RNA into GRAV coat protein and thereby assists in aphid transmission. GRAV and GRV can replicate autonomously, but satRNA totally depends on GRV for replication. Variants of the GRV-satRNA are responsible for the two different major disease symptom types, chlorotic and green rosette (Murant & Kumar, 1990). GRAV or GRV on their own cause mild mottle symptoms resulting in limited yield losses. Hence, each agent has a role to play in disease development and epidemiology. All the three agents of GRD are transmitted in nature by aphid (Aphis craccivora) in a persistent manner but are not transmitted by seed. GRV and Satellite RNA can be mechanically transmitted under experimental condition (Waliyar et al., 2007). The number of plants that possess all the three agents plays a crucial role in the secondary spread of the disease in a field, while the number of plants that show typical GRD symptoms influence yield (Hema et al., 2014).
Tobacco bushy top disease
Tobacco (Nicotiana tabacum) is widely cultivated in Africa for its leaves used in product processing. The major growing countries include Zimbabwe, Mozambique, Malawi, Tanzania, Uganda and Ethiopia. Of the several viruses reported on tobacco worldwide, disease complexes caused by umbraviruses (genus Umbravirus, family Tombusviridae) and associated viral and sub-viral agents cause significant losses in parts of SSA (Tolin, 2008). The most damaging of these is tobacco bushy top disease (TBTD) known to occur in Zimbabwe, Malawi, Zambia and South Africa since 1960’s (Gates, 1962) and recently reported from Ethiopia where it causes a reduction in harvestable leaf yield of up to 60% in infected plants (Abraham et al., 2014). Symptoms include stunted growth, leaf distortion and curling, early formation of lateral shoots on which other shoots were produced resulting characteristic bushy top appearance. A similar disease is known to occur in China since 1990’s (Mo et al., 2002). Earlier research in southern Africa based on symptomatology and transmission characteristics showed that TBTD is caused by two viruses; tobacco bushy top virus and tobacco vein distorting virus, the former being aphid-transmissible only when it is together with the latter which acts as a helper (Cole, 1962; Gates, 1962). In China, the association of four components, tobacco bushy top virus (TBTV, genus umbravirus, family Tombusviridae), tobacco vein distorting virus (TVDV, genus Polerovirus, family Solemoviridae), TBTV satellite RNA (TBTV-satRNA) and TVDV-associated RNA (TVDV-aRNA) were described (Chen et al., 2022). TBTD in Ethiopia was shown to be caused by at least three agents quite distinct from those in China: a previously unreported umbravirus named Ethiopian tobacco bushy top virus (ETBTV), a novel satellite RNA associated with it (ETBTV-satRNA) and potato leaf roll virus (PLRV) (Abraham et al., 2014). It was further shown that ETBTV and its satRNA are associated with TBTD in Zimbabwe and Malawi (Abraham et al., 2014; Udagawa et al., 2020). In addition, an unpublished partial PLRV sequence from TBTD-infected tobacco in Zimbabwe deposited in the public database (GenBank Acc No. MW113251) indicates that PLRV is associated with TBTD there presumably as a helper to ETBTV. No sequence identical to those of TBTV or TVDV described from China was reported in African TBTD-infected tobacco samples analyzed so far. This suggests that ETBTV and TBTV with their associated helper viruses and satellite RNA molecules are responsible for symptomatically the same tobacco disease in different geographical regions of Africa and China respectively. The International Committee on Taxonomy of Viruses (ICTV) currently recognizes TBTV and ETBTV as two distinct species in the genus Umbravirus (ICTV Release, 2022). Interestingly, sequence analysis shows that ETBTV is phylogenetically more closely related to GRV infecting groundnut in Africa than TBTV from tobacco in China (Abraham et al., 2014; Udagawa et al., 2020).
All the three agents involved in TBTD in Ethiopia are efficiently transmitted from bushy top-diseased tobacco by field-collected red tobacco aphid (Myzus persicae nicotianae) to healthy tobacco plants including Virginia variety and several other plant species (Abraham et al., 2014). Hence, this aphid which commonly colonizes tobacco is believed to be responsible for the field spread of the disease. While ETBV and ETBV-satRNA are mechanically transmissible, PLRV is required for transmission of all the viral agents by the aphid in the laboratory experiment. Two other subviral species with 3 kb and 1 kb size observed in dsRNA extractions from Ethiopian samples (Abraham et al., 2014) are presumed to represent an associated RNA and additional satellite RNA respectively like those described from China (Chen et al., 2022) but further molecular characterization is necessary. In addition, future studies should focus on the geographical distribution and relative importance and further analysis of umbravirus-polerovirus-satellite complex in tobacco growing regions of SSA as well as the biological role of each agent in disease symptom and virus perpetuation.
Mechanisms of viral synergism
Plants have a robust immunity based on silencing virus-derived small-interfering (si)RNAs to defend themselves from invading viruses. Viruses counter this plant defense by encoding viral suppressor of RNA silencing (VSRs) proteins (Pumplin & Voinnet, 2013). Research in the last decades has shown that in the majority of the cases of mixed viral infection that results in synergistic interaction, VSRs play a key role (Ghosh et al., 2021; Mascia & Gallitelli, 2016). VSRs-encoded proteins of one virus may act by assisting the other virus to cope with the already weakened host’s antiviral response resulting in more virus accumulation, leading to a state of synergism. This phenomenon was first demonstrated in a synergistic mixed infection of potato virus X (PVX) and potato virus Y (PVY) where potyviral P1/helper component protease (HC-Pro) proteins suppressed plant RNA silencing in potato enhancing PVX multiplication and hence symptom severity (Vance et al., 1995; Pruss et al. 1997). Several other studies on viral synergistic interactions were shown to have similar mechanisms (Ghosh et al., 2021; Mascia & Gallitelli, 2016). In MLND for example, HC-Pro encoded by the potyvirus SCMV was implicated in VSRs activity. Mutation of an active motif of HC-Pro protein of SCMV impaired MCMV accumulation and synergistic interaction in mixed infection of MCMV and SCMV in maize (Xu et al., 2020). Coinfection of MCMV and SCMV also remarkably increased the accumulation of MCMV-derived siRNAs suggesting its role in synergistic interaction (Xia et al. 2016). In synergistic interaction of SPFMV and SPCSV leading to SPVD, however, VSRs was shown to be mediated by the RNase3 protein encoded by SPCSV, the non-potyviral component (Cuellar et al., 2009). It was suggested that RNase3 may synergize SPFMV and other viruses by targeting a specific host component via interference with small-RNA biogenesis. In severe mosaic disease of cassava caused by synergistic mixed infection of two related begomoviruses, African cassava mosaic virus-Cameroon (ACMV-CM) and East African cassava mosaic Cameroon virus (EACMCV), both viruses benefit from each other in terms of viral replication (Vanitharani et al., 2004). AC4 protein encoded by ACMV-CM was alone able to enhance the accumulation of EACMCV, and similarly, AC2 encoded by the counterpart, EACMCV, could increase the viral titer of ACMV-CM and these two proteins (ACMV-AC4 and EACMCV-AC2) act as suppressors of post transcriptional gene silencing (Vanitharani et al., 2004). These examples indicate that diverse types of virus-encoded proteins are involved in VSRs in different combinations of synergistic virus-virus interactions.
Mechanisms other than VSRs are also known to mediate viral synergism (Ghosh et al., 2021; Mascia & Gallitelli, 2016). These include situations where a virus makes the other virus replication competent, or a virus facilitates the systemic movement of another virus. In one of such examples involving two begomoviruses, the C2 protein encoded by beet curly top virus induces the expression of cell cycle genes and renders the infected cell replication-competent, which promotes its synergistic partner, tomato yellow leaf curl Sardinia virus, to replicate more (Caracuel et al., 2012). In another example in which virus movement is enhanced, the P3N-PIPO protein encoded by the potyvirus white clover yellow vein virus (WClYVV) synergistically interacts with the potexvirus white clover mosaic virus (WClMV) and facilitates its systemic spread in pea, without enhancing its accumulation per cell (Hisa et al., 2014). This result suggests that PN3-PIPO is involved in the synergy between WClMV and WClYVV by facilitating virus movement, but not via a suppression of RNA silencing.
Helper-dependence is a particularly interesting example of virus synergism that occurs when a “dependent” virus that lacks genes encoding for certain protein with essential functions can utilize complementary proteins encoded by a co-infecting “helper” virus. In many instances, such co-infections also result in significantly enhanced symptom development in the host and significantly increased accumulation of one or more of the co-infecting viruses (Erickson & Falk, 2023). In complex helper-dependent synergism like that of umbraviruses, poleroviruses and satellite RNAs exemplified by GRD and TBTD, the polerovirus provides coat protein for the umbravirus and satellite RNA to assist in encapisidation and aphid transmission (Naidu et al., 1999; Chen et al., 2022). The umbravirus helps the satellite RNA in replication as the latter does not code for replicase protein. Satellite RNAs on the other hand are largely responsible for symptom type or severity as demonstrated in GRD (Murant et al., 1988) and more recently in TBTD (Chen et al., 2022). The mechanisms responsible for increased accumulation of some viruses in umbravirus-polerovirus co-infection aren’t precisely known. However three main ways are suggested (Erickson & Falk, 2023): (a) an increase in the replication of one or more co-infecting viruses, resulting in more viral copies per cell; (b) an interaction of umbravirus encoded movement proteins with co-infecting heterologous viral RNAs to impart them with cell-to-cell and systemic movement within the plant thereby resulting in more cells being infected with the dependent virus; and (c) the possibility that one or more of the co-infecting viruses have different host defense mechanisms that can suppress host defense systems against which the other co-infecting virus(es) may be susceptible.
Taken together, mixed virus infections can cause disease synergism due to VSRs of host defense mechanism, an increase in viral replication, or viral movement. It is likely that a combination of these various viral functions and other possible unknown mechanisms interplay to produce synergistic interaction in various virus-virus and virus-host combinations. Despite the existence a large body of knowledge on the essential role of VSRs in virus synergism, the exact mechanisms on how VSRs induce and exacerbate the development of more severe symptoms is still poorly understood (Ghosh et al., 2021; Mascia & Gallitelli, 2016). More research focusing on molecular characterization of viral synergistic interactions will not only lead to better understanding of the molecular basis of synergism but also enable virologists to devise a suitable innovative antiviral control strategy based for instance on siRNA.
Detection and diagnosis
A fast and accurate identification of the causal agent(s) of a disease complex is crucial to devise suitable management practices. Disease complexes caused by multiple viruses are obviously more difficult to diagnose and manage than single infections and demand additional efforts and resources. Recent developments in methods used for plant virus diagnosis have been reviewed by Boonham et al. (2014) and Mehetre et al. (2021). Serological methods, particularly enzyme-linked immunosorbent assay (ELISA), polymerase chain reaction (PCR) or reverse transcription (RT)-PCR and next generation sequencing are the most versatile and commonly used diagnostic techniques. To reduce the cost, improve speed and increase efficiency, various modifications of these methods were adopted for simultaneous detection of different viruses in a mixed infection in one test. Some of the commonly used modifications are briefly described below.
Various ELISA formats are routinely used to specifically detect cassava, maize, sweet potato and cowpea viruses using commercially available virus-specific monoclonal and polyclonal antibodies. Individual assays conducted using different virus-specific antibodies using separate solid supports (e.g. microtiter plates or nitrocellulose membranes) in parallel for the single sample have been commonly used to reveal mixed infection if it exists. Sometimes, broad spectrum monoclonal antibodies detecting viruses in certain taxonomic groups are employed in ELISA to capture different viruses in mixed infections in one test when the mixture belongs to a broader group. The mixture may include a novel virus belonging to that group. Examples are potyvirus-specific monoclonal antibody 2-3H5 known to detect up to 73 species (Menzel & Winter, 2021) and monoclonal antibody 2-5G4 that detects many members of genera Polerovirus and Luteovirus (Abraham et al., 2006; Katul, 1992). In a recent development, cocktail ELISA in which recombinant polyclonal antibodies are raised against the bacterial expressed fused coat protein of different viruses were successfully used for multiple virus detection in plant samples (Kapoor et al., 2014). The major drawback of ELISA method when used for detecting mixed infection is the need to have specific antibodies of a suspected “known” virus which precludes viruses for which specific antibody is not available, including novel species. Other limitations include its being less amenable to multiplexing, its failure to differentiate between some related viruses due to cross reaction (e.g., cassava begomoviruses), its inability to detect umbraviruses and satellite elements which do not have coat protein and its lesser sensitivity compared modern molecular techniques such as PCR.
Among several PCR modifications, multiplex PCR (or reverse transcription (RT)-PCR) has become a fast, cost-effective, and more efficient method for the detection of multiple viruses in mixed infections. Several sets of specific or generic PCR primer pairs designed for the diagnosis and detection of the different viral components in disease complexes are currently available for use single or multiplex PCR, including all the viral agents covered in this review. In many cases, PCR-based specific diagnosis is confirmed by sequencing the corresponding amplicons or restriction fragment length polymorphism (RFLP) analysis. Multiplex PCR/RT-PCR tests have also been developed to detect all members of a specific genus or genera by designing degenerate or specific primers that target highly conserved regions of the virus genome (Pallas et al., 2018). In some cases, such an approach has led to the discovery of previously undescribed viruses involved in disease etiology (e.g., Abraham et al., 2006). For some applications, multiplex quantitative (q)PCR and RT-qPCR enable the quantitative analysis of viruses in epidemiological studies, synergistic interaction, or changes in expression of viral or host gene in virus-virus or plant-virus interaction (Pallas et al., 2018, Mehetre et al., 2021). The multiplex detection via qPCR has been limited to a few targets due to the number of florescent targets that can be used, for example the number reporter and quencher dyes for TaqMan probes or melting curve analysis (Pallas et al., 2018).
Next generation sequencing (NGS) also known as high throughput sequencing, is the most powerful technique currently available for multiplex detection of unlimited number of viruses in mixed infections. NGS is highly sensitive and has the potential to detect the full spectrum of viruses infecting a given host, including known and novel viruses regardless of their genome nature or structure. This unique property of unbiased and non-targeted detection makes NGS a method of choice for simultaneous detection of multiple viruses in mixed infection in disease complexes. NGS provides a high level of multiplexing without the need to use virus-specific reagents (Boonham et al., 2014). In addition, it has made characterization of complete viral genome, or its component much easier. Many recent studies on virus disease complexes in SSA have benefitted significantly from applications of NGS. Relevant examples from SSA include the diagnosis of the viral components of MLND in east Africa (Adams et al., 2013; Guadie et al., 2019), sweet potato viruses in eastern and southern Africa (Kreuze et al., 2009; Mulabisana et al. 2019), sequencing of genomes of GRV and GRAV in groundnut (Jones et al., 2020; Wainaina et al., 2018) and potyviruses and other novel viruses in cowpea (Palanga et al., 2016), and gene expression analysis of mosaic-infected cassava (Bizabani et al., 2012). The two major challenges in the use of NGS for virus disease diagnosis in SSA are its high cost which makes it unaffordable to most poorly funded laboratories and the need for advanced bioinformatics skills and facilities to analyze complex and massive sequence data. On the technical side, determining the link between the viruses inferred from sequence data and the symptoms of disease in the samples from which they were sequenced is sometimes difficult especially when novel or unexpected viruses are discovered. This has recently been observed in SSA where maize and cowpea samples suspected of MLND in Kenya and mosaic symptoms in Burkina Faso, respectively, were found to contain novel and previously unreported viruses with potyvirus-like, polerovirus-like and tombusvirus-like sequences whose contribution to the disease symptoms of the samples largely remained unclear (Palanga et al., 2016; Wamaitha et al., 2018).
Disease management
The most effective way to manage plant virus diseases is to use agricultural practices that prevent virus multiplication or spread since plants cannot be cured once infected. This is best achieved when there is a sound knowledge of disease epidemiology. Commonly used approaches in virus disease management are the use of resistant crop varieties, phytosanitation, chemical control of vectors and cultural practices. A single measure is often inadequate and thus a combination of two or more is used depending on the type of virus disease. The use of virus-resistant crop varieties, when available, is the best and easiest way to control virus disease complexes. Resistant varieties are developed and in wide use in different countries to control MLND in maize (Prasanna et al. 2020, Regassa et al., 2021b), CMD in cassava (Fondong, 2017), GRD in groundnut (Waliyar et al., 2007), SPVD in sweet potato (Mwanga et al., 2003) and CPMD in cowpea (Hampton et al., 2003; Legg et al., 2019). At the same time, research efforts are being made to develop virus-resistant varieties using traditional or modern approaches. Resistant varieties however may not give complete control and have sometimes to be supplemented with other measures, or they may not be available to farmers in some geographical areas. Hence, other control measures such as phytosanitation must be used in combination with resistance varieties or alone. The major feature of phytosanitation is crop hygiene, the use of virus-free planting materials being the focus. For control of diseases of vegetatively propagated crops like CMD in cassava and SPVD in sweet potato, emphasis was given to obtain virus-free plant cuttings (vines or stems) which may be selected by farmers from healthy looking plants or obtained by eliminating the viruses by meristem culture often followed by thermotherapy and chemotherapy (Gibson et al., 1997; Maruthi et al., 2019; Mashilo et al., 2013). For virus diseases transmitted via seeds such as MCMV in maize and CABMV in cowpea, the use of virus-tested and free seeds will minimize crop losses as contaminated seed can act as initial source of inoculum for further spread by insect vectors (Hema et al., 2014; Legg et al., 2019).
Crop hygiene also involves the removal of crop residues or debris and surviving plants from the previous crops to reduce carrying diseased inoculum to new crops and rogueing infected plants early in the growing season. Cultural practices such as planting date adjustment and crop rotation and the use of early maturing cultivars are known to reduce incidence of diseases such as MLND and GRD (Legg et al., 2015; Phillips et al., 1982). For TBTD, there is no resistant tobacco variety available to date (Udagawa et al., 2020). In Malawi and Zimbabwe, combining the use of insecticide such as Imidacloprid (Confidor 70 WG), Thiamethoxam (Actara 25 WG or Acetamiprid (Acetamark 20 SP) to control the aphid vectors, employing cultural practices like avoiding planting during the time with high aphid pressure, suitable adjustment in crop calendar and safe distance between seedbed and standing crops reduce yield losses considerably (Mainjeni et al., 2016). Insecticides applied as foliar sprays or seed treatments are also effective against the spread of many vector-borne viruses including GRD, MLND and CMD (Legg et al., 2015; Naidu et al., 1999; Redinbaugh & Stewart, 2018). However, most small holders in SSA cannot afford pesticides and do not use them, their use mainly being limited to research and quarantine purposes. Overall, a single measure is often inadequate to control a virus disease and combining two or more options tailored to specific virus pathosystem, agronomic practices, and geographical regions in an integrated manner are the most effective.
Conclusion and perspectives
Synergistic virus disease complexes of the major crops grown in SSA covered in this review comprise some of the most important constraints to agricultural production in the region. Research conducted on these disease complexes in the region has led to the description of the main characteristics of several viral or subviral agents involved and associated disease symptoms, estimation of economic importance and crop losses incurred, understanding of the mode of spread and the development of diagnostic tools and management options. Diseases such as MLND, SPVD, CMD are caused by two or more independent viruses that occur together in mixed infection in which at least one virus benefits the other. In the case of cowpea mosaic viruses, multiple virus infections are common but there is no adequate information on the existence of synergistic interaction highlighting the need for more studies. The consequence of synergistic interaction of independent viruses in the different pathosystems also varies between disease complexes. For diseases like MLND and SPVD, synergistic interaction is a primary factor contributing to yield reduction whereas for CMD, single infection can also cause substantial yield losses while mixed infection can aggravate the disease causing more crop losses. On the other hand, GRD on groundnut and TBTD on tobacco are good examples of helper-dependent synergism. An interesting observation with the causal agents of these two diseases in SSA is that their umbravirus components, GRV and ETBTV, are phylogenetically closer to each other than any other virus known (Abraham et al., 2014; Udagawa et al., 2020). An earlier study has shown that based on coat protein sequence, GRAV (which is the assistor to GRV) is phylogenetically most closely related to chickpea chlorotic stunt virus (CpCSV), a polerovirus first described from Ethiopia (Abraham et al., 2006). Another study (Abraham et al., 2009) indicated that among several CpCSV isolates analyzed, those from Ethiopia and Sudan (SSA) are phylogenetically more closely related to GRAV than those from North Africa and West Asia. It has been suggested that the groundnut rosette disease agents including GRAV and GRV have coevolved with indigenous plants other than groundnut in Africa and later infected groundnut when it was introduced in the 16th century in a new encounter phenomenon (Naidu et al., 1998; Jones, 2020). Since tobacco was also introduced to Africa in 16th century, similar phenomenon can be proposed on the introduction and coevolution of tobacco and viruses like ETBTV which infect it in Africa. Because ETBTV, GRV, GRAV and CpCSV have an overlapping geographical distribution in SSA, it is plausible to conclude that ETBTV and GRV on one hand, and CpCSV and GRAV on the other, had common umbravirus- and polerovirus-like ancestors, respectively in SSA from which they have diverged by adaptation to different hosts like tobacco and legumes. It is also possible that the ancestral virus then migrated to Asia where they have evolved to viruses such as TBTV in geographical isolation.
Each virus disease complex described has its own characteristics and has a variety of factors affecting its epidemiology that must be considered when devising diagnosis and management methods. Moreover, climate change can exacerbate the situation further resulting in disease outbreaks due to the increased activity of vectors and enhanced viral multiplication with rising temperatures, although studies are required to determine what the overall effects could be. In terms of improving management practices, modern technologies such as genetic modification can potentially complement traditional breeding for virus resistance. However, despite previous research efforts to introduce virus-resistance by genetic transformation in crops like sweet potato (Sivparsad & Gubba, 2014), cassava (Fondong, 2017) and cowpea (Cruz & Aragão, 2014), no genetically engineered virus resistant crop variety has been released and commercialized in SSA. This is attributed mainly to technical barriers and biosafety regulatory concerns. In this connection, the potential of novel, low-cost and easy-to adopt non-transgenic genetic modification techniques such as spray-induced gene silencing of dsRNA molecules and CRISPR/CAS that targets viral genome or to edit susceptibility disease (Mitter et al., 2017; Tatineni & Hein, 2023) should be explored as alternative strategies. These technologies appear to be technically less demanding, more acceptable to the public and easier to apply for smaller holders in SSA. Finally, there is a need to generate further information on viruses in less studied crops such as cowpea and tobacco particularly in eastern and southern Africa where little research attention has been given so far. In addition, since many reports from virus surveys of crop plants show multiple virus infections (Moreno & López-Moya, 2020), future studies should focus on the interaction between these viruses. These studies may include widespread and economically important viruses in SSA that are not covered in this review such as maize streak viruses in maize and cassava brown streak viruses in cassava.
Data availability
Not applicable.
References
Abraham, A. D., Menzel, W., Lesemann, D. E., Varrelmann, M., & Vetten, H. J. (2006). Chickpea chlorotic stunt virus: A new polerovirus infecting cool-season food legumes in Ethiopia. Phytopathology, 96, 43–446.
Abraham, A. D., Menzel, W., Varrelmann, M., & Vetten, H. J. (2009). Molecular, serological and biological variation among chickpea chlorotic stunt virus isolates from five countries of North Africa and West Asia. Archive of Virology, 154, 791–799.
Abraham, A., Menzel, W., Bekele, B., & Winter, S. (2014). A novel combination of a new umbravirus species, a new satellite RNA and potato leafroll virus causes tobacco bushy top disease in Ethiopia. Archive of Virology, 159, 3395–3399.
Adams, I. P., Miano, D. W., Kinyua, Z. M., Wangai, A., Kimani, E., Phiri, N., Reeder, R., Harju, V., Glover, R., Hany, U., Souza-Richards, R., Deb-Nath, P., Nixon, T., Fox, A., Barnes, A., Smith, J., Skelton, A., Thwaites, R., Mumford, R., & Boonham, N. (2013). Use of next-generation sequencing for the identification and characterization of maize chlorotic mottle virus and sugarcane mosaic virus causing maize lethal necrosis in Kenya. Plant Pathology, 62, 741–749.
Aritua, V., Adipala, E., Carey, E. E., & Gibson, R. W. (1998). The incidence of sweet potato virus disease and virus resistance of sweet potato grown in Uganda. Annals of Applied Biology, 132, 399–411.
Bizabani, C., Rogans, S. J., & Rey, M. E. (2012). Differential miRNA profiles in South African cassava mosaic virus-infected cassava landraces reveal clues to susceptibility and tolerance to cassava mosaic disease. Virus Research, 303, 19840.
Boonham, N., Kreuze, J., Winter, S., van der Vlugt, R., Bergervoet, J., Tomlinson, J., & Mumford, R. (2014). Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Research, 186, 20–31.
Brunt, A. A., Crabtree, K., Dallwitz, M. J., Gibbs, M. J., & Watson, L. (2000). Viruses of Plants. Description and Lists from the VIDE Database. pp. 1484.
Cabanas, D., Watanabe, S., Higashi, C. H. V., & Bressan, A. (2013). Dissecting the mode maize chlorotic mottle virus transmission (Tombusviridae, Machlomovirus) by Frankiniella Williamsi (Thysanoptera: Thripidae). Journal of Economic Entomology, 106, 16–24.
Caracuel, Z., Lozano-Duran, R., Huguet, S., Arroyo-Mateos, M., Rodríguez-Negrete, E. A., & Bejarano, E. R. (2012). C2 from Beet curly top virus promotes a cell environment suitable for efficient replication of geminiviruses, providing a novel mechanism of viral synergism. New Phytologist, 194, 846–858.
Carey, E. E., Gibson, R. W., Fuentes, S., Machmud, M., Mwanga, R. O. M., Turyamureeba, G., Zhang, L., Ma, D., El-Abbas, A., El‐Bedewy, F., R., & Salazar, L. F. (1999). The causes and control of virus diseases of sweet potato in developing countries: is sweet potato virus disease the main problem? In C. Arthur, P. Fergusson, & B. Smith (Eds.), CIP Program Report 1997–98 (pp. 241–248). International Potato Center.
Castillo, J., & Hebert, T. T. (1974). New virus disease affecting maize in Peru. (Nueva enfermedad virosa afectando al maiz en el Peru.) Fitopatologia. Fitopatologia, 9, 79–84.
Chen, X., Luo, H., Zhang, J., Ma, Y., Li, K., Xiong, F., Yang, Y., Yang, J., Lan, P., Wei, T., Xu, Y., Chen, H., & Li, F. (2022). Synergism among the four tobacco bushy top disease casual agents in symptom induction and aphid transmission. Frontiers in Microbiology, 13, 6857. https://doi.org/10.3389/fmicb.2022.846857
Clark, C. A., Davis, J. A., Abad, J. A., Cuellar, W. J., Fuentes, S., Kreuze, J. F., Gibson, R. W., Mukasa, S. B., Tugume, A. K., Tairo, F. D., & Valkonen, J. P. T. (2012). Sweetpotato viruses: 15 years of progress on understanding and managing complex diseases. Plant Disease, 96, 168–185.
Cole, J. S. (1962). Isolation of tobacco vein distorting virus from tobacco plants infected with aphid-transmissible bushy-top. Phytopathology, 52, 1312.
Cruz, A. R. R., & Aragão, F. J. L. (2014). RNAi-based enhanced resistance to cowpea severe mosaic virus and cowpea aphid-borne mosaic virus in transgenic cowpea. Plant Pathology, 63, 831–837. https://doi.org/10.1111/ppa.12178
Cuellar, W. J., Kreuze, J. F., Rajamaki, M. L., Cruzado, K. R., Untiveros, M., & Valkonen, J. P. T. (2009). Elimination of antiviral defense by viral RNase III. Proceedings of the National academy of Sciences of the United States of America, 106, 10354–10358.
Erickson, A., & Falk, B. W. (2023). Dissecting dynamic plant virus synergism in mixed infections of poleroviruses, umbraviruses, and tombusvirus-like associated RNAs. Frontiers in Microbiology, 14, 1223265. https://doi.org/10.3389/fmicb.2023.1223265
Fisher, H. U., & Lockhart, B. E. (1976). A strain of cowpea aphid-borne mosaic virus isolated from cowpeas in Morocco. Phytopathol Z, 85, 43–48.
Fondong, V. N. (2017). The search for resistance to cassava mosaic geminiviruses: How much we have accomplished, and what lies ahead. Frontiers in Plant Science, 8, 408.
Fondong, V. N., Pita, J. S., Rey, M. E., de Kochko, A., Beachy, R. N., & Fauquet, C. M. (2000). Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. Journal of General Virology, 81, 287–297.
Gates, L. F. (1962). A virus causing axillary bud sprouting of tobacco in Rhodesia and Nyasaland. The Annals of Applied Biology, 50, 169–174.
Ghosh, D., Malavika, M., & Chakraborty, S. (2021). Impact of viral silencing suppressors on plant viral synergism: A global agro-economic concern. Applied Microbiology and Biotechnology, 105, 6301–6313.
Gibson, R. W., & Krueze, J. K. (2015). Degeneration in sweet potato due to viruses, virus-cleaned planting material and reversion: A review. Plant Pathology, 64, 1–15.
Gibson, R. W., Mwanga, R. O. M., Kasule, S., Mpembe, I., & Carey, E. E. (1997). Apparent absence of viruses in most symptomless field-grown sweet potato in Uganda. Annals of Applied Biology, 130, 481–490.
Goldberg, K. B., & Brakke, M. K. (1987). Concentration of maize chlorotic mosaic virus increased in mixed infections with maize dwarf mosaic virus strain B. Phytopathology, 77, 162–167.
Guadie, D., Tesfaye, K., Knierim, D., Winter, S., & Abraham, A. (2019). Survey and geographical distribution of maize viruses in Ethiopia. European Journal of Plant Pathology, 153, 271–281.
Hampton, R. O., Thottappilly, G., & Rossel, H. W. (2003). Viral diseases of cowpea and their control by resistance-conferring genes. In B. B. Singh, D. R. Mohan Raj, K. E. Dashiell, & L. E. N. Jackai (Eds.), Advances in Cowpea Research (pp. 159–175). IITA.
Harrison, B. D., Zhou, X., Otim-Nape, G. W., Liu, Y., & Robinson, D. J. (1997). Role of a novel type of double infection in the geminivirus induced epidemic of severe cassava mosaic in Uganda. Annals of Applied Biology, 131, 437–448.
Hema, M., Sreenivasulu, P., Patil, B. L., Kumar, P. L., & Reddy, D. V. R. (2014). Tropical food legumes: Virus diseases of economic importance and their control. Advances in Virus Research, 90, 431–505.
Hisa, Y., Suzuki, H., Atsumi, G., Choi, S. H., Nakahara, K. S., & Uyeda, I. (2014). P3NPIPO of Clover yellow vein virus exacerbates symptoms in pea infected with White clover mosaic virus and is implicated in viral synergism. Virology, 449, 200–206.
ICTV Release (2022). International Committee of Taxonomy of Viruses. https://ictv.global/taxonomy. Accessed 22 June 2023.
Jones, R. A. C. (2020). Disease pandemics and major epidemics arising from new encounters between indigenous viruses and introduced crops. Viruses, 12, 1388. [CrossRef].
Jones, S., Cowan, G., MacFarlane, S., Mukoye, B., Mangenic, B. C., Were, H., & Torrance, L. (2020). RNA sequence analysis of diseased groundnut (Arachis hypogaea) reveals the full genome of groundnut rosette assistor virus (GRAV). Virus Research, 277, 197837. https://doi.org/10.1016/j.virusres.2019.197837
Kapoor, R., Mandal, B., Paul, P. K., Chigurupati, P., & Jain, R. K. (2014). Production of cocktail of polyclonal antibodies using bacterial expressed recombinant protein for multiple virus detection. Journal of Virological Methods, 196, 7–14.
Karyeija, R. F., Kreuze, J. F., Gibson, R. W., & Valkonen, J. P. (2000). Synergistic interactions of a potyvirus and a phloem-limited crinivirus in sweet potato plants. Virology, 269, 3826–3836.
Katul, L. (1992). Serological and molecular characterization of bean leaf roll virus and faba bean necrotic yellows virus. Ph.D. thesis, University of Göttingen, pp. 115, (in German).
Kimani, E. N., Kiarie, S. M., Micheni, C., Muriki, L. G., Miano, D. W., Macharia, I., Munkvold, G. P., Muiru, W. M., Prasanna, B. M., & Wangai, A. (2021). Maize seed contamination and seed transmission of Maize Chlorotic Mottle Virus in Kenya. Plant Health Progress. https://doi.org/10.1094/PHP-02-21-0018-RS
Kreuze, J. F., Perez, A., Untiveros, M., Quispe, D., Fuentes, S., Barker, I., & Simon, R. (2009). Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: A generic method for diagnosis, discovery and sequencing of viruses. Virology, 388, 1–7. https://doi.org/10.1016/j.virol.2009.03.024
Kreuze, J., Cuellar, W., & Low, J. (2021). Challenge of virus disease threats to ensuring sustained uptake of vitamin-A-rich sweetpotato in Africa. In: Scott P. (et al. (Eds.)) Plant diseases and food security in the 21st Century ( pp. 73–94). Springer Nature.
Kulkarni, H. Y. (1973). Notes on east African plant viruses: 5. Identification and economic importance of sugarcane mosaic virus on maize in East Africa. East African Agricultural and Forestry Journal, 39, 56–64.
Legg, J. P., Kumar, L., Makeshkumar, P., Ferguson, T., Kanju, M., Ntawuruhunga, E., Tripathi, P., & Cuellar, W. (2015). Cassava virus diseases: Biology, epidemiology and management. Advances in Virus Research, 91, 85–142.
Legg, J. P., Kumar, L. P., Mahuku, G., Wosula, E., Stavalone, L., Terry, E., & Bosque-Perez, N. (2019). Viruses affecting African crops and their vectors. P. 1–40 In: Neuenschwander, P. & Tamò, M. (Eds.), Critical issues in plant health: 50 years of research in African agriculture, Burleigh Dodds Science Publishing, (ISBN: 978 1 78676 232 0; www.bdspublishing.com
Lencho, A., Abraham, A., Bekele, B., & Tessera, M. (1997). Identification of viruses from maize and grass hosts. Pest Management Journal of Ethiopia, 1, 73–76.
Mahuku, G., Lockhart, B. E., Wanjala, B., Jones, M. W., Kimunye, J. N., Stewart, L. R., Cassone, B. J., Sevgan, S., Nyasani, J. O., Kusia, E., Kumar, P. L., Niblett, C. L., Kiggundu, A., Asea, G., Pappu, H. R., Wangai, A., Prasanna, B. M., & Redinbaugh, M. (2015). Maize lethal necrosis (MLN), an emerging threat to maize-based food security in sub-saharan Africa. Phytopathology, 105, 956–965.
Mainjeni, C., Kamangira, D., & Chilumpha, R. (2016). Pest management decision guide: Green and Yellow, Bushy Top on Tobacco. Plantwise (www.plantwise.com), CAB International. Published under a CC-BY-SA. Accessed 22 June 2023.
Maruthi, M. N., Whitfield, E. C., Otti, G., Tumwegamire, S., Kanju, E., Legg, J. P., Mkamilo, G., Kawuki, R., Benesi, I., Zacarias, A., Munga, T., Mwatuni, F., & Mbugua, E. (2019). A method for generating virus-free cassava plants to combat viral disease epidemics in Africa. Physiological and Molecular Plant Pathology, 105, 77–87.
Mascia, T., & Gallitelli, D. (2016). Synergies and antagonisms in virus interactions. Plant Science, 252, 176–192.
Mashilo, J., van Niekerk, R., & Shanahan, P. (2013). Combined thermotherapy and meristem tip culture for efficient elimination of feathery mottle virus in sweet potato (Ipomoea batatas). Acta Horticulture, 1007, 719–725. https://doi.org/10.17660/ActaHortic.2013.1007.83
Mehetre, G. T., Leo, V. V., Singh, G., Sorokan, A., Maksimov, I., Yadav, M. K., Upadhyaya, K., Hashem, A., Alsaleh, A. N., Dawoud, T. M., Almaary, K. S., & Singe, B. P. (2021). Current developments and challenges in plant viral diagnostics: A systematic review. Viruses, 13, 412.
Menzel, W., & Winter, S. (2021). Reassessing the suitability of a monoclonal antibody for the generic serological detection of potyviruses. Acta Horticulture, 1316, 121–126.
Mih, A. M., Atiri, G. I., & Thottappilly, G. (1991). Relationships between co-infection with cowpea aphid-borne and cucumber mosaic viruses and yield of cowpea lines with varying resistance to these viruses. Phytoparasitica, 19, 65–72.
Mitter, N., Worrall, E. A., Robinson, K. E., Xu, Z. P., & Carroll, B. J. (2017). Induction of virus resistance by exogenous application of double-stranded RNA. Current Opinion in Virology, 26, 49–55. https://doi.org/10.1016/j.coviro.2017.07.009
Mo, X. H., Qin, X. Y., Wu, J., Wu, J. Y., Li, T. F., & Chen, H. R. (2002). First report of tobacco bushy top disease in China. Plant Disease, 86, 74.
Moreno, A. B., & López-Moya, J. J. (2020). When viruses play team sports: Mixed infections in plants. Phytopathology, 110, 29–48.
Mukasa, S. B., Rubaihayo, P. R., & Valkonen, J. P. T. (2006). Interactions between a crinivirus, an ipomovirus and a potyvirus in co-infected sweetpotato plants. Plant Pathology, 55, 458–467.
Mulabisana, M., Cloete, M., Laurie, S., Mphela, W., Maserumule, M., Nhlapo, T., Cochrane, N., Oelofse, D., & Rey, M. (2019). Yield evaluation of multiple and co-infections of begomoviruses and potyviruses on sweet potato varieties under feld conditions and confrmation of multiple infection by NGS. Crop Protection, 119, 102–112.
Murant, A. F., & Kumar, I. K. (1990). Different variants of the satellite RNA of groundnut rosette virus are responsible for the chlorotic and green forms of groundnut rosette disease. The Annals of Applied Biology, 117, 85–92.
Murant, A. F., Rajeshwari, R., Robinson, D. J., & Rashke, J. H. (1988). A satellite RNA of groundnut rosette virus that is largely responsible for symptoms of groundnut rosette disease. Journal of General Virology, 69, 1479–1486.
Mwanga, R. O. M., Odongo, B., Turyamureeba, G., Alajo, A., Yencho, G. C., Gibson, R. W., Smit, N. E. J. M., & Carey, E. E. (2003). Release of six sweetpotato cultivars (‘NASPOT 1 to NASPOT 6’) in Uganda. HortScience, 38, 475–476.
Naidu, R. A., Bottenberg, H., Subrahmanyam, P., Kimmins, F. M., Robinson, D. J., & Thresh, J. M. (1998). Epidemiology of groundnut rosette virus disease: Current status and future research needs. The Annals of Applied Biology, 132, 525–548.
Naidu, R. A., Kimmins, F. M., Deom, C. M., Subrahmanyam, P., Chiyembekeza, A. J., & van der Merwe, P. J. A. (1999). Groundnut rosette: A virus disease affecting groundnut production in sub-saharan Africa. Plant Disease, 83, 700–709.
Nault, L. R., Styer, W. E., Coffey, M. E., Gordon, D. T., Negi, L. S., & Niblett, C. L. (1978). Transmission of maize chlorotic mottle virus by chrysomelid beetles. Phytopathology, 68, 1071–1074.
Olorunju, P. E., Kuhn, C. W., Demski, J. W., Misari, S. M., & Ansa, O. A. (1991). Disease reactions and yield performance of peanut genotypes grown under groundnut rosette and rosette-free field environments. Plant Disease, 75, 1269–1273.
Owor, B., Legg, J. P., Okao-Okuja, G., Obonyo, R., & Ogenga‐Latigo, M. W. (2004). The effect of cassava mosaic geminiviruses on symptom severity, growth and root yield of a cassava mosaic virus disease‐susceptible cultivar in Uganda. The Annals of Applied Biology, 145, 331–337.
Palanga, E., Filloux, D., Martin, D. P., Fernandez, E., Gargani, D., Ferdinand, R., Zabre, J., Bouda, Z., Neya, J. B., Sawadogo, M., Traore, O., Peterschmitt, M., & Roumagnac, P. (2016). Metagenomic-based screening and molecular characterization of cowpea-infecting viruses in Burkina Faso. PLoS ONE, 11, e0165188.
Pallas, V., Sanchez-Navarro, J. A., & James, D. (2018). Recent advances on the multiplex molecular detection of plant viruses and viroids. Frontiers in Microbiology, 9, 2087.
Phillips, N. J., Uyemoto, J. K., & Wilson, D. L. (1982). Maize chlorotic mottle virus and crop rotation: Effect of sorghum on virus incidence. Plant Disease, 6, 376–379.
Pio-Ribeiro, G., Wyatt, S. D., & Kuhn, C. W. (1978). Cowpea stunt: A disease caused by the synergistic interaction of two viruses. Phytopathology, 68, 1260–1265. https://doi.org/10.1094/Phyto-68-1260
Pita, J. S., Fondong, V. N., Sangaré, A., Otim-Nape, G. W., Ogwa, S., & Fauquet, C. M. (2001). Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. Journal of General Virology, 82, 655–665.
Prasanna, B. M., Suresh, L. M., Mwatuni, F., Beyene, Y., Makumbi, D., Gowda, M., Olsen, M., Hodson, D., Worku, M., Mezzalama, M., Molnar, T., Dhugga, K., Wangai, A., Gichuru, L., Angwenyi, S., Alemayehu, Y., Grønbech-Hansen, J., & Lassen, P. (2020). Maize lethal necrosis (MLN): Containing the spread and impact of a devastating transboundary disease in sub-saharan Africa. Virus Research, 282, 197943. https://doi.org/10.1016/j.virusres.2020.197943
Pruss, G., Ge, X., Shi, X. M., Carrington, J. C., & Bowman Vance, V. (1997). Plant viral synergism: The potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. The Plant Cell, 9, 859–868.
Pumplin, N., & Voinnet, O. (2013). RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nature Reviews Microbiology, 11, 745–760. https://doi.org/10.1038/nrmicro3120
Rao, G. P., & Reddy, M. G. (2020). Overview of yield losses due to plant viruses. In L. P. Awasthi (Ed.), Applied Plant Virology: Advances, detection, and antiviral strategies (pp. 531–562). Academic. https://doi.org/10.1016/B978-0-12-818654-1.00038-4
Redinbaugh, M. G., & Stewart, L. R. (2018). Maize lethal necrosis: An emerging, synergistic viral disease. Annual Review of Virology, 5, 301–322. https://doi.org/10.1146/annurev-virology-092917-04341
Regassa, B., Abraham, A., Fininsa, C., & Wakgary, D. (2021a). Alternate hosts and seed transmission of maize lethal necrosis in Ethiopia. Journal of Phytopathology, 169, 303–315.
Regassa, B., Wakgari, D., Fininsa, C., & Abraham, A. (2021b). Screening maize genotypes for resistance to Maize Lethal Necrosis Disease in Ethiopia. Trop Plant Pathol, 46, 583–595.
Regassa, B., Abraham, A., Fininsa, C., Wegary, D., & Wolde-Hawariat, Y. (2022). Transmission and persistence of maize lethal necrosis in infested soil and infected maize residue. European Journal of Plant Pathology, 162, 263–273.
Schaefers, G. A., & Terry, E. R. (1976). Insect transmission of sweet potato disease agents in Nigeria. Phytopathology, 66, 642–645.
Sivparsad, B. J., & Gubba, A. (2014). Development of transgenic sweet potato with multiple virus resistance in South Africa (SA). Transgenic Research, 23, 377–388.
Stewart, L. R., Willie, K., Wijeratne, S., Redinbaugh, M. G., Massawe, D., Niblett, C. L., Kiggundu, A., & Asiimwe, T. (2017). Johnsongrass mosaic virus contributes to Maize lethal necrosis in East Africa. Plant Disease, 101, 1455–1462.
Syller, J. (2012). Facilitative and antagonistic interactions between plant viruses in mixed infections. Molecular Plant Pathology, 13, 204–216.
Tatineni, S., & Hein, G. L. (2023). Plant viruses of agricultural importance: Current and future perspectives of virus disease management strategies. Phytopathology, 113, 117–141.
Thottappilly, G., & Rossel, H. W. (1992). Virus diseases of cowpea in tropical Africa. Tropical Pest Management, 38, 337–348.
Thresh, J. M. (2003). Control of plant virus diseases in sub-saharan Africa: The possibility and feasibility of an integrated approach. African Crop Science Journal, 11, 199–223.
Tolin, S. A. (2008). Tobacco viruses. In W. J. Brian, Mahy, Marc H. V. van Regenmortel (eds.), DeskEncyclopedia of Plant and Fungal Virology (pp. 471–478).
Udagawa, H., Koga, K., Shinjo, A., Kitashiba, H., & Takakura, Y. (2020). Reduced susceptibility to a tobacco bushy top virus Malawi isolate by loss of function in host eIF(iso)4E genes. Breeding Science, 70, 313–320. https://doi.org/10.1270/jsbbs.19135
Untiveros, M., Fuentes, S., & Salazar, L. F. (2007). Synergistic interaction of sweet potato chlorotic stunt virus (Crinivirus) with Carla-, Cucumo-, Ipomo-, and Potyviruses Infecting Sweet Potato. Plant Disease, 91, 669–676.
Vance, V. B., Berger, P. H., Carrington, J. C., Hunt, A. G., & Shi, X. M. (1995). 5’ proximal potyviral sequences mediate potato virus X/potyviral synergistic disease in transgenic tobacco. Virology, 206, 583–590.
Vanitharani, R., Chellappan, P., Pita, J. S., & Fauquet, C. M. (2004). Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. Journal of Virology, 78, 9487–9498.
Wainaina, J. M., Harvey, J., Ateka, E., Makori, T., Karanja, D., Kehoe, M. A., & Boykin, L. M. (2018). Genomic characterisation and evolutionary relationships of groundnut rosette virus from the western highlands of Kenya. Trop Plant Pathol, 43, 583–585.
Waliyar, F., Kumar, P. L., Ntare, B. R., Monyo, E., Nigam, S. N., Reddy, A. S., Osiru, M., & Diallo, A. T. (2007). A century of research on groundnut rosette disease and its management. Information Bulletin no. 75 (p. 40). Technical Report. International Crops Research Institute for the Semi-Arid Tropics.
Wamaitha, M. J., Nigam, D., Maina, S., Stomeo, F., Wangai, A., Njuguna, J. N., Holton, T. A., Wanjala, B. W., Wamalwa, M., Lucas, T., Djikeng, A., & Garcia-Ruiz, H. (2018). Metagenomic analysis of viruses associated with maize lethal necrosis in Kenya. Virology Journal, 15, 90.
Wangai, A. W., Redinbaugh, M. G., Kinyua, Z. M., Mahuku, G., Sheets, K., & Jeffers, D. (2012). First report of Maize chlorotic mottle virus and maize lethal necrosis in Kenya. Plant Disease, 96, 1582.
Warburg, O. (1894). Die kulturpflanzen Usambaras. Mitteilungen aus den Deutschen Schutzgebieten, 7, 131.
Xia, Z., Zhao, Z., Chen, L., Li, M., Zhou, T., Deng, C., Zhou, Q., & Fan, Z. (2016). Synergistic infection of two viruses MCMV and SCMV increases the accumulations of both MCMV and MCMV-derived siRNAs in maize. Scientific Reports, 6, 20520.
Xu, X. J., Li, H. G., Cheng, D. J., Liu, L. Z., Geng, C., & Tian, Y. P. (2020). A spontaneous complementary mutation restores the RNA silencing suppression activity of HC-Pro and the virulence of sugarcane mosaic virus. Frontiers in Plant Science, 11, 1279. https://doi.org/10.3389/fpls.2020.01279
Author information
Authors and Affiliations
Contributions
The author conceived the idea, searched literature, wrote the draft, prepared Table 1, made revisions, and submitted the manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abraham, A. Synergistic crop virus disease complexes in Sub-saharan Africa: causes, consequences and control. Phytoparasitica 52, 27 (2024). https://doi.org/10.1007/s12600-024-01143-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12600-024-01143-9