Abstract
Plant-specific remorin genes have been identified in angiosperms, gymnosperms, ferns, and mosses. Several remorin genes are highly conserved in plant genomes, and their basic characteristics have been determined. Remorins have multiple biological functions, including in antibacterial defense, signal transduction, damage repair, and resistance to environmental stresses. In the present study, a full-length cDNA clone of the StREMa4 remorin gene was isolated from Ralstonia solanacearum-infected potato (Solanum tuberosum L.) cultivar ‘ED13’ plants through the rapid amplification of cDNA ends. Sequence analyses revealed that StREMa4 comprised 803 bp, including a 591 bp open reading frame that encoded a protein consisting of 197 amino acids. The StREMa4 protein was highly homologous to remorins from potato and other Solanaceae species. Real-time PCR analyses revealed that in addition to being up-regulated by Ralstonia solanacearum, StREMa4 expression was induced by exogenous hormones (i.e., salicylic acid, methyl jasmonate, and abscisic acid), with some differences in the expression patterns. Tissue localization analyses indicated that StREMa4 expression was tissue-specific, occurring primarily in the phloem of stem and leaf tissues.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Remorins are plant-specific proteins (Raffaele et al. 2007) common in angiosperms, gymnosperms, ferns, and mosses (Checker and Khurana 2013). In 1989, the first remorin was identified in potato (Solanum tuberosum L.), and was named pp34 (i.e., phosphorylated protein) (Farmer et al. 1989). Remorins are hydrophilic and associated with plasma membranes (Reymond et al. 1996; Rajendran and Simons 2005). Owing to its ability to attach to the plasma membrane, these proteins were later renamed remorins (Jacinto et al. 1993). Thus far, remorins have been detected in leek (Allium porrum L.) seedlings (Laloi et al. 2007), Arabidopsis thaliana (L.) seedlings (Bhat et al. 2005; Yue et al. 2014), Bright Yellow 2 cells (Morel et al. 2006), tobacco (Nicotiana tabacum L.) leaves (Mongrand et al. 2004), and barrel medic (Medicago truncatula Gaertn.) roots (Lefebvre et al. 2007). Each remorin contains a conserved C-terminal region and a variable N-terminal region, which is responsible for the structural and functional differences among remorins (Marin and Ott 2012). In addition to its structure and distribution, the functions of remorins during plant growth and development have been investigated.
In 2000, Reymond et al. (Reymond et al. 2000) reported that the production of the remorin in A. thaliana is induced by drought stress, and that remorin is similar to the late embryogenesis abundant protein, which plays a major role in drought and salt stress responses. In 2002, Kreps et al. (Kreps et al. 2002) investigated gene expression changes in A. thaliana and observed elevated levels of remorin gene expression during exposures to salt, osmotic, or cold stresses. Additionally, Yue et al. (Yue et al. 2014) determined that remorins can enhance salinity tolerance in A. thaliana. Lefebvre et al. (Lefebvre et al. 2010) revealed that remorins attach to the host plasma membrane surrounding bacteria to protect plants from infection. They also interact with symbiotic receptors to acquire signaling molecules from bacteria. S. Li et al. (S. Li et al. 2013) reported that remorins influence wood strength and other properties in poplar trees. Moreover, remorins have been confirmed to interact with potato virus X by directly binding to triple gene block protein 1 (Raffaele et al. 2009a, b).
Potato is the fourth largest food crop in the world in terms of cultivated area, after wheat, rice, and corn. However, fungal, viral, and bacterial diseases can severely damage potato crops and considerably limit yield. Therefore, investigations of the molecular mechanisms regulating potato growth and development are necessary. However, to the best of our knowledge, investigations of potato remorins have focused only on their distributions and structures, while their specific functions have not been studied.
In the present study, we cloned the full-length cDNA of a new potato remorin gene using a pathogen-induced suppression subtractive hybridization (SSH) cDNA library combined with the rapid amplification of cDNA ends (RACE) technique. We also analyzed the sequence, evolutionary characteristics, and induced expression patterns of the potato remorin gene.
Materials and methods
Plant culture and inoculation
Potato (Solanum tuberosum L.) plants of the bacterial wilt-resistant cultivar ‘ED13’ were kindly provided by the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China). Seedlings were transplanted to pots (diameter: 10 cm) containing peat/vermiculite (3:1, w/w) and grown under standard greenhouse conditions (light intensity: 2000–4000 lx; photoperiod: 16 h day/8 h night; daytime temperature: 25–27 °C; nighttime temperature: 16–20 °C; and relative humidity: 70–80 %).
Ralstonia solanacearum strain PO41 of race 3 (biovar 2) was provided by the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences. The bacterium was used to inoculate potato seedlings at the 9–10-leaf stage using the root injury and drenching method of He et al. (He et al. 1983). After root injury, a 30 mL bacterial suspension (108 colony-forming units/mL) was added to each pot. Mock inoculations with water were used as the controls. Stem samples were collected after 6, 12, 24, 36, 48, 72, and 96 h, and immediately frozen in liquid nitrogen and stored at −80 °C (Li et al. 2014). Each treatment consisted of three replicates.
Chemical treatments
Healthy seedlings at the 9–10 leaf stage were subjected to the following chemical treatments. Salicylic acid (SA) was dissolved in water to final concentration of 5 mM. Methyl-jasmonate (MeJA) was first dissolved in 100 % ethanol to a stock concentration of 100 mM, and then diluted in water to final concentration of 100 μM. Tween 20 was added (0.01 %) as a surfactant. Final solutions were sprayed onto leaves until droplets formed. Control plants were sprayed with water for SA and Tween 20 plus 0.1 % ethanol for MeJA and allowed to dry in the same manner as the corresponding treatment. ABA (abscisic acid, Sigma, St. Louis, USA) was dissolved to 20 mM in 100 % ethanol. This stock solution was then diluted with water to a final concentration of 100 μM and sprayed evenly over the plants before sealing as described for the MeJA treatment. Control plants were sprayed evenly with 0.1 % (w/v) ethanol solution. Plants were immediately sealed in black plastic bags and incubated at room temperature for 24 h. The treatments were repeated once daily for 5 consecutive days (Sawano et al. 2008). Leaf and stem tissues were then sampled after treatments, immediately frozen in liquid nitrogen for subsequent RNA extraction.
RNA preparation
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The purified RNA was reverse transcribed using the Clontech SMART PCR cDNA Synthesis Kit (Clontech, PaloAlto, CA, USA) following the manufacturer’s instructions.
Isolation of full-length cDNA
In an earlier experiment (Gao et al. 2008), we extracted total RNA from leaf samples and synthesized the first-strand cDNA. An SSH cDNA library was constructed and 384 positive cDNA clones were screened to construct the cDNA library. Gene-specific primer (5′-CTGAAAAACAAGATTGCTTTAGTCC-3′) based on the target gene screened from the SSH cDNA library was designed according to the 3′-RACE method with the SMART-RACE cDNA Amplification Kit (Clontech). The cDNA sequence was compared with the sequences available in the Potato Genome Sequence Database (Lingle and Dyer 2001). The gene was subsequently named StREMa4 and deposited in the GenBank database (Accession No: EU057713.1).
Sequence analysis of StREMa4
After isolated of full-length cDNA, gene sequencing performed by Sangon Technologies (Shanghai, China), and the sequence was BLAST in GenBank, EMBL, DDBJ database. We analyzed StREMa4 and determined the encoded amino acid sequence using the BioEdit v5.0 software (Hall 1999). Homologous gene and amino acid sequences were searched for, and comparisons were made using the BLAST program from the National Center for Biotechnology Information website. Phylogenetic trees were constructed using the neighbor-joining method of MEGA 5.0 (Tamura et al. 2011). The isoelectric point and hydrophilicity/hydrophobicity of StREMa4 were analyzed using the ProtParam (http://web.expasy.org/protparam/) and ProtScale (http://web.expasy.org/protscale/) online tools. Phosphorylation sites were predicted using the NetPhos2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/). Signal peptides were analyzed with the SignalP4.1 Server (http://www.cbs.dtu.dk/services/SignalP), while the TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM) was used to investigate the transmembrane region.
Protein subcellular localization was predicted with WoLF PSORT (Horton et al. 2007) and the TargetP1.1 Server (http://www.cbs.dtu.dk/services/TargetP/). The protein was classified based on Gene Ontology categories (http://geneontology.org/) and the Protfun2.2 Server (http://www.cbs.dtu.dk/services/ProtFun/).Protein disulfide bonds were analyzed using the SCRATCH Protein Predictor online program (http://scratch.proteomics.ics.uci.edu/).The secondary and tertiary structures were predicted using the PRABI–Lyon–Gerland database (https://npsa-prabi.ibcp.fr) and the SWISS-MODEL online server (http://www.swissmodel.expasy.org/), respectively.
Real-time PCR
Real-time PCR analysis was completed according to the method of Jain et al. (Jain et al. 2006). The qRT-PCR analysis was performed using the SYBR green PCR master mix (Applied Biosystems) and an ABI Prism 7500 sequence detector (Applied Biosystems, Foster City, CA, USA). The relative expression levels and reaction specificities were calculated according to a published method (Pfaffl 2001). We used the following gene-specific primers: forward, 5′-AGGCTGAGCTAAAAAGGACTGA-3′; reverse, 5′-GACATTTTGGTGCCATC TCCT-3′. The following primers were used to amplify an actin gene (GenBank Accession: X55747), which served as the internal control: forward, 5′-TGTCCTCCTAACTGAA GCACCT-3′; reverse, 5′-CCACTG GCATACAGCGAAA-3′. The PCR program was as follows: 95 °C for 2 min; 40 cycles of 94 °C for 5 min, 95 °C for 10 s, and 60 °C for 40 s. Uninoculated plants served as controls. The real-time PCR experiment was repeated three times.
In situ hybridization of StREMa4 gene
In situ hybridizations were conducted using paraffin sections of potato leaf tissue (10 μm thick) and digoxigenin-labeled RNA probes according to the method of Elorza et al. (Elorza et al. 2004). Positive hybridization signals are visualized by violet staining using a digoxigenin-labelled RNA immunodetection system. Probes were synthesized using the StREMa4 PCR product. Antisense probe and sense probe was labeled with digoxigenin-UTP using the DIG RNA Labeling Kit (SP6/T7; Roche). The primer sequence was 5′-TTTGGTGCCATC TCCTCTGCGGTAA-3′.
Results
Cloning and analysis of StREMa4 gene
We constructed a pathogen-induced potato stem-specific SSH cDNA library in an earlier experiment. The target gene clone was screened from the SSH cDNA library, and its full-length cDNA sequence (Accession: EU057713.1) was amplified using a 3′-RACE method. A BLAST analysis revealed the corresponding gene sequence was highly similar (up to 98 % identical) to the putative remorin sequence cloned previously from potato with the accession number NM_001288026.1. Thus, the gene was considered to encode a putative remorin a4-e8. Its full-length sequence consisted of 803 bp, which comprised a 591 bp open reading frame encoding 197 amino acids (Fig. 1).
The similarity between the StREMa4 and potato remorin a3b4 gene sequences suggested that the former is a homologous sequence of the latter. Moreover, StREMa4 was also highly similar to the Solanum pennellii remorin-like LOC107023866 (Accession: XM_015224685.1) and Solanum lycopersicum remorin LOC101266641 (Accession: XM_004240689.2) sequences. The 5′ and 3′ ends of these sequences were generally conserved. At the amino acid level, putative remorin a4-e8 (Accession: NP_001275297.1) and remorins from other Solanaceae species contained highly conserved sequences.
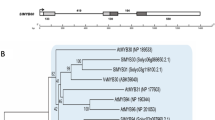
A phylogenetic tree based on 16 amino acid sequences was constructed using the neighbor-joining method (Fig. 2). In terms of the node position and branch length, StREMa4 was located in the same clade with one remorin of potato and two remorins of tomato (Solanum lycopersicum), which may have a common origin. The second closest phylogenetic relationship was observed with a tobacco remorin. In contrast, there was a relatively large phylogenetic distance between StREMa4 and tomato remorin 1 (Accession: NM_001247302.2).
Phylogenetic relationships of the potato StREMa4 based on the sequence alignments of the encoded proteins. An neighbor-joining tree was constructed using MEGA 5.0 software. The bootstrap values are indicated at the nodes of the tree and expressed as percentages. The scale bar indicates the distance calculated from the multiple alignment
The predicted StREMa4 protein (pI approximately 6.70) was structurally similar to the other Solanaceae remorins. Hydrophilic and hydrophobic amino acids were unevenly distributed throughout the deduced amino acid sequence. Additionally, there were considerably more hydrophilic amino acids, indicating StREMa4 was highly hydrophilic. Thus, StREMa4 was predicted to be a water-soluble protein. Regarding its tertiary structure, it was predicted to be a non-spherical molecule. Subcellular localization analysis revealed that the protein was located on the plasma membrane, and likely involved in immune responses.
Phosphorylation and dephosphorylation are important processes affecting intracellular signal transduction. A total of 12 phosphorylation sites were identified in the StREMa4 sequence. The predicted tertiary structure suggested that the Cys residues at positions 181 and 195 formed a disulfide bond that introduced a fold in the peptide chain to stabilize the protein structure. Additionally, the protein was predicted to contain five α-helices, with a long and short α-helix at the N and C terminals, respectively.
Ralstonia solanacearum-induced StREMa4 expression pattern
To reveal the spatiotemporal expression pattern of StREMa4 during the early interaction between potato plants and R. solanacearum, we analyzed the StREMa4 expression level in inoculated potato stem tissues at different time points using a real-time PCR assay (Fig. 3). The bacterial pathogen induced a relatively rapid increase in the StREMa4 expression level beginning 6 h after inoculation. The highest expression level occurred 72 h after inoculation (i.e., more than 2-fold higher than the expression level at the first time point). The expression level rapidly decreased 96 h after inoculation to almost undetectable levels. The mock inoculations did not induce significant changes to the StREMa4 expression level, which remained low. These observations implied that StREMa4 was expressed at low levels under normal conditions to regulate basic metabolic activities. However, the pathogen-induced up-regulation of StREMa4 expression suggested that the putative remorin was closely related to immune responses associated with bacterial wilt resistance in potato plants.
Remorin gene expression is affected by plant hormones
All hormone treatments, including SA, MeJA, and ABA, up-regulated StREMa4 expression to varying degrees (Fig. 4a–c). The StREMa4 expression levels peaked at 5, 3, and 2 days after treatment with SA, MeJA, and ABA, respectively. Of the three tested plant hormones, the highest and lowest StREMa4 expression levels were induced by ABA and SA, respectively. The ABA and MeJA induced StREMa4 expression levels that decreased after peaking, while the SA-induced StREMa4 expression level remained relatively low. The peak StREMa4 expression level induced by ABA was approximately 3-fold higher than that induced by MeJA, and occurred 1 day earlier. In other words, ABA affected StREMa4 expression more rapidly than MeJA. This finding implied the mechanism regulating the induction of StREMa4 expression differed between ABA and SA or MeJA. The ABA mechanism enabled a more rapid and efficient induction of StREMa4 expression. Consequently, 2 days after hormone solutions were applied, the StREMa4 transcript abundance was greater in ABA treated plants than in SA or MeJA treated plants. Thus, ABA may be a more effective regulator of StREMa4 expression than SA or MeJA.
Localization of remorin gene expression
We conducted in situ hybridizations to determine in which plant tissues StREMa4 was expressed. In the samples collected 3 days after inoculation with R. solanacearum, StREMa4 mRNA was mainly distributed in the vascular bundles of leaves (Fig. 5b) and phloem of stems (Fig. 5d), while extremely weak hybridization signals were detected in the stem epidermal cells. Hybridization signals were not detected in the uninoculated control plants (Fig. 5a and c). StREMa4 was not expressed in the roots of inoculated and control plants. These results indicated that StREMa4 was mainly expressed in the stem and leaf vascular tissues (although not in the xylem). This tissue specificity was likely in that R. solanacearum is a pathogen that targets plant vascular tissues which has been confirmed by previous work, in which environmental scanning electron micrographs showed colonizing bacteria in the vascular tissue (Gao et al. 2009).
NISH localization of StREMa4 mRNAs in potato stem and leaf tissues. a Leaves mocked with water. b Leaves inoculated with R. solanacearum. c Stems mocked with water. d Stems inoculated with R. solanacearum. The StREMa4 RNA probe were hybridized with the consecutive 10 μm transverse sections of leaves and stems. Transverse sections were probed with digoxigenin-labelled antisense mRNA and view under brightfield. Positive hybridization signals are visualized by violet staining using a digoxigenin-labelled RNA immunodetection system. P phloem, X xylem, UE upper epidermis, LE lower epidermis. Scale bar = 50 μm
Discussion
In this study, we cloned a new remorin gene, StREMa4, using the RACE technique, and systematically analyzed its structural characteristics, functional attributes, and expression properties. Our results provide clues regarding the biological functions of remorins. Analyses of phosphorylation sites indicated StREMa4 contains seven Ser phosphorylation sites, four Thr phosphorylation sites, and one Tyr phosphorylation site. The relative abundance of phosphorylation sites in StREMa4 suggests they are important for the activation of this remorin, which is consistent with the results of a previous study (Reymond et al. 1996). Subcellular localization experiments revealed that StREMa4 is located on the plasma membrane, which is in agreement with the findings of a confocal microscopy study involving α-130 antibody labeling and tissue cell assays (Bariola et al. 2004). According to our phylogenetic tree, StREMa4 and a pathogen-induced remorin identified by Perraki et al. (Perraki et al. 2014) belong to the same family, implying that StREMa4 may be associated with defense responses against pathogens. This is supported by the observed pathogen-induced expression patterns and previous results indicating that remorins participate in disease resistance responses in other plants (Bozkurt et al. 2014; Gui et al. 2014; S. Li et al. 2013; Son et al. 2014; Yue et al. 2014; Jamann et al. 2016). Furthermore, analyses of the conserved region, functional site, and phylogenetic evolution of the StREMa4 amino acid sequence indicated that this protein is a new remorin that shares a common ancestor with the S. lycopersicum (XM_004240689.2) and S. pennellii (XM_015224685.1) remorins.
Recently, several studies have concluded that remorin production in plants is often associated with defense signaling molecules (Bray 2002; Wu et al. 2006; Anderson et al. 2004; Chen and Charles-An 2006). A study examining ABA-induced gene expression in A. thaliana plants exposed to drought stress, determined that remorin gene expression levels are also elevated (Bray 2002). Moreover, a genetic study concerning rice responses to exogenous ABA, concluded that a remorin homolog is up-regulated, implying the remorin is involved in the ABA signal transduction pathway (Lin et al. 2003). In the present study, the StREMa4 expression level was regulated by SA, MeJA, and ABA, indicating that StREMa4 is part of a complex regulatory network affecting plant host interactions with pathogens. In-depth investigations of the molecular mechanisms regulating StREMa4 expression are required. These future studies will help characterize the early interactions between potato plants and R. solanacearum, and may identify new targets relevant for the control of bacterial wilt.
We constructed an R. solanacearum-induced potato stem tissue-specific SSH cDNA library and designed a 3′-end-specific primer for 384 up-regulated expressed sequence tags. The full-length StREMa4 cDNA sequence was then cloned from the SSH cDNA library using the 3′-RACE technique. StREMa4 consists of 591 bp, and encodes a mature protein with 197 amino acids. Its genetic structure shares similarities with remorins. Additionally, this gene is phylogenetically related to other Solanaceae remorins. The first StREMa4 expression peak is induced by the early interactions between the host potato plant and R. solanacearum, while a second expression peak occurs 72 h after infection, resulting in a rare bimodal expression pattern. The first expression peak likely stimulates the second expression peak, but the specific mechanism regulating this stimulation requires further study. Treatments with SA, MeJA, and ABA influence StREMa4 expression, which is most responsive to ABA. Furthermore, StREMa4 expression is tissue-specific, and mainly occurs in the phloem of stems and leaves, but not in the roots. Future investigations on StREMa4 activities and the interacting genes and proteins will greatly expand our understanding of the molecular mechanisms regulating potato growth and development.
References
Anderson, J. P., Badruzsaufari, E., Schenk, P. M., Manners, J. M., Desmond, O. J., Ehlert, C., et al. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell, 16(12), 3460–3479. doi:10.1105/tpc.104.025833.
Bariola, P. A., Retelska, D., Stasiak, A., Kammerer, R. A., Fleming, A., Hijri, M., et al. (2004). Remorins form a novel family of coiled coil-forming oligomeric and filamentous proteins associated with apical, vascular and embryonic tissues in plants. Plant Molecular Biology, 55(4), 579–594. doi:10.1007/s11103-004-1520-4.
Bhat, R. A., Miklis, M., Schmelzer, E., Schulze-Lefert, P., & Panstruga, R. (2005). Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proceedings of the National Academy of Sciences of the United States of America, 102(8), 3135–3140. doi:10.1073/pnas.0500012102.
Bozkurt, T. O., Richardson, A., Dagdas, Y. F., Mongrand, S., Kamoun, S., & Raffaele, S. (2014). The plant membrane-associated REMORIN1.3 accumulates in discrete perihaustorial domains and enhances susceptibility to phytophthora infestans. Plant Physiology, 165(3), 1005–1018. doi:10.1104/pp.114.235804.
Bray, E. A. (2002). Abscisic acid regulation of gene expression during water-deficit stress in the era of the Arabidopsis genome. Plant, Cell and Environment, 25(2), 153–161.
Checker, V. G., & Khurana, P. (2013). Molecular and functional characterization of mulberry EST encoding remorin (MiREM) involved in abiotic stress. Plant Cell Reports, 32(11), 1729–1741. doi:10.1007/s00299-013-1483-5.
Chen, K., & Charles-An, Y.-Q. (2006). Transcriptional responses to gibberellin and abscisic acid in barley aleurone. Journal of Integrative Plant Biology, 48(5), 591–612.
Elorza, A., Leon, G., Gomez, I., Mouras, A., Holuigue, L., Araya, A., et al. (2004). Nuclear SDH2-1 and SDH2-2 genes, encoding the iron-sulfur subunit of mitochondrial complex II in Arabidopsis, have distinct cell-specific expression patterns and promoter activities. Plant Physiology, 136(4), 4072–4087. doi:10.1104/pp.104.049528.
Farmer, E. E., Pearce, G., & Ryan, C. A. (1989). In vitro phosphorylation of plant plasma membrane proteins in response to the proteinase inhibitor inducing factor. Proceedings of the National Academy of Sciences of the United States of America, 86(5), 1539–1542.
Gao, G., Ren, C. H., Jin, L. P., Xie, K. Y., & Qu, D. Y. (2008). Cloning, expression and characterization of a non-specific lipid transfer protein gene from potato. Acta Agronomica Sinica, 34(9), 1510–1517. doi:10.3321/j.issn:0496-3490.2008.09.004.
Gao, G., Jin, L. P., Xie, K. Y., & Qu, D. Y. (2009). The potato StLTPa7 gene displays a complex Ca-associated pattern of expression during the early stage of potato-Ralstonia solanacearum interaction. Molecular Plant Pathology, 10(1), 15–27. doi:10.1111/j.1364-3703.2008.00508.x.
Gui, J., Liu, C., Shen, J., & Li, L. (2014). Grain setting defect1, encoding a remorin protein, affects the grain setting in rice through regulating plasmodesmatal conductance. Plant Physiology, 166(3), 1463–1478. doi:10.1104/pp.114.246769.
Hall, T. A. (1999). BioEdit. A user-friendly biological sequence alignment and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series, 41(41), 95–98.
He, L. Y., Sequeira, L., & Kelman, A. (1983). Characteristics of strains of pseudomonas solanacearum from China. Plant Disease, 67(12), 1357–1361. doi:10.1094/pd-67-1357.
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Research, 35(Web Server issue), W585–587. doi:10.1093/nar/gkm259.
Jacinto, T., Farmer, E. E., & Ryan, C. A. (1993). Purification of potato leaf plasma membrane protein pp 34, a protein phosphorylated in response to oligogalacturonide signals for defense and development. Plant Physiology, 103(4), 1393–1397.
Jain, M., Kaur, N., Tyagi, A. K., & Khurana, J. P. (2006). The auxin-responsive GH3 gene family in rice (Oryza sativa). Functional and Integrative Genomics, 6(1), 36–46. doi:10.1007/s10142-005-0142-5.
Jamann, T. M., Luo, X., Morales, L., Kolkman, J. M., Chung, C. L., & Nelson, R. J. (2016). A remorin gene is implicated in quantitative disease resistance in maize. Theoretical and Applied Genetics, 129(3), 591–602. doi:10.1007/s00122-015-2650-6.
Kreps, J. A., Wu, Y., Chang, H. S., Zhu, T., Wang, X., & Harper, J. F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiology, 130(4), 2129–2141. doi:10.1104/pp.008532.
Laloi, M., Perret, A. M., Chatre, L., Melser, S., Cantrel, C., Vaultier, M. N., et al. (2007). Insights into the role of specific lipids in the formation and delivery of lipid microdomains to the plasma membrane of plant cells. Plant Physiology, 143(1), 461–472. doi:10.1104/pp.106.091496.
Lefebvre, B., Furt, F., Hartmann, M. A., Michaelson, L. V., Carde, J. P., Sargueil-Boiron, F., et al. (2007). Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiology, 144(1), 402–418. doi:10.1104/pp.106.094102.
Lefebvre, B., Timmers, T., Mbengue, M., Moreau, S., Herve, C., Toth, K., et al. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proceedings of the National Academy of Sciences of the United States of America, 107(5), 2343–2348. doi:10.1073/pnas.0913320107.
Li, S., Su, X., Zhang, B., Huang, Q., Hu, Z., & Lu, M. (2013). Molecular cloning and functional analysis of the Populus deltoides remorin gene PdREM. Tree Physiology, 33(10), 1111–1121. doi:10.1093/treephys/tpt072.
Li, H., Xue, Z., Yanping, Z., & Gao, G. (2014). Cloning and expression analysis of the protease inhibitor StPIa 3 gene from potato. International Journal of Plant Research, 27(2), 199.
Lin, F., Xu, S. L., Ni, W. M., Chu, Z. Q., Xu, Z. H., & Xue, H. W. (2003). Identification of ABA-responsive genes in rice shoots via cDNA macroar. Cell Research, 13(1), 59–68. doi:10.1038/sj.cr.7290151.
Lingle, S. E., & Dyer, J. M. (2001). Cloning and expression of sucrose synthase-1 cDNA from sugarcane. Journal of Plant Physiology, 158(1), 129–131. doi:10.1078/0176-1617-00266.
Marin, M., & Ott, T. (2012). Phosphorylation of intrinsically disordered regions in remorin proteins. Frontiers in Plant Science, 3, 86. doi:10.3389/fpls.2012.00086.
Mongrand, S., Morel, J., Laroche, J., Claverol, S., Carde, J. P., Hartmann, M. A., et al. (2004). Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. Journal of Biological Chemistry, 279(35), 36277–36286. doi:10.1074/jbc.M403440200.
Morel, J., Claverol, S., Mongrand, S., Furt, F., Fromentin, J., Bessoule, J. J., et al. (2006). Proteomics of plant detergent-resistant membranes. Molecular and Cellular Proteomics, 5(8), 1396–1411. doi:10.1074/mcp.M600044-MCP200.
Perraki, A., Binaghi, M., Mecchia, M. A., Gronnier, J., German-Retana, S., Mongrand, S., et al. (2014). StRemorin1.3 hampers potato virus X TGBp1 ability to increase plasmodesmata permeability, but does not interfere with its silencing suppressor activity. FEBS Letters, 588(9), 1699–1705. doi:10.1016/j.febslet.2014.03.014.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research, 29(9), e45.
Raffaele, S., Mongrand, S., Gamas, P., Niebel, A., & Ott, T. (2007). Genome-wide annotation of remorins, a plant-specific protein family: evolutionary and functional perspectives. Plant Physiology, 145(3), 593–600. doi:10.1104/pp.107.108639.
Raffaele, S., Bayer, E., Lafarge, D., Cluzet, S., German Retana, S., Boubekeur, T., et al. (2009a). Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell, 21(5), 1541–1555. doi:10.1105/tpc.108.064279.
Raffaele, S., Bayer, E., & Mongrand, S. (2009b). Upregulation of the plant protein remorin correlates with dehiscence and cell maturation: a link with the maturation of plasmodesmata? Plant Signaling & Behavior, 4(10), 915–919.
Rajendran, L., & Simons, K. (2005). Lipid rafts and membrane dynamics. Journal of Cell Science, 118(Pt 6), 1099–1102. doi:10.1242/jcs.01681.
Reymond, P., Kunz, B., Paul-Pletzer, K., Grimm, R., Eckerskorn, C., & Farmer, E. E. (1996). Cloning of a cDNA encoding a plasma membrane-associated, uronide binding phosphoprotein with physical properties similar to viral movement proteins. Plant Cell, 8(12), 2265–2276. doi:10.1105/tpc.8.12.2265.
Reymond, P., Weber, H., Damond, M., & Farmer, E. E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell, 12(5), 707–720.
Sawano, Y., Hatano, K., Miyakawa, T., Komagata, H., Miyauchi, Y., Yamazaki, H., et al. (2008). Proteinase inhibitor from ginkgo seeds is a member of the plant nonspecific lipid transfer protein gene family. Plant Physiology, 146(4), 1909–1919. doi:10.1104/pp.107.111500.
Son, S., Oh, C. J., & An, C. S. (2014). Arabidopsis thaliana remorins interact with SnRK1 and play a role in susceptibility to beet curly Top virus and beet severe curly top virus. Plant Pathology Journal, 30(3), 269–278. doi:10.5423/PPJ.OA.06.2014.0061.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5 : molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), 2731–2739.
Wu, C. Q., Hu, H. H., Zeng, Y., Liang, D. C., Xie, K. B., Zhang, J. W., et al. (2006). Identification of novel stress-responsive transcription factor genes in rice by cDNA array analysis. Journal of Integrative Plant Biology, 48(10), 1216–1224.
Yue, J., Li, C., Liu, Y., & Yu, J. (2014). A remorin gene SiREM6, the target gene of SiARDP, from foxtail millet (Setaria italica) promotes high salt tolerance in transgenic Arabidopsis. PLoS One, 9(6), e100772. doi:10.1371/journal.pone.0100772.
Acknowledgments
We would like to thank Bo Yu (Cell Laboratory, Shanxi normal university) for technical services; thank the institutes of Vegetables & Flowers and Plant Protection of the Chinese Academy of Agricultural Sciences for materials. This work was supported by the project of National Natural Science Foundation of China (31271774).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest was declared.
Rights and permissions
About this article
Cite this article
Kong, CY., Luo, Yp., Duan, TT. et al. Potato remorin gene StREMa4 cloning and its spatiotemporal expression pattern under Ralstonia solanacearum and plant hormones treatment. Phytoparasitica 44, 575–584 (2016). https://doi.org/10.1007/s12600-016-0536-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12600-016-0536-z