Abstract
Soil samples were collected from different rice fields of Singur, Hooghly, West Bengal, India. Spore forming bacteria were isolated from the soil samples and among them, two isolates (BUSNC25 and BUSNC26) were larvicidal against third, fourth and fifth instar larvae of rice leaf folder, Cnaphalocrocis medinalis. The phenotypic, biochemical characterization and 16S rDNA analysis of the two isolates were done. On the basis of phenotypic, biochemical and phylogenetic analysis, the selected bacterial isolates (BUSNC25 and BUSNC26) were identified as Bacillus thuringiensis. The antibiotic sensitivity tests of these two isolates against selected doses of some standard antibiotics were done. Against the 3rd, 4th and 5th instar larvae of C. medinalis, the LC50 values of BUSNC25 were 2.45 × 104, 1.325 × 104 and 2.35 × 104 cfu/ml and of BUSNC26 were 3.375 × 104, 1.9 × 104 and 3.325 × 104 cfu/ml, respectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Rice is the staple food of more than one-third population of the earth (Zhang et al. 2013) and is seriously attacked by different insect pests causing huge economical loss to many rice growing countries. Annually, worldwide pest accounted rice loss is more than 5 % of the total production. And in India, grain yield loss of rice due to insect pest is almost 21–51 % varying from area to area depending on the variation in the agro climatic condition (Kalode et al. 1995). There are more than 100 species of insects, known to attack rice crop and among them the rice leaf folder (RLF), Cnaphalocrocis medinalis (Guenee) is an important one. The rice leaffolder, considered as a sporadic insect pest of rice is widely distributed in rice growing areas in Asia and Oceania, Northeast Australia and Madagascar. The leaffolder was previously considered as a minor pest of rice until recently increasing in importance in areas where modern high yielding varieties were grown (Bautista et al. 1984). In India and almost all over the world, synthetic insecticides are still the primary way to control leaffolder menace. Indiscriminate use of synthetic pesticides has resulted in damage to the environment, pest resurgence, pest resistance to insecticides, and lethal effects on non target organisms. Biopesticides, either botanically derived insecticides or microbial pesticides are inherently less harmful than conventional pesticides. They are clearly and mostly target specific in contrast to broad spectrum conventional chemical pesticides and are often quickly biodegradable and thus ecologically acceptable (Weinzierl and Henn 1991; Senthil and Kalaivani 2005). It is evident that biopesticides can play an important role either as principal or as supplementary system in the control of agricultural pest (Ramarethinam 2002). The common trend in the past two decades toward using biopesticides for controlling agricultural pests has made Bacillus thuringiensis a worldwide desirable alternative of chemical pesticide (Gill et al. 1992). Singur is an important crop production area in Hooghly district of West Bengal. The aim of the present piece of work is to study on the microbial diversity and functionality of the agricultural soils and entomopathogens to understand soil health conditions and to select potent microbial pathogens to augment soil nutritional status or develop broad spectrum bio-control agents and to select most effective indigenous Bt against serious rice pests for the development of cost effective, environment friendly and safe control of the pests.
Materials and Methods
Soil Collection
The soil samples were collected from rice fields of six different village areas adjoining to Singur, Hooghly, West Bengal. [Anandanagar (22°51′27.58″, 88°16′04.94″E), Apurbapur (22°48′22.00″N, 88°14′08.52″E), Bajemelia (22°50′28.52″N, 88°12′53.27″E), Beraberi (22°51′50.15″N, 88°12′50.47″E), Gopalnagar (22°45′37.61″N, 87°51′27.73″E), Ratanpur (22°49′12.46″N, 88°13′58.31″E), Environmental temperature 27 ± 1 °C]. The top most soil (1 cm) was scrapped off and then about 100 g soils (pH 6.9–7.1) from each area were collected in sterile polythene bags sealed with rubber bands.
Isolation of Bacteria
A portion of soil was blotted to optimum dryness within the sterile filter papers. One g soil was suspended in 9 ml sterile water and diluted serially up to 10−3 level. A 100 μl portion was mixed with 100 ml nutrient agar (NA) (g/l: peptone 5, beef extract 3, agar 2, pH 7), distributed in five plates and incubated at 30 ± 0.1 °C in the BOD incubator for 24 h. The colonies were checked under a phase-contrast microscope and those having spores were purified on NA plates and the pure cultures were maintained at 4 ± 0.1 °C on NA slants.
Characterization of Bacterial Isolate
Morphological, physiological and biochemical characters of the bacteria were studied following standard methods (Collee and Miles 1989; Smibert and Krieg 1995; Lacey 1997; Logan and de Vos 2009). Antibiotic sensitivity was tested using different antibiotic discs viz. kanamycin (30 µg/disc), nalidixic acid (30 µg/disc), rifampicin (5 μg/disc), doxycycline (30 µg/disc), gatifloxacin (10 µg/disc), vancomycin (30 µg/disc), gentamycin (10 µg/disc), ampicillin (10 µg/disc), ofloxacin (5 μg/disc), levofloxacin (5 μg/disc), streptomycin (10 µg/disc) following Brown (2004). The bacterium was phenotype according to Logan and de Vos (2009).
Scanning Electron Microscopy of Bacterial Isolate
Bacteria were grown for 3 days on NA plates, smears were prepared on cover glasses, heat fixed over a flame for 1–2 s followed by 2.5 % glutaraldehyde (aqueous) for 45 min. The slides were then dehydrated passing through 50, 70, 90 and 100 % alcohol for 5 min each. The specimens were gold coated and observed under a SEM (Hitachi-S-530).
Extraction and Electrophoresis of Genomic DNA
Genomic DNA from the bacteria was isolated following standard method (Wilson 2001). The organisms were grown for 6–8 h at 30 ± 0.1 °C in 2 ml of nutrient broth (NB). The cultures were centrifuged at 10,000 rpm for 10 min at 4 ± 0.1 °C; pellet was washed with 8.5 % (w/v) NaCl solution followed by sterile water, the supernatant was discarded and the pellet was suspended in 576 μl TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) by repeated pipetting. To it, 30 μl of 10 % SDS and 3 μl of proteinase K (20 mg/ml in 0.5 % SDS) were mixed and incubated for 1 h at 37 ± 0.1 °C. To the reaction mixture, 100 μl of 5 M NaCl and 80 μl of CTAB/NaCl solution (10 % CTAB in 0.7 M NaCl) were mixed sequentially, incubated for 10 min at 65 ± 0.1 °C. To the reaction mixture 0.7–0.8 ml chloroform-isoamyl alcohol (24:1 v/v) was mixed, centrifuged at 5000 rpm for 4–5 min at 4 ± 0.1 °C. The aqueous and viscous upper phase was removed in a fresh microcentrifuge tube, equal volume of phenol–chloroform-isoamyl alcohol (25:24:1, v/v/v) was added, mixed by gently inverting the tubes and spun for 5 min at 4 ± 0.1 °C and 7000 rpm. The supernatant was taken in a fresh tube, added 0.6 volume of isopropyl alcohol, inverted back and forth until a stringy white precipitate of nucleic acid was clearly visible. The precipitate was pelleted by spinning for 20–30 s at 7000 rpm at 4 ± 0.1 °C. The DNA pool was washed with 70 % ethanol to remove residual CTAB, spun for 5 min at 7000 rpm at 30 ± 0.1 °C. The supernatant was removed and the pellet was dried in a lyophilizer. The DNA samples were revived in sterile water and electrophoresed through 0.8 % agarose gel at a constant 5 V for 30 min and subsequently 5 V/cm for the required time according to the gel size. The DNA profile was visualized under a UV transilluminator (312 nm), documented and analyzed using Photocapt software Photocapt software (PhotoCaptMW (G) Serial no. 1149).
Amplification and Sequencing of 16SrRNA Gene and Phylogenetic Analysis
The ~1.5 kbp rDNA fragment was amplified using high-fidelity PCR polymerase. The PCR product was sequenced bi-directionally through a genetic analyzer using the forward primer (5′-AGAGTRTGATCMTYGCTWAC-3′) and reverse primer (5′-CGYTAMCTTWTTACGRCT-3′). The sequence data were aligned using the ClustalW submission form (http://www.ebi.ac.uk/clustalw) and analyzed by ClustalW software (Thompson et al. 1994). Evolutionary distances were calculated using the method of Jukes and Cantor (1969) and the topology was inferred using the neighbor-joining method (Saitou and Nei 1987). Phylogenetic trees were constructed following Tamura et al. (2007).
Cut Leaf Assay
The bacteria (BUSNC25 and BUSNC26) were grown separately in 100 ml sterilized NB (NA without agar) on a rotary shaker (100 ± 1 rpm, 30 ± 0.1 °C, 12 h) in 500 ml flasks, pelleted at 8000 g (10 min, 4 ± 0.1 °C) and washed three times with sterilized distilled water. Each pellet was suspended in sterilized distilled water and colony forming units (cfu) were assessed by bulk plating (five plates) 2 µl suspensions mixed with 100 ml NA. The suspensions were adjusted to 105 bacteria/ml water, diluted logarithmically and assayed for virulence. The bacterial suspensions and sterilized distilled water (control) were simultaneously used for bioassay.
Rice leaf pieces (10 cm each) were soaked for 10 min in water containing 0.001 % Tween 80, washed three times with sterilized distilled water and then surface sterilized for 5 min with 1.6 % sodium hypochlorite (NaOCl). The leaf pieces were further washed three times with sterilized water and blotted dry on sterilized blotting paper. Ten leaf pieces were soaked separately in each bacterial (spore crystal forming and asporogenous) suspension (of different concentrations; 102, 103, 104, 105, 106 cfu/ml) and sterile distilled water (control) for 15 min. Excess inoculum was drained off and the leaf pieces were placed on sterile moist filter paper in a sterile (150 × 20 mm diameter) test tube covered with an insect proof nylon cloth. Healthy larvae (reared in the net house) were surface sterilized with 1.6 % NaOCl for 5 min, washed three times with sterile water and 10 larvae of C. medinalis were placed into the tubes. The experiments were repeated three times. All these operations were performed under a laminar flow hood. The tubes were maintained in a controlled room at 30 ± 0.1 °C, 85 ± 3 % RH, and 12 h alternating light and dark phases. Fresh leaf pieces (10 cm) were surface sterilized with 1.6 % NaOCl for 5 min, washed 3 times with sterile distilled water and replaced (without bacteria) the original leaves within the tubes at 48 h intervals.
The potted rice seedlings with 4–5 leaves were washed thoroughly with sterile distilled water containing 0.001 % Tween 80 followed by sterile distilled water for 15 min to remove the Tween 80 and then air dried. The bacterial suspensions (of different concentrations) and sterile distilled water (control) were sprayed separately on the plants (3 replications) until the inoculum droplets were formed. On each plant, ten larvae of 3rd, 4th and 5th instar (reared in the net house) were released and maintained in the controlled room.
Results
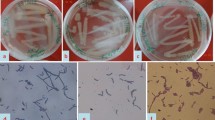
These two bacteria were identified and characterized by following morphological, biochemical and molecular attributes. Both of them showed circular, elevated and gummy colony. But they showed difference in colour and margins of colonies. BUSNC25 showed light brown coloured colonies with entire margin. Whereas BUSNC26 had white metallic colonies with erose type of margin (Table 1). The individual bacteria were observed under a compound, as well as, scanning electron microscope. Vegetative bodies of both of the bacteria were rod shaped and motile. Both were Gram positive, spore and crystal forming. The crystals of BUSNC25 were spherical with 1 µm (mean) diameter. The crystals of BUSNC26 were polymorphic/bipyramidal in shape (Figs. 1, 2). BUSNC25 was able to tolerate and grow at nutrient agar added with 15 % NaCl and BUSNC26 with 10 % NaCl (Table 1). Both of them were able to tolerate 60 °C temperature. They were able to utilize glucose, fructose, and sucrose as carbon sources but not mannose. BUSNC25 was able to produce extracellular enzymes to digest gelatin, cholesterol, Tween 80, but not casein. BUSNC26 was able to digest gelatin and casein both. But it was unable to hydrolyse fat. Both of them were able to produce amylase enzyme to digest carbohydrate (Table 1). Both isolates were able to produce catalase and showed positive reactions to methyl red test, Voges-Proskauer test and nitrate test. BUSNC26 showed positive reactions to urease test and oxidase test whereas BUSNC25 showed negative results to these tests. Both the isolates showed negative results to indole production test and citrate utilization test (Table 1). Antibiotic sensitivity test showed that BUSNC25 was resistant to the recommended doses of the following antibiotics viz., penicillin G (10 U), ampicillin (10 mg), nystatin (100 U), nalidixic acid (30 mcg), doxycycline hydrochloride (30 mcg). BUSNC26 was resistant to penicillin G (10 U), ampicillin (10 mg), nystatin (100 U), amoxycillin (10 μg), trimethoprim (30 mg), triple sulphas (300 mg) and sensitive to vancomycin, polymyxin B (300 U), norfloxacin (10 mg), bacitracin (10 U), erythromycin (15 mg), gentamycin (10 μg), tetracycline (30 μg), amoxycillin (10 μg), chlortetracycline (30 μg), kanamycin (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), rifampicin (5 μg), streptomycin (10 μg), levofloxacin (5 mcg), gatifloxacin (5 mcg), ofloxacin (5 mcg). BUSNC26 was sensitive to vancomycin (30 mg/disc), polymyxin B (300 U), norfloxacin (10 mg/disc), bacitracin (10 U), erythromycin (15 mg), gentamycin (10 μg), tetracycline (30 μg), chlortetracycline (30 μg), kanamycin (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), rifampicin (5 μg), streptomycin (10 μg), levofloxacin (5 mcg), gatifloxacin (5 mcg), ofloxacin (5 mcg), nalidixic acid (30 mcg), doxycycline hydrochloride (30 mcg) (Table 1). The phylogenetic study showed that BUSNC25 and BUSNC26 branched with the cluster containing different species of Bacillus (Fig. 3). On the basis of the morphophysiological characters, the two isolates were identified as B. thuringiensis. The LC50 of these two bacterial isolates against the RLF was determined. Against the 3rd, 4th and 5th instar larvae of C. medinalis, the LC50 values of BUSNC25 were 2.45 × 104, 1.325 × 104 and 2.35 × 104 cfu/ml and of BUSNC26 were 3.375 × 104 cfu/ml, 1.9 × 104 cfu/ml and 3.325 × 104 cfu/ml respectively (Table 2).
Discussion
The lethal concentration of the bacterial isolates varied with the stages of larval instar of the RLF which matched with the previous work of Tabashnik and Carrie`re (2004), where they showed that the lethal dose of Bt is instar dependent and the susceptibility of mature larvae is very low. The Bt-exposure to C. medinalis larvae in the laboratory reduced digestive enzyme activity (Senthil 2000) which was related to the physiological condition and also had effect on the absorption, digestion, and positive transport of nutrients in the midgut of the rice leaffolder. The application of Bt on larval instar of C. medinalis caused damages in the midgut epithelial cells through its parasporal bodies that release the active toxin after digestion by serine proteases under the alkaline conditions in the intestinal fluid which ultimately decreased the digestive enzyme activities (Eguchi et al. 1972; Mathavan et al. 1989; Smirle et al. 1996). Furthermore Bt crystals causes intestinal paralysis and perforation of the gut wall which leads to the imbalance of the ions and osmotic balance of gastrointestinal cells and get the insect killed (Zhang et al. 2013). The use of this Bt crystals against the leaf folder in the field is safe as they do not have any effect on non target organisms like mammals, birds, fishes and other insects. BT proteins can be dissolved completely in a few seconds under the effect of acidic gastric juice after it reaches into the stomach of mammals. It can be dissolved easily and has little environment impact on the soil.
Conclusion
From our study we can conclude that the Bt isolates BUSNC25 and BUSNC26 have significant effects on the larvae of RLF C. medinalis. These two isolates can be used as potential biocides against the RLF as an alternative of chemical insecticides which would be beneficial in terms of economy as well as environmental safety.
References
Bautista, R.C., E.A. Heinrichs, and R.S. Rejesus. 1984. Economic injury levels for the rice leaffolder Cnaphalocrocis medinalis (Lepidoptera: Pyralidae): Insect infestation and artificial leaf removal. Environmental Entomology 13(2): 439–443.
Brown, A. 2004. Benson’s microbiological applications: Laboratory manual in general microbiology. New York: McGraw-Hill.
Collee, J.G., and P.S. Miles. 1989. Tests for identification of bacteria. In Practical medical microbiology, 13th ed, ed. J.G. Collee, J.P. Duguid, A.G. Fraser, and B.P. Marmion. New York: Churchil Livingstone.
Eguchi, M., M. Sawaki, and Y. Suzuki. 1972. Multiple forms of midgut alkaline phosphatase in the silkworm: New band formation and the relationship between the midgut and digestive fluid. Insect Biochemistry 2: 297–304.
Gill, S.S., E.A. Cowles, and P.V. Pietrantonio. 1992. The mode of action of Bacillus thuringiensis endotoxins. Annual Review of Entomology 37: 615–635.
Jukes, T.H., and C.R. Cantor. 1969. Evolution of protein molecules. In Mammalian protein metabolism, ed. H.N. Munro. New York: Academic Press.
Kalode, M.B., Pasalu, I.C., Krishnaiah, N.V., and J.S. Bentur, 1995. Changing insect pest complex in relation to cropping system of rice. In Proceedings of th national seminar on changing pest situation in the current agriculture scenario of India. New Delhi: ICAR.
Lacey, L.A. 1997. Manual of techniques in insect pathology. New York: Academic Press.
Logan, N.S., and P. de Vos. 2009. Genus I. Bacillus. In Bergey’s manual of systematic bacteriology, vol. 3, 2nd ed, ed. P. de Vos, G.M. Garrity, D. Jones, N.R. Krieg, W. Ludwing, F.A. Rainey, K.H. Schleifer, and W.B. Whitman. New York: Springer.
Mathavan, S., P.M. Sudha, and S.M. Pechimuthu. 1989. Effect of Bacillus thuringiensis israelensison the midgut cells of Bombyx morilarvae: A histopathological and histochemical study. Journal of Invertebrate Pathology 53: 217–227.
Ramarethinam, S. 2002. Biopesticides—An overview. Resource management in plant protection, vol. I, 167–179. NPPTI Campus, Rajendranagar, Hyderabad: Plant Protection Association of India.
Saitou, N., and M. Nei. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425.
Senthil, N.S., and K. Kalaivani. 2005. Efficacy of nucleopolyhedrovirus (NPV) and azadirachtin on Spodoptera litura Fabricius (Lepidoptera: Noctuidae). Biological Control 34: 93–98.
Senthil, N.S. 2000. Studies on the synergistic effect of Bacillus thuringiensis (Berliner) Sub. Sp. Kurstaki, Azadirachta indicaand Vitex negundoon the feeding, growth, reproduction and biochemical changes of Cnaphalocrocis medinalis(Guene´e) (Rice leaffolder) (Insecta: Lepidoptera: Pyralidae). PhD thesis, Bharathiar University, Coimbatore, Tamilnadu, India, 1–90.
Smibert, R, and N.R. Krieg.1995. Phenotypic characterization. In Methods for general and molecular bacteriology, 607–654. Washington: American Society of Microbiology.
Smirle, M.J., D.T. Lowery, and C.L. Zurowski. 1996. Influence of neem oil on detoxication enzyme activity in the oblique banded leafroller, Choristoneura rosaceana. Pest Biochemistry and Physiology 56: 220–230.
Tabashnik, B.E., and Y. Carrie`re. 2004. Bt transgenic crops do not have favorable effects on resistant insects. Journal of Insect Science 4: 1–4.
Tamura, K., Dudley, J., Nei, M., and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution.
Thompson, J.D., D.G. Higgins, and T.J. Gibson. 1994. ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Residence 22: 4673–4680.
Weinzierl, R, and T. Henn. 1991. Alternatives in insect management: Biological and biorational approaches. North Central Regional Extension Publication 401. Cooperative Extension Service, University of Illinois at Urbana-Champaign.
Wilson, K. 2001. Preparation of genomic DNA from bacteria. Current protocols in molecular biology. Hoboken: Wiley. doi:10.1002/0471142727.mb0204s56.
Zhang, Q., L. Cong, L. Shao-kui, L. Dong, Q. Qing-ming, and L. Chuan-gen. 2013. Breeding and identification of insect-resistant rice by transferring two insecticidal genes, sbk and sck. Rice Science 20(1): 19–24.
Acknowledgements
The authors are grateful to the Department of Agriculture (RKVY), Govt. of West Bengal for their kind financial assistance, and The University of Burdwan for providing laboratory facilities required for the present study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Rights and permissions
About this article
Cite this article
Chatterjee, S., Azmi, S.A., Ghosh, T.S. et al. Characterization of the Bacillus thuringiensis Isolates Virulent Against Rice Leaf Folder, Cnaphalocrocis medinalis (Guenee). Proc Zool Soc 70, 74–80 (2017). https://doi.org/10.1007/s12595-015-0161-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12595-015-0161-8