Abstract
Antimicrobial peptides/proteins (AMPs), the important host-defence molecules with antimicrobial and immunomodulatory properties, effectively ward off pathogenic organisms. The responses of such molecules of Atlantic salmon during Aeromonas salmonicida infection that causes losses to salmon farms are not clearly described. Two trials were carried out to understand the regulation of two known AMP genes (cathelicidins, cath1, 2) and five newly cloned AMP genes (beta-defensins, defb1-4 and l-amino acid oxidase, lao); in vivo responses following intraperitoneal injection of A. salmonicida vaccine and in vitro responses upon exposure of gill tissues to A. salmonicida. The vaccine injection induced cath1, cath2 and lao expression in the gills at 24 h post injection. The live A. salmonicida exposure of Atlantic salmon gill tissue explant revealed the upregulation of the cath1 and cath2 within 48 h. The defb genes were not altered by either injection or exposure to A. salmonicida. Based on the evidence we suggest that the cathelicidins (cath1 and cath2) are A. salmonicida-inducible AMPs and could be possible biomarkers of infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A robust innate immune system is essential for fishes because their adaptive counterpart takes considerable time to respond. The innate immune system of fishes compensates for the lack of a sophisticated adaptive immune system (Ellis 2001). In addition, the immune tissues of fishes and mammals show some similarities, but the lymphoid organs are less complex compared to mammals (Tort et al. 2003). Notably, the fish body surface tissues such as the skin and gills are tightly protected because of their susceptibility to pathogen invasion and related infection. The protection is provided by, among others, a variety of bioactive molecules such as complements, immunoglobulins, lectins, protease inhibitors, lysozyme, alkaline phosphatase and antimicrobial peptides (AMPs). The AMPs, which are participants of the first line of immune defence in fishes, are powerful, rapid killers of pathogens. Furthermore, their receptor-independent mechanisms such as direct killing and membrane disruption (Rakers et al. 2010, 2013) make them safe agents for keeping pathogens in check in aquatic systems.
AMPs, that are generally positively charged 2–6 kDa peptides with disulfide bond/s, can kill bacteria by interfering with the bacterial cell membranes (Shai 1999, 2002). The fish body surface AMPs are roughly classified into amphiphilic peptides and positively charged peptides. The amphiphilic nature and their positive charge are necessary for making pores in bacterial cell membranes and causing leakage of the cell contents. AMPs are present in the skin of fishes; examples are pardaxin in Moses sole Pardachirus marmoratus (Primor and Tu 1980; Shai et al. 1988), pleurocidin in winter flounder Pleuronectes americanus (Cole et al. 1997) and gramistins in goldenstriped soapfish Grammistes sexlineatus and spotted soapfish Pogonoperca punctata (Sugiyama et al. 2005; Shiomi et al. 2000). Positively charged AMPs include defensins (Zou et al. 2007; Ruangsri et al. 2013) and cathelicidins (Basañez et al. 2002; Chin-I Chang et al. 2006). The three mammalian defensin subgroups are alpha-, beta- and theta-defensins. In fishes, only beta-defensins-like proteins are found, and the precursors of these antibacterial peptides consist of 60–77 amino acid residues, and the mature peptide has 38–45 amino acids (Masso-Silva and Diamond 2014). The defensins of Atlantic salmon Salmo salar are not yet cloned from the body surface tissues such as skin and gills. The mature peptide contains six conserved cysteine residues and forms disulfide bridges to stabilize the structure, and their net positive charge may help to attach and penetrate the target bacterial cell body. The cathelicidins — also known as LL-37 in humans — are cationic 37-amino acid residues; they have disulfide bonds and a molecular weight of 4 kDa (Zanetti et al. 1995). In Atlantic salmon, two cathelicidins have in their precursors 202 or 207 amino acid residues and the mature peptides have 53 or 63 amino acid residues (Chang et al. 2006). In addition to the above-mentioned AMPs, the l-amino acid oxidase (LAO) that is categorized as an antibacterial protein in rockfish Sebastes schlegelii, great sculpin Myoxocephalus polyacanthocephalus, starry flounder Platichthys stellatus and rabbitfish Siganus oramin (Kitani et al. 2007; Nagashima et al. 2009; Kasai et al. 2010; Wang et al. 2011) is a glycosylated flavoenzyme with hydrogen peroxide-dependent antibacterial activity (Du and Clemetson 2002). Beta-defensins, cathelicidins and LAO possess antibacterial activity against Gram-negative and positive bacterial species (Ruangsri et al. 2013; Chang et al. 2006; Kitani et al. 2008; Kasai et al. 2015). Furthermore, AMPs are known to exhibit immunomodulatory functions and they are effective molecules of the humoral immune system (Romo et al. 2016). As in mammals, fish AMPs are secreted when there is a wound or upon pathogen contact (Rakers et al. 2013). However, the cause of induction and the mode of action of these microbicidal molecules, especially their innate responses and immunomodulatory functions in farmed fishes, are not fully elucidated. Understanding the regulation of different AMPs when Atlantic salmon comes in contact with a particular pathogen will help identify molecules that respond to that pathogen. In addition, the body surface epidermal cells or skin mucus can be collected easily for diagnostics. The AMPs in these samples can be used as non-invasive diagnostic markers and could be helpful in disease prevention, thereby hindering the spread of infectious disease in the salmon aquaculture industry.
In this study, we newly cloned the beta-defensins 1–4 and l-amino acid oxidase genes, and then clarified the defence capacity of these AMPs as well as the already cloned cathelicidins (Chang et al. 2006) in Atlantic salmon, an economically important farmed fish species. We describe the alterations in the expressions of AMP genes in vivo upon stimulation with Aeromonas salmonicida vaccine antigen and in vitro after exposure to live A. salmonicida.
Materials and methods
Fish
Hatchery produced Atlantic salmon (AquaGen strain), procured as pre-smolts from Cermaq, Bodø, Norway and maintained on commercial feeds in the indoor rearing facilities of the Research Station, Nord University, Bodø, Norway were used for all the studies. The water temperature of the flow-through seawater system was 7–8 °C and the oxygen saturation was above 90%. The specimens were handled by authorized personnel and the procedures were in accordance with the guidelines of the Norwegian Animal Research Authority (FDU approval number: 5617).
Cloning of AMP genes, target gene selection, and primer designing
The beta-defensins 1–4 and l-amino acid oxidase genes were cloned for this work. Total RNA was purified from the skin tissues of adult Atlantic salmon using the spin column RNA purification system (EZNA Total RNA kit 1, Omega Biotek, Norcross, GA, USA), following the manufacturer’s protocol. The cDNA was synthesized using SuperScript III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) and oligo-dT adaptor primer (Online Resource, Table S1). To amplify the AMP gene fragments, the gene-specific primers were designed based on the Atlantic salmon genome sequence and comparing the known fish AMP gene sequences (Online Resource, Table S1). After cloning the Atlantic salmon AMP genes, quantitative PCR (qPCR) primers were designed around intron–exon boundaries using PerlPrimer Ver. 1.1.21 (Marshall 2004). The genes targeted for expression studies were cathelicidin 1 (cath1: AY728057), cathelicidin 2 (cath2: AY360357), beta-defensin 1 (defb1: LC387973), beta-defensin 2 (defb2: LC387974), beta-defensin 3 (defb3: LC387975), beta-defensin 4 (defb4: LC387976) and l-amino acid oxidase (lao: AB831259). The primer set for interleukin 1 beta gene (il1b: AY617117, Vasanth et al. 2015) was used to evaluate the inflammatory response of in vitro bacterial exposure experiment. The reference genes were ribosomal protein S29 (rps29: NM_001139600), ribosomal protein L13 (rpl13: BT058587), elongation factor alpha 1 (ef1a: BG933853), ubiquitin (ubi: AB036060) and hypoxanthine phosphoribosyltransferase 1 (hprt1: BT043501). Primers for the reference gene amplification (rps29, rpl13, hprt1) were obtained from other literature (Valente et al. 2012; Guiry et al. 2010; Løvoll et al. 2011). The sequence information of the primers is summarized in Online Resource, Table S1.
LAO activity determination
The LAO activities of Atlantic salmon skin and gills were determined. Briefly, the skin mucus was collected from adult fish and centrifuged at 18,000×g for 30 min at 4 °C. The gill tissues were also collected from adult fish and the extract was prepared as follows; 100 mg of tissue in 900 µl of 0.1 M phosphate buffer (pH 7.0) was disrupted with ceramic beads using a MagNA Lyzer (Roche, Basel, Switzerland) at 6000 rpm for 40 s. The homogenate was centrifuged (18,000×g for 30 min at 4 °C) as described above. The supernatant samples obtained were employed for determining the LAO activity. The amino acid substrate specificity of the skin mucus and gill tissue extract was examined using glycine and 19 different l-amino acids by an o-phenylenediamine/horseradish peroxidase method that is described in our previous report (Kitani et al. 2010).
Vaccine injection study
To understand the alterations of the AMP genes to vaccine components, an injection trial was carried out. Unvaccinated Atlantic salmon pre-smolts (average weight 38.8 g) were individually tagged prior to the start of the experiment (described later). They were maintained in 1-m3 tanks that were part of a freshwater flow-through system. The rearing temperature was 11–12 °C and oxygen saturation was maintained above 90%. The fish were anaesthetized with 80 mg/l of MS 222 (tricaine methanesulphonate, Argent Chemical Laboratories, Redmond, USA) before tagging/injection as well as before sampling. Either vaccine or adjuvant (A. salmonicida vaccine and the adjuvant were supplied by PHARMAQ AS, Skøyen, Oslo, Norway) or phosphate buffered saline (PBS) was administered to 10 fish per treatment, by intraperitoneal injections. After the injection, samples were collected from 10 fish per treatment at 2 time points: (1) at 24 h post vaccination (24 hpv) and (2) at 7 weeks post vaccination (7 wpv; i.e., after approximately 500 day-degrees). The skin (dorsal part of fish) and gills (a portion of the first gill arch including gill filaments) were collected at each sampling point and fixed in RNAlater (Thermo Fisher Scientific). The fixed samples were stored at − 20 °C until use. RNA extraction, cDNA synthesis and qPCR analysis condition are described in the section of RNA extraction, cDNA synthesis and quantitative PCR.
In vitro pathogen exposure study on gills
Based on the results from the vaccination experiment, in vitro pathogen exposure was carried out as described below. Gill tissues were obtained from 12 fish (average weight 476 g) and the first gill arch was collected and transferred into the L15 medium (Sigma-Aldrich, MO, USA) with 500 unit/ml of penicillin (Sigma-Aldrich). The gill arches were split into pairs of gill filaments with the cartilage. The separated gill filaments were placed into the wells of a microplate (24 wells; Nunc, NY, USA) containing 2 ml of L15 medium (pH 7.8) with 10% fetal bovine serum (FBS, Sigma-Aldrich) and antibiotics (5 unit/ml of penicillin, 5 mg/ml of streptomycin and 2.5 µg/ml of amphotericin B, Sigma-Aldrich) and incubated for 16 h at 8 °C. After the primary incubation, the medium was discarded, and the tissues were washed twice with 2 ml of PBS (pH 7.8) by pipetting. The washed tissues were exposed to the suspension of the bacterial pathogen A. salmonicida (107 CFU/ml) in the same PBS at 8 °C for 2 h. A control PBS group (unexposed group) was also included in this study. Later, the bacteria-exposed gills as well as unexposed gills were washed and replaced into the fresh L15 with 10% FBS without antibiotics. The tissues were collected at 12, 24, 48 and 96 h after exposure. All samples obtained were fixed in RNAlater and stored at − 20 °C until used for examining the expression of the target genes. RNA extraction, cDNA synthesis and qPCR analysis conditions are described in the next section.
RNA extraction, cDNA synthesis and quantitative PCR
Total RNA from 10 mg of RNAlater-fixed fish skin or gills was extracted using an EZ96 Total RNA kit 1 (Omega Biotek) with vacuum manifold, following the manufacturer’s protocols. Briefly, the tissue samples were disrupted with 400 µl of lysis buffer and three 2.8-mm diameter zirconium oxide beads in a 2.0-ml screw-cap tube using a MagNA Lyzer (3000 rpm, 180 s, Roche). This homogenate was centrifuged (18,000×g, 20 min, 20 °C) and the supernatant was mixed with an equal volume of 70% ethanol. The mixture was applied to the RNA binding plate (Omega Biotek). After the washing step, the total RNA was eluted with 100 µl of RNase free water, after which it was centrifuged using a microplate centrifuge (3000×g, 15 min, 4 °C). Total RNA (500 ng per reaction) was reverse-transcribed using a Quantitect RT cDNA synthesis kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The gene expression profiling of the AMP genes was carried out using the qPCR instrument (LightCycler 96, Roche) and FastStart Universal SYBR Green Master (Roche) with each gene-specific primer set having a 0.2 µM working concentration (Online Resource, Table S1). No-template and no-reverse transcriptase controls were included for each primer pair. The thermal cycling profile for qPCR was 95 °C for 1 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. The specificity of PCR amplification was determined by the agarose gel electrophoresis, the melting curve analysis that runs at a temperature range of 60–90 °C with a ramp speed of 0.2 °C/s, and further confirmed by Sanger sequencing.
Sequence data analysis, target gene expression and statistical assessment
In silico analyses of the molecular characteristics were done using ExPASy Compute pI/Mw (http://web.expasy.org/compute_pi/), SOSUI signal (http://harrier.nagahama-i-bio.ac.jp/sosui/sosuisignal/sosuisignal_submit.html), NCBI Conserved Domain search tool (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/) and Clustal Omega Multiple Sequence Alignment tool (https://www.ebi.ac.uk/Tools/msa/clustalo/). Phylogenetic analysis was performed to understand the phylogenetic relationship of the cloned AMPs of Atlantic salmon. Phylogenetic trees of known defensins and LAOs were generated by Bayesian inference using a mixed model of amino acid substitution in MrBayes version 3.2.6 (Ronquist et al. 2012) for 1.0 × 106 or 2.5 × 105 generations and burning the first 2500 or 650 trees, respectively.
The expression levels of AMP genes were estimated by the relative standard curve method and normalized against the geometric mean determined by GeNorm (Vandesompele et al. 2002). The normalization factor was calculated based on the two most stable reference genes judged by the GeNorm algorithm. For the injection study, the expression from the vaccine/adjuvant-injected fish was divided by the average value of the PBS-injected group. For the in vitro pathogen exposure study on gills, alteration of the gene expression of interleukin-1 beta, beta-defensins, cathelicidins and LAO were evaluated; the results are presented as the ratio of the exposure group to PBS group (Fig. 4a). Statistical significance of the differences in these data was evaluated using 2-way ANOVA followed by Sidak’s multiple comparison test in the case of parametric data, except for the non-parametric data for which one-way ANOVA was employed, followed by Dunn’s test. In the case of the pathogen exposure study, we detected the significant differences by performing one-way ANOVA followed by Tukey’s HSD test. Values of P < 0.05 are considered to be significant.
Results
Cloning of beta-defensin genes
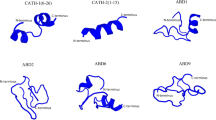
Four beta-defensin genes were successfully cloned from Atlantic salmon skin (the DDBJ accession numbers are LC387973, LC387974, LC387975 and LC387976 for beta-defensin 1, 2, 3 and 4, respectively). All the Atlantic salmon beta-defensins consisted of a signal peptide, a mature peptide domain and conserved cysteine residues (Online Resource, Fig. S1). The estimated mature peptide length (aa, amino acid residues), molecular weight (Mw) and theoretical isoelectric point (pI) of the defensins were: beta-defensin 1, Defb1-42 aa, 4.5 kDa and 8.35; beta-defensin 2, Defb2-43 aa, 5.1 kDa and 9.08; beta-defensin 3, Defb3-39 aa, 4.1 kDa and 8.95 and beta-defensin 4, Defb4-42 aa, 4.5 kDa and 7.79, respectively. Amino acid sequence identities of the mature beta-defensins are given in Table 1; the peptide identities ranged between 28.6 and 50.0%. Phylogenetic analysis of the known defensin precursors (Fig. 1) indicated that all of the Atlantic salmon beta-defensins are related to other vertebrate defensins. Atlantic salmon beta-defensins 1, 2 and 4 clustered together with the respective fish beta-defensins, while beta-defensin 3 was observed in an independent clade.
Phylogenetic tree of fish defensins. The phylogenetic tree was constructed by Bayesian inference with the following invertebrate and vertebrate defensin protein sequences: Atlantic salmon (Defb1; BBE43043.1, Defb2; BBE43044.1, Defb3; BBE43045.1, Defb4; BBE43046.1), rainbow trout (Defb1; CBB12546.1, Defb2; CBB12547.1, Defb3; CBB12548.1, Defb4; CBB12549.1), Atlantic cod (Defb; AEB69787.1), channel catfish (Defb1-like; NP_001352159.1), Chinese perch (Defb; AEO45759.1), Chinese loach (Defb2; AGH10110.1), striped beakfish (AJA33388.1), Wuchang bream (Def2; AGX24925.1), grass carp (Defb1; AOG20790.1), zebrafish (Def1-like; CAJ57442.1, Def2-like; CAJ57443.1, Def3-like; CAJ57444.1), tiger pufferfish (Def1-like; CAJ57646.1), orange-spotted grouper (Defb; AET25528.1), Japanese flounder (Defb1-1; ADA84138.1, Defb1-2; ADA84139.1, Defb1-3; ADA84140.1, Defb1-4; ADA84141.1, Defb1-5; ADA84142.1), bovine tracheal antimicrobial peptide (TAP; AAD01521.1), human alpha-defensin 1 (DEFA1; NM_004084) and human beta-defensins (DEFB1; NM_005218, DEFB4; NM_004942). Scarab beetle Anomala cuprea defensin A (DefA; AB176885) was used as an outgroup. Bayesian posterior probability values are indicated on the tree nodes
Cloning of LAO gene and LAO enzyme activities
The LAO gene of Atlantic salmon was also cloned from skin tissue (the DDBJ accession number is AB831259). The predicted LAO consisted of 509 amino acids including a signal peptide (1–26 residues), co-enzyme binding motifs (dinucleotide binding motif and GG motif) and four N-glycosylation sites (Asn-X-Ser or Thr, X is any amino acid except proline) on amino acid positions 218, 276, 294 and 310 (Online Resource, Fig. S2). The mature protein has an estimated molecular weight of 53.9 kDa and an isoelectric point of 6.16. The identity of predicted Atlantic salmon LAO protein sequence to other animal LAO sequences is summarized in Table 2. Phylogenetic analysis revealed that the Atlantic salmon LAO clustered with zebrafish, medaka and catfish LAOs (Fig. 2).
Phylogenetic tree of animal LAOs. The phylogenetic tree was constructed by Bayesian inference with the following LAO protein sequences: Atlantic salmon (AB831259), Atlantic cod (AB828203), great sculpin (AB299273.1), rockfish (AB218876.1), yellow perch (HQ206474.1), chub mackerel (AJ400871.1), starry flounder (AB495360.1, AB495361.1, AB495362.1), tiger pufferfish (XM_003978704.1), white-spotted rabbitfish (HQ540313.1), medaka (XM_004075791.1, XM_004079594.1), zebrafish (XM_003200032), catfish (FD292921), mouse milk LAO (AB034801), mouse B cell LAO (Fig1 protein; U70429) and human interleukin 4 induced 1 (IL4I1; BC131625). Sea hare LAO (aplysianin A, NM_001204595) was used as an outgroup. Bayesian posterior probability values are indicated on the tree nodes
Substrate-specific LAO enzyme activity was not detected in the Atlantic salmon skin mucus and gill tissue extracts (data not shown).
Alterations of the AMP genes in skin and gills following vaccine injection
The response of the seven AMP genes upon injection with Aeromonas antigen was studied in the skin and gills. The expression of the target genes, namely cath1, cath2, defb1, defb4 and lao were detected and quantified by qPCR.
In the skin, cath2 expression tended upwards at 1 day after vaccine injection, while the expression of cath1, defb1, defb4 and lao were not altered (Fig. 3a). At 7 weeks after vaccine injection, cath2 (P < 0.05) and lao (P > 0.05) expressions were 0.3- or 0.5-fold lower compared to the PBS-injected control group (Fig. 3a), while the other genes were not altered.
Alteration of the AMP genes by the vaccination. Gene expression in a skin, b gills. Y-axes show the gene expression values of AMP normalized by the normalization factor calculated by the GeNorm algorithm and then converted to the ratio of the exposure group to the PBS group. Empty bars; PBS injection control group, light grey bars; adjuvant injection control group, dark grey bars; vaccine injection group. The results are expressed as mean value ± standard error (n = 8–10). Different letters indicate significant difference at P < 0.05
In the gills, cath1, cath2 and lao were significantly upregulated at 1 day after vaccine injection (Fig. 3b). The expression of cath1, cath2 and lao were, respectively, 4.5, 25 and 2.5 times significantly higher compared to both PBS and adjuvant injection groups (P < 0.05). However, such alterations in the genes were not observed at 7 weeks. At 1 day and 7 weeks, there was a slight upregulation of defb1 following the adjuvant injection, but not with the antigenic stimulation.
The defensins defb2 and defb3 were not detected in this experiment (data not shown).
Alterations in the AMP genes in the gills upon in vitro exposure to bacteria
The differences in the expression of AMP genes in gill tissues following an in vitro exposure to pathogenic bacteria A. salmonicida were examined (Fig. 4). The il1b expression in the gill tissue increased corresponding to the tissue incubation time; by 96 h it was significantly different (4 times; P < 0.05) from the expression at the initial time point. The cathelicidins, cath1 and cath2 were also significantly upregulated (Fig. 4c and d); at 96 h cath1 expression was 1.6 times higher and at 48 h cath2 expression was 1.9 times higher compared to the expression at the initial point. A tendency for upregulation was noted in the case of defb1 expression (2 times higher at 96 h; Fig. 4e), as well as for the lao expression (slightly upregulated at 48 h; Fig. 4f). However, defb2, defb3 and defb4 were not detected in this experiment (data not shown).
Aeromonas salmonicida exposure of gill tissue explant. a Schematic diagram of the experimental design, b−f alteration of the il1b and AMP genes (c; cath1, d; cath2, e; defb1 and f; lao). Y-axes show the gene expression ratio of the experimental group to the PBS control group. The results are expressed as mean value ± standard error (n = 9–12). Different letters indicate significant difference at P < 0.05
Discussion
Antimicrobial peptides and proteins are the front-line defence molecules against pathogen invasion, and they possess immunomodulatory functions (Katzenback 2015; Rakers et al. 2013). Cathelicidins and defensins are present in different tissues of fishes and mammals (Katzenback 2015; Lehrer 2004), and LAO is a new member of the antimicrobial molecules in fish species. They are known to ward off pathogens effectively. However, less is known about the defence potential of fish AMPs compared to those of mammals. As a first step to understanding the functions of these molecules in Atlantic salmon, we conducted A. salmonicida vaccination and exposure trials. We employed the already cloned cath1 and cath2 genes (Chang et al. 2006), and other AMP genes that we cloned for this work, viz defb1, defb2, defb3, defb4 and lao. We found that the cathelicidin genes in the gills were quickly upregulated by both injection and exposure to the pathogen. We did not find such significant expressional changes for defb1, defb2, defb3, defb4 or lao.
Cathelicidin genes were upregulated by the A. salmonicida vaccine injection but not by the mock or adjuvant injections. This result suggests that the genes respond to the A. salmonicida cell body or its surface molecules. Interestingly, the intraperitoneal injection was able to invoke the expression of cathelicidins even on external tissues such as the skin of Atlantic salmon. This remote response of both cath1 and cath2 suggests that their expression is under the immunoregulatory system of the fish. Cathelicidins are known to be induced by some chemical and biological agents. Human cathelicidin (hCAP18) was induced in various types of cells by the addition of vitamin D3; via the promoter sequence containing vitamin D response elements (Xhindoli et al. 2016). In the case of fish cathelicidins, lipopolysaccharide injection in ayu Plecoglossus altivelis larvae (Nsrelden et al. 2017) and zymosan exposure of rainbow trout Oncorhynchus mykiss intestine cells (Schmitt et al. 2015) stimulated cathelicidin gene expression. An in vitro study also reported the response of cathelicidin genes; Brucella pinnipedialis infection induced expression of this gene at 24 h in the Atlantic cod Gadus morhua head kidney derived monocyte/macrophage like-cells (Nymo et al. 2016).
Furthermore, cathelicidin was induced by the exposure of rainbow trout to Lactococcus garvieae, Yersinia ruckeri, A. salmonicida and Flavobacterium psychrophilum (Furlan et al. 2018). The above-mentioned pathogen challenge study using rainbow trout reported the induction of both cath1 and cath2 within 24 h in several tissues; the response of cath2 was strong in all experiments (Furlan et al. 2018). These results are similar to our observations: quick and drastic responses of cath2 expression after pathogen exposure.
The acute phase reaction molecules in fishes are triggered by the bacterial infection. The il1b is one of the interleukins that was characterized early and is produced by a wide range of fish cells after stimulation by pathogen-associated molecular patterns (Zou and Secombes 2016). Il1b protein drives (in 4 h) the expression of the acute phase protein, serum amyloid A, in head kidney leucocytes of salmon (Lee et al. 2017). On the other hand, the il1b gene expression was initiated in Moritella viscosa-infected Atlantic salmon only after 7 days (Løvoll et al. 2009). In our gill tissue explant cath2 was stimulated prior to il1b, suggesting that cath2 can be a potential early phase marker of infection.
Although we have not compared the expressions in different tissues, we would like to direct the reader’s focus to the responses in the examined tissues. At 24 h after injection, we did not find any alteration in the expression of skin cath1, but skin cath2 was slightly upregulated, though the level was lower compared to gills (Fig. 3a). Despite the fact that this change is not confirmed through statistical analysis, the gill lamella may be an efficient target for pathogens because of the large surface area for gas exchange (Koppang et al. 2010; Haugarvoll et al. 2008; Rombout et al. 2014). Both cath1 and cath2 were induced in the gills by the A. salmonicida exposure; we observed a time-dependent increase in expressions. This result suggests that local responses of cathelicidin genes could be evoked by pathogen exposure. Based on both the injection and exposure experiments, it is suggested that the cathelicidins are involved in both local and systemic immune responses. In an earlier study on Atlantic salmon, both cath1 and cath2 were induced in the gills following Yersinia ruckeri challenge (Bridle et al. 2011). This aligns with the results from the present experiment using explanted tissue. Furthermore, it was reported that this AMP gene was induced in the liver of ayu in response to lipopolysaccharide injection (Nsrelden et al. 2017). In addition, cathelicidins drive the induction of interleukin-8 in peripheral blood leucocytes of Atlantic salmon (Bridle et al. 2011). As it is clear that cathelicidins respond to bacterial pathogens and function as immunomodulants, it would be worth studying the associated mechanisms further.
Cathelicidins are cationic antimicrobial peptides found in both aquatic and land animals, e.g. hagfish (Uzzell et al. 2003), salmonids (Chang et al. 2006) and several mammals (Zanetti 2004). The structural differences between Atlantic salmon cathelicidins has been described previously (Chang et al. 2006). The mature peptides of cathelicidin 1 and 2 in this fish share only 29% sequence identity. On the other hand, for the cathelin domains of their pro-peptides, the identity value is 75%. The cathelicidin pro-domain itself does not have antibacterial activity in humans (Pazgier et al. 2013); the present study did not specifically examine the cathelicidin mature peptide and cathelin domains. However, in both the injection and pathogen exposure experiments, the magnitude of responses of the two cathelicidins was slightly different — cath2 responded strongly. These differences could be due to the structural and functional differences of the genes present in the skin and gills and merits further attention; the focus should be on the cathelicidin promoter sequence, and their peptide molecules using antibody approaches or mass spectrometry.
Defensins from Atlantic salmon skin tissue were cloned for this study. The predicted amino acid sequences of Atlantic salmon beta-defensins were of low identity (Table 1). Phylogenetic analysis revealed that the changes in the Defb3 sequence occurred earlier than those in other defensins (Defb1, 2 and 4), and Defb3 clustered together with rainbow trout Defb3. This result suggests that Defb3 structure and functions are specific to salmonids. In our study, beta-defensins were not significantly altered in the skin and gills by A. salmonicida injection and exposure. The pathogen-unrelated response of beta-defensin genes suggests that the inducers of these molecules are not necessarily bacterial substances and may not be linked to any bacterial infection-stimulated immunological pathways such as those starting with the toll-like receptor (Stafford et al. 2003). In addition, the direct exposure of gill tissues to the pathogen invoked only a weak response of defb1; further suggesting that defb1 is not affected by the bacterial pathogen (at least by A. salmonicida). The Atlantic cod beta-defensin — which closely clusters with Atlantic salmon defb1 (Fig. 1) — was upregulated in the head kidney but not in skin or gills as a response to Vibrio anguillarum H610 challenge (Ruangsri et al. 2013). Taken together, these results suggest that the body surface beta-defensins are not impacted by certain bacteria and their constitutive expression might aid in protection.
In addition to the defensins, the lao gene was successfully cloned from the Atlantic salmon skin tissue. The first fish LAO was identified in the viscera of chub mackerel and is known as an apoptosis-inducing protein (Jung et al. 2000). Furthermore, several studies have suggested it to be a new family of antibacterial protein in fishes (Kitani et al. 2007, 2008; Kasai et al. 2010; Nagashima et al. 2009; Wang et al. 2011; Kitani et al. 2010). The biological activity of LAO is mainly caused by the hydrogen peroxide that is generated from oxidative deamination of the substrate l-amino acids (Du and Clemetson 2002). This oxidation is caused by electron-transfer via flavin adenine dinucleotide (Izidoro et al. 2014). Our result showed that the amino acid sequence of the Atlantic salmon LAO included the dinucleotide binding motif and GG motif for flavin adenine dinucleotide binding (Online Resource, Fig. S2) (Vallon 2000). The sequence identity of amino acid residues of Atlantic salmon LAO and the marine fish LAOs were around 53–57%, and the identity values of freshwater fish species were higher, around 67–73% (Table 2). The phylogenetic analysis also points out that the Atlantic salmon LAO sequence and other freshwater fish LAOs belong to the same clade (Fig. 2). The diversity of the fish LAOs in both marine and freshwater fishes and the structure-habitat relationships of the LAO molecules merits further investigation.
The lao gene expression was upregulated in the injection study (Fig. 3b). Immunological functions of fish LAO are still unclear; there is only a report regarding the induction of Atlantic cod LAO at 48 h post infection in the skin, gills, spleen and head kidney after V. anguillarum challenge (Kitani et al. 2015). In the present study, bacterial pathogen exposure did not significantly upregulate lao in the gills of Atlantic salmon (Fig. 4f), so parhaps lao indirectly regulates against bacterial infection, unlike cathelicidins. The reasons for LAO inactivity despite the expressional changes of the gene might be indicative of low protein expression, low translation efficiency or the need for an unknown substrate. It should be noted that enzymatic characterization is necessary to understand the influence of LAO on fish immunity.
In conclusion, the body surface tissue cathelicidins of Atlantic salmon responded quickly to the Gram-negative fish-pathogen A. salmonicida. Both the injection and exposure experiments suggest that the cathelicidins are involved in local and systemic immune responses. In addition, the beta-defensins were not altered by A. salmonicida, suggesting the specificity of their regulation or protection via their constitutive expression. Moreover, the differential regulation of lao gene after vaccination and exposure indicates that the gene has an indirect regulatory system against bacterial infection, unlike cathelicidins. Taken together, we suggest that cathelicidins can be used as non-invasive early phase diagnostic markers of bacterial infection in salmon, and could therefore be adopted for disease management practices. Although the results from our study and previous reports indicate the suitability of cathelicidins as diagnostic markers, further in-depth investigations are required to confirm the sensitivity and dependability of the markers.
References
Basañez G, Shinnar AE, Zimmerberg J (2002) Interaction of hagfish cathelicidin antimicrobial peptides with model lipid membranes. FEBS Lett 532:115–120
Bridle A, Nosworthy E, Polinski M, Nowak B (2011) Evidence of an antimicrobial-immunomodulatory role of Atlantic salmon cathelicidins during infection with Yersinia ruckeri. PLoS One 6:e23417. https://doi.org/10.1371/journal.pone.0023417
Chang C-I, Zhang YA, Zou J, Nie P, Secombes CJ (2006) Two cathelicidin genes are present in both rainbow trout (Oncorhynchus mykiss) and Atlantic salmon (Salmo salar). Antimicrob Agents Chemother 50:185–195. https://doi.org/10.1128/AAC.50.1.185-195.2006
Cole AM, Weis P, Diamond G (1997) Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J Biol Chem 272:12008–12013
Du XY, Clemetson KJ (2002) Snake venom l-amino acid oxidases. Toxicon 40:659–665
Ellis AE (2001) Innate host defense mechanisms of fish against viruses and bacteria. Dev Comp Immunol 25:827–839. https://doi.org/10.1016/S0145-305X(01)00038-6
Furlan M, Rosani U, Gambato S, Irato P, Manfrin A, Mardirossian M, Venier P, Pallavicini A, Scocchi M (2018) Induced expression of cathelicidins in trout (Oncorhynchus mykiss) challenged with four different bacterial pathogens. J Pept Sci 24:e3089. https://doi.org/10.1002/psc.3089
Guiry A, Flynn D, Hubert S, O’Keeffe AM, LeProvost O, White SL, Forde PF, Davoren P, Houeix B, Smith TJ, Cotter D, Wilkins NP, Cairns MT (2010) Testes and brain gene expression in precocious male and adult maturing Atlantic salmon (Salmo salar). BMC Genom 11:211. https://doi.org/10.1186/1471-2164-11-211
Haugarvoll E, Bjerkås I, Nowak BF, Hordvik I, Koppang EO (2008) Identification and characterization of a novel intraepithelial lymphoid tissue in the gills of Atlantic salmon. J Anat 213:202–209. https://doi.org/10.1111/j.1469-7580.2008.00943.x
Izidoro LF, Sobrinho JC, Mendes MM, Costa TR, Grabner AN, Rodrigues VM, da Silva SL, Zanchi FB, Zuliani JP, Fernandes CF, Calderon LA, Stábeli RG, Soares AM (2014) Snake venom l-amino acid oxidases: trends in pharmacology and biochemistry. Biomed Res Int 2014:1–19. https://doi.org/10.1155/2014/196754
Jung SK, Mai A, Iwamoto M, Arizono N, Fujimoto D, Sakamaki K, Yonehara S (2000) Purification and cloning of an apoptosis-inducing protein derived from fish infected with Anisakis simplex, a causative nematode of human anisakiasis. J Immunol 165:1491–1497
Kasai K, Ishikawa T, Komata T, Fukuchi K, Chiba M, Nozaka H, Nakamura T, Sato T, Miura T (2010) Novel l-amino acid oxidase with antibacterial activity against methicillin-resistant Staphylococcus aureus isolated from epidermal mucus of the flounder Platichthys stellatus. FEBS J 277:453–465. https://doi.org/10.1111/j.1742-4658.2009.07497.x
Kasai K, Ishikawa T, Nakamura T, Miura T (2015) Antibacterial properties of l-amino acid oxidase: mechanisms of action and perspectives for therapeutic applications. Appl Microbiol Biotechnol 99:7847–7857. https://doi.org/10.1007/s00253-015-6844-2
Katzenback B (2015) Antimicrobial peptides as mediators of innate immunity in teleosts. Biology (Basel) 4:607–639. https://doi.org/10.3390/biology4040607
Kitani Y, Tsukamoto C, Zhang G, Nagai H, Ishida M, Ishizaki S, Shimakura K, Shiomi K, Nagashima Y (2007) Identification of an antibacterial protein as l-amino acid oxidase in the skin mucus of rockfish Sebastes schlegeli. FEBS J 274:125–136. https://doi.org/10.1111/j.1742-4658.2006.05570.x
Kitani Y, Kikuchi N, Zhang G, Ishizaki S, Shimakura K, Shiomi K, Nagashima Y (2008) Antibacterial action of l-amino acid oxidase from the skin mucus of rockfish Sebastes schlegelii. Comp Biochem Physiol B Biochem Mol Biol 149:394–400
Kitani Y, Ishida M, Ishizaki S, Nagashima Y (2010) Discovery of serum l-amino acid oxidase in the rockfish Sebastes schlegeli: isolation and biochemical characterization. Comp. Biochem Physiol B Biochem Mol Biol 157:351–356. https://doi.org/10.1016/j.matchemphys.2015.05.051
Kitani Y, Fernandes JMO, Kiron V (2015) Identification of the Atlantic cod l-amino acid oxidase and its alterations following bacterial exposure. Dev Comp Immunol 50:116–120. https://doi.org/10.1016/j.dci.2015.02.007
Koppang EO, Fischer U, Moore L, Tranulis MA, Dijkstra JM, Köllner B, Aune L, Jirillo E, Hordvik I (2010) Salmonid T cells assemble in the thymus, spleen and in novel interbranchial lymphoid tissue. J Anat 217:728–739. https://doi.org/10.1111/j.1469-7580.2010.01305.x
Lee PT, Bird S, Zou J, Martin SAM (2017) Phylogeny and expression analysis of C-reactive protein (CRP) and serum amyloid-P (SAP) like genes reveal two distinct groups in fish. Fish Shellfish Immunol 65:42–51. https://doi.org/10.1016/j.fsi.2017.03.037
Lehrer RI (2004) Primate defensins. Nat Rev Microbiol 2:727–738. https://doi.org/10.1038/nrmicro976
Løvoll M, Wiik-Nielsen CR, Tunsjø HS, Colquhoun D, Lunder T, Sørum H, Grove S (2009) Atlantic salmon bath challenged with Moritella viscosa pathogen invasion and host response. Fish Shellfish Immunol 26:877–884. https://doi.org/10.1016/j.fsi.2009.03.019
Løvoll M, Austbø L, Jørgensen JB, Rimstad E, Frost P (2011) Transcription of reference genes used for quantitative RT-PCR in Atlantic salmon is affected by viral infection. Vet Res 42:8. https://doi.org/10.1186/1297-9716-42-8
Marshall OJ (2004) PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20:2471–2472. https://doi.org/10.1093/bioinformatics/bth254
Masso-Silva JA, Diamond G (2014) Antimicrobial peptides from fish. Pharmaceuticals 7:265–310. https://doi.org/10.3390/ph7030265
Nagashima Y, Tsukamoto C, Kitani Y, Ishizaki S, Nagai H, Yanagimoto T (2009) Isolation and cDNA cloning of an antibacterial l-amino acid oxidase from the skin mucus of the great sculpin Myoxocephalus polyacanthocephalus. Comp Biochem Physiol B Biochem Mol Biol 154:55–61. https://doi.org/10.1016/j.cbpb.2009.05.006
Nsrelden RM, Horiuchi H, Furusawa S (2017) Expression of ayu antimicrobial peptide genes after LPS stimulation. J Vet Med Sci 79:1072–1080. https://doi.org/10.1292/jvms.16-0609
Nymo IH, Seppola M, Al Dahouk S, Bakkemo KR, Jiménez de Bagüés MP, Godfroid J, Larsen AK (2016) Experimental challenge of Atlantic cod (Gadus morhua) with a Brucella pinnipedialis strain from hooded seal (Cystophora cristata). PLoS One 11:e0159272. https://doi.org/10.1371/journal.pone.0159272
Pazgier M, Ericksen B, Ling M, Toth E, Shi J, Li X, Galliher-Beckley A, Lan L, Zou G, Zhan C, Yuan W, Pozharski E, Lu W (2013) Structural and functional analysis of the pro-domain of human cathelicidin, LL-37. Biochemistry 52:1547–1558. https://doi.org/10.1021/bi301008r
Primor N, Tu AT (1980) Conformation of pardaxin, the toxin of the flatfish Pardachirus marmoratus. Biochim Biophys Acta Protein Struct 626:299–306. https://doi.org/10.1016/0005-2795(80)90124-5
Rakers S, Gebert M, Uppalapati S, Meyer W, Maderson P, Sell AF, Kruse C, Paus R (2010) “Fish matters”: the relevance of fish skin biology to investigative dermatology. Exp Dermatol 19:313–324. https://doi.org/10.1111/j.1600-0625.2009.01059.x
Rakers S, Niklasson L, Steinhagen D, Kruse C, Schauber J, Sundell K, Paus R (2013) Antimicrobial peptides (AMPs) from fish epidermis: perspectives for investigative dermatology. J Invest Dermatol 133:1140–1149. https://doi.org/10.1038/jid.2012.503
Rombout JHWM, Yang G, Kiron V (2014) Adaptive immune responses at mucosal surfaces of teleost fish. Fish Shellfish Immunol 40:634–643. https://doi.org/10.1016/j.fsi.2014.08.020
Romo RM, Pérez-Martínez D, Ferrer CC (2016) Innate immunity in vertebrates: an overview. Immunology 148:125–139. https://doi.org/10.1111/imm.12597
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Ruangsri J, Kitani Y, Kiron V, Lokesh J, Brinchmann MF, Karlsen BO, Fernandes JMO (2013) A novel beta-defensin antimicrobial peptide in Atlantic cod with stimulatory effect on phagocytic activity. PLoS One 8:e62302. https://doi.org/10.1371/journal.pone.0062302
Schmitt P, Wacyk J, Morales-Lange B, Rojas V, Guzmán F, Dixon B, Mercado L (2015) Immunomodulatory effect of cathelicidins in response to a β-glucan in intestinal epithelial cells from rainbow trout. Dev Comp Immunol 51:160–169. https://doi.org/10.1016/j.dci.2015.03.007
Shai Y (1999) Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta 1462:55–70
Shai Y (2002) Mode of action of membrane active antimicrobial peptides. Biopolymers 66:236–248. https://doi.org/10.1002/bip.10260
Shai Y, Fox J, Caratsch C, Shih YL, Edwards C, Lazarovici P (1988) Sequencing and synthesis of pardaxin, a polypeptide from the Red Sea Moses sole with ionophore activity. FEBS Lett 242:161–166
Shiomi K, Igarashi T, Yokota H, Nagashima Y, Ishida M (2000) Isolation and structures of grammistins, peptide toxins from the skin secretion of the soapfish Grammistes sexlineatus. Toxicon 38:91–103
Stafford JL, Ellestad KK, Magor KE, Belosevic M, Magor BG (2003) A toll-like receptor (TLR) gene that is up-regulated in activated goldfish macrophages. Dev Comp Immunol 27:685–698
Sugiyama N, Araki M, Ishida M, Nagashima Y, Shiomi K (2005) Further isolation and characterization of grammistins from the skin secretion of the soapfish Grammistes sexlineatus. Toxicon 45:595–601. https://doi.org/10.1016/j.toxicon.2004.12.021
Tort L, Balasch JC, Mackenzie S (2003) Fish immune system. A crossroads between innate and adaptive responses. Inmunologia 22:277–286
Uzzell T, Stolzenberg ED, Shinnar AE, Zasloff M (2003) Hagfish intestinal antimicrobial peptides are ancient cathelicidins. Peptides 24:1655–1667. https://doi.org/10.1016/j.peptides.2003.08.024
Valente LMP, Bower NI, Johnston IA (2012) Postprandial expression of growth-related genes in Atlantic salmon (Salmo salar L.) juveniles fasted for 1 week and fed a single meal to satiation. Br J Nutr 108:2148–2157. https://doi.org/10.1017/S0007114512000396
Vallon O (2000) New sequence motifs in flavoproteins: evidence for common ancestry and tools to predict structure. Proteins 38:95–114
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. https://doi.org/10.1186/gb-2002-3-7-research0034
Vasanth G, Kiron V, Kulkarni A, Dahle D, Lokesh J, Kitani Y (2015) A microbial feed additive abates intestinal inflammation in Atlantic salmon. Front Immunol https://doi.org/10.3389/fimmu.2015.00409
Wang F, Li R, Xie M, Li A (2011) The serum of rabbitfish (Siganus oramin) has antimicrobial activity to some pathogenic organisms and a novel serum l-amino acid oxidase is isolated. Fish Shellfish Immunol 30:1095–1108. https://doi.org/10.1016/j.fsi.2011.02.004
Xhindoli D, Pacor S, Benincasa M, Scocchi M, Gennaro R, Tossi A (2016) The human cathelicidin LL-37—a pore-forming antibacterial peptide and host-cell modulator. Biochim Biophys Acta Biomembr 1858:546–566. https://doi.org/10.1016/j.bbamem.2015.11.003
Zanetti M (2004) Cathelicidins, multifunctional peptides of the innate immunity. J Leukoc Biol 75:39–48. https://doi.org/10.1189/jlb.0403147
Zanetti M, Gennaro R, Romeo D (1995) Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett 374:1–5
Zou J, Secombes CJ (2016) The function of fish cytokines. Biology 5:23. https://doi.org/10.3390/biology5020023
Zou J, Mercier C, Koussounadis A, Secombes C (2007) Discovery of multiple beta-defensin like homologues in teleost fish. Mol Immunol 44:638–647. https://doi.org/10.1016/j.molimm.2006.01.012
Acknowledgements
We would like to thank the staff at the Research Station, Nord University, Norway for maintenance of the fish. This research was supported by the Researcher Exchange Program between the Japan Society for the Promotion of Science and The Research Council of Norway (Project number 219006).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kitani, Y., Hieu, D.Q. & Kiron, V. Cloning of selected body surface antimicrobial peptide/protein genes of Atlantic salmon and their responses to Aeromonas salmonicida. Fish Sci 85, 847–858 (2019). https://doi.org/10.1007/s12562-019-01331-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12562-019-01331-1