Abstract
The origin and maintenance of sexual gametogenesis might provide insights into eukaryote evolution. Epigenetic variation of eukaryote well-being would provide clues that could relate ecological and evolutionary processes. To investigate the relationship between gametogenic variability and epigenetic polymorphism in the Pacific oyster Crassostrea gigas, DNA methylation differences were represented by analyzing genome-wide methylation patterns using the fluorescent-labeled methylation sensitive-amplified polymorphism (F-MSAP) technique. The C. gigas genome was widely methylated and marked variations in the pattern of methylation were shown during female and male gonad development. Statistically, the total methylation level increased from early developing stage to resorbing stage, from 28.86 to 34.53% in ovaries, while it increased from 31.20 to 35.46% in this period in testes. The unweighted pair group method with arithmetic mean and principal coordinate analysis separated the C. gigas during female and male gonad development remarkably well, indicating remarkable differences across gametogenesis between ovaries and testes. Variation was higher in testes than in ovaries by the frequency based and multivariate analyses above. Both multivariate analyses and a significantly different population epigenetic structure suggested that a striking difference might exist across gametogenesis, reflecting a turning point of the methylation mechanism during reproductive progress. This study provides some information on the role of DNA methylation in the potential mechanism during female and male gonad development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gene spreading of survival and offspring could be useful information for eukaryote evolution, and reproductive organs are the tools for chance to evolve out the new reproduction strategy at the time (Guo et al. 1998). Gametogenesis in sexual reproduction, as an important link in the life cycle, is a biological prelude in which ploidy or haploid precursor cells originated. The concept is also critical for the prevalence of sexual differentiation, which is highly variable among eukaryotes (Torres-Maldonado et al. 2002). Generally, a primary signal initiates the onset of a molecular cascade of transcriptional or mRNA splicing factors, allowing the final differentiation of the gonad into testis or ovary (Naimi et al. 2009b). The sex mechanism could be affected by genetic, cytoplasmic, behavioral and environmental factors. The genetic control of sex determination has been determined in higher organisms (Wilhelm et al. 2007; Devlin and Nagahama 2002) and some arthropods, such as Drosophila (Caudy et al. 1988). The origin and maintenance of sexual gametogenesis might provide clues to the evolution of eukaryotes.
As a well-characterized mechanism of epigenetics, DNA methylation plays a central role in the matter of gene regulation and phenotype variation (Klose and Bird 2006). In the eukaryote genome, the pattern of DNA methylation can modify the phenotype without entailing changes in nucleotide sequences (Nicotra et al. 2015). By the same token, it is presumed to be an ancient evolutionary mechanism in all multicellular life (Ponger and Li 2005). DNA methylation has been therefore proved to be critical in response to environmental conditions and integrating signals in organisms (Feil and Fraga 2012). In recent decades, methylation changes have turned out to be essential for embryo viability (Razin and Shemer 1995), sexual differentiation, and determination (Reik 2007); additionally, the genomic methylation pattern in the germ line has been proved to be different for male and female gametes (Sanford et al. 1987; La Salle et al. 2004), implying that DNA methylation in mammals plays an important role in the regulation of sexual dimorphism. However, a big part of the epigenetic disparity between vertebrates and invertebrates is that methylation is absent in major model invertebrates and/or the methylation level is too low (Yi and Goodisman 2009). This could lead to few studies focused on the role of genome-wide DNA methylation in the regulation of gametogenesis sexual dimorphism in invertebrates. Consequently, genome-wide DNA methylation profiles in the phase of invertebrate gonad establishment still remain questionable.
The Pacific oyster Crassostrea gigas is a eurythermal and euryhaline species which has been distributed and cultured worldwide. As an attractive model of invertebrates, C. gigas is a marine bivalve mollusk whose development occurs in three main periods, embryo, larva, and spat (Guo et al. 1998; Naimi et al. 2009a). And the life cycle of adult C. gigas comprised two phases: gametogenesis (including ripeness and spawning) and sexual repose during the rest of the year (Dridi et al. 2007). An increasing number of studies have demonstrated that DNA methylation is of great importance for development, metabolism and stress response (Dheilly et al. 2012; Riviere et al. 2013; Gavery and Roberts 2014; Olson and Roberts 2014), which also contributes a comprehensive characterization for the distribution of DNA methylation in the adult gamogenetic cycle by regulating gene expression. Therefore, determining the epigenetic mechanism of regulation of C. gigas sexual dimorphism will provide information for further exploration.
In this study, we assessed genome-wide DNA methylation differentiation during C. gigas gametogenesis using the fluorescent-labeled methylation-sensitive amplified polymorphism (F-MSAP) technique. The objectives were to characterize the pattern and extent of DNA methylation and to determine the occurrence of epigenetic changes associated with male and female gonad development.
Materials and methods
Experimental oysters collection and DNA extraction
C. gigas populations were collected from Rushan, Shandong Province, China, in March, May, July and October, 2016, respectively, when the gonadal development of the oysters was at early development, late development, spawned, and spent/resorbing stages, respectively (Deng 2016). Their parents were both from the selective line of Shandong Province. In 2015, the rearing of larvae and spat was carried out using standard practices. When the spat reached 2–3 mm in shell height, they were put into cages and cultured in Rushan Bay. The next year, 2016, C. gigas individuals were selected every sample time for further analysis. Upon arrival, body weight, shell length, shell width, and shell height of each individual were weighed and measured (Table 1). And then paraffin sections and photomicrographic techniques were used to identify their sex and gonadal development.
The gonad tissue samples (digestive gland not included) were rapidly isolated from oysters and stored at − 80 °C until further analysis. Genomic DNA was extracted using a modified phenol–chloroform protocol (Li et al. 2006) and dissolved in 1× TE buffer. The integrity and concentration of DNA was detected using NanoDrop-2000 and by 1% agarose gel electrophoresis.
F-MSAP assay
The F-MSAP procedure was carried out to detect methylation polymorphism as described by Jiang et al. (2016). A sample of 100 ng of each genomic DNA was digested with 2 U EcoRI and 2 U HpaII (Thermo). The second reaction comprised digestion with 2 U of EcoRI and 2 U of MspI under the same conditions as the first reaction. The DNA digestion fragments were ligated with T4 DNA ligase (Thermo) at 16 °C, overnight. The pre-selective amplification was performed in 10 μl, an initial denaturation at 72 °C for 2 min, 20 PCR cycles of 20 s at 94 °C, 30 s at 56 °C, and 2 min at 72 °C were performed; followed by 60 °C incubation for 30 min. Selective amplification was performed on 20 times diluted pre-selective amplification products, as a volume of 10 μl, with the following profile: 2 min of denaturing at 94 °C, then 10 cycles of 20 s at 94 °C, 30 s at 66 °C, and 2 min at 72 °C, with a 1 °C decrease in the annealing temperature of each cycle, followed by 20 cycles of 20 s at 94 °C, 30 s at 56 °C, and 2 min at 72 °C, with 60 °C incubation for 30 min. The EcoRI and HpaII/MspI adapters and pre-selective and selective amplification primers were used as described by Reyna-López et al. (1997) and Xu et al. (2000), synthesized by Sangon, and the sequences are shown in Table 2. Moreover, EcoRI primers were 5′-labeled using a 6-FAM reporter molecule. PCR products were loaded simultaneously with a GeneScan™-500 LIZ™ Size Standard into an ABI 3130 Genetic Analyzer (Applied Biosystems). Data was performed using GeneMapper v.4.0 software (Applied Biosystems). DNA fragments less than 50 bp in length, or longer than 500 bp were excluded from the analysis due to low levels of reproducibility. Lastly, the data were obtained in the form of an Excel table for further scoring.
Data analyses
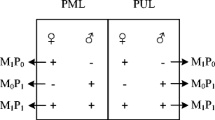
According to the differential sensitivity to DNA methylation and electrophoresis profiles, the MSAP patterns digested with the isoschizomers were divided into four types: type I (1, 1) present in both EcoRI/HpaII and EcoRI/MspI products, denoted no methylation state (Fig. 1); type II (1, 0) present only in EcoRI/HpaII products, denoted a hemi-methylation state (Fig. 1); type III (0, 1) present only in either EcoRI/MspI products, and absent from EcoRI/HpaII, denoted a fully methylated state (Fig. 1); type IV was absent from both enzyme combinations (0, 0), and was regarded as genetic polymorphism or hyper-methylation (Keyte et al. 2006), which was considered to be an uninformative state. In this research, types II and III together correspond to a methylated state. MSAP profiles, collected from all twelve primer combinations, were analyzed using the R (ver.3.2.3) package msap (Pérez-Figueroa 2013; http://msap.r-forge.r-project.org). Individual fragments were classified as either methylation-susceptible loci (MSL) or non-methylated loci (NML), depending on whether the observed proportion of discordant HpaII-MspI scores suggestive of methylation exceeded a 5% threshold, rounding the repeatability value obtained before. Only those fragments showing polymorphism, with at least two occurrences of each state, were used for subsequent analyses. Analysis in MSAP followed a band-based strategy (Bonin et al. 2007) and MSL were used to assess epigenetic variation. Epigenetic differentiation among tissues and maturity stages was assessed by means of principal coordinate analysis (PCoA), and then followed by analyses of molecular variance (AMOVA; Excoffier et al. 1992).
Examples of MSAP banding patterns in Crassostrea gigas. H lane means digested by EcoRI/HpaII; M lane means digested by EcoRI/MspI; Type I indicated absence of methylation due to the presence of bands in both EcoRI/HpaII and EcoRI/MspI digestion; Type II bands appeared only in EcoRI/HpaII digestion but not in the EcoRI/MspI digestion; Type III generated bands obtained in EcoRI/MspI digestion but not in EcoRI/HpaII digestion
Results
Global DNA methylation changes during gonad development
Gametogenesis of C. gigas in Rushan began in March, after a period of sexual inactivity. To assess the differences from proliferating stage to recovering stage between female and male gonads, different states of cytosine at CCGG sites were reflected by analyzing the presence of banding pattern at CCGG sites between EcoRI + HpaII and EcoRI + MspI. Figure 2 shows the general cytosine methylation changes of each type. Considering only the tissue gonad, twelve primer combinations amplified a total of 1678–3655 bands obtained from all timing materials (data not shown). In this regard, 401–633 loci were detected as MSL and 193–419 (48.13–66.19%) could be regarded as polymorphic. The extent of DNA methylation ranged from 28.86% (March, female) to 35.46% (October, male) of gonad samples. Overall, remarkable differences for total DNA methylation were observed. The total DNA methylation rate was increasing and the extent of the male was higher than that of the female, except in May (Fig. 2). The hemi-methylation ratios in the ovaries increased gradually from 11.23% in March to a maximum value of 15.38% in July and decreased afterwards. In contrast, the hemi-methylation ratios in the testes decreased from 12.77% at first, and then to a maximum value of 14.76% in July. The common point is that hemi-methylation reached a maximum in both ovaries and testes in July. The full methylation ratios in the tissue gonad increased gradually from March, decreased in July, then reached a maximum in October. The full methylation ratios were always higher than the levels of hemi-methylation, and the full methylation levels of males were also higher than those of females.
Extents and patterns of DNA methylation at different developmental stages
To assess methylation differences in CCGG sites during gametogenesis, methylation patterns between ovaries and testes were compared for early development, late development, spawned, and resorbing stages. If the differential gene expression occurring during gametogenesis is mediated by methylation, we would expect differences among developmental stages. Just two independent analyses were done; one for female individuals, and the other for males. The number of loci classified as MSL by the msap package was 592 and 520 for females and males, respectively. The number of loci classified as polymorphic MSL was 499 and 443, respectively. Accordingly, the frequency of polymorphic MSL was approximately 88.80 and 85.19%. AMOVA results (female φST = 0.2136, P < 0.0001, male φST = 0.2467, P < 0.0001) revealed a significant difference between female and male gonad development, and the testes showed more epigenetic differentiation than the ovaries. Additionally, this finding also held for the pairwise analyses of MSL differentiation (Table 3). In ovaries, the pairwise φST between late development and spawned stage was the highest (0.2374) whereas the lowest difference was between spawned and resorbing stages (0.1805), and all differences were highly significant. In testes, the observed pairwise differences between sampling points were higher than in ovaries and all differences were also highly significant. Again, spawned stages showed the lowest difference compared to October (0.1501); however, the greatest difference was between early and late development stages (0.4057). Generally, the pairwise φST values in testes were also higher than those in ovaries (Table 3). All these results showed there might exist extensive and significant epigenetic differentiation between testes and ovaries.
However, PCoA showed some different types of epigenetic variation for females and males. The output plots of the two first principal coordinates are shown in Fig. 3. The second coordinate separated the ovaries samples (Fig. 3a) into three groups (10.6% of variance explained), one of them includes early and late development stages, and the others are spawned and resorbing stages, respectively. This result indicated no clear grouping could be observed between early and late development stages, which was different from that of AMOVA. And in testes, the first coordinate, accounting for 17.4% of the total variance (Fig. 3b), also showed three clusters of testes: one of them includes July and October, and the others are early and late development stages, respectively. In general, the overall clustering patterns confirmed the AMOVA results to a degree.
Representation of principal coordinate analysis (PCoA) results for epigenetic differentiation during gametogenesis. The first two coordinates (C1 and C2) are demonstrated with the percentage of variance explained in parentheses. Scores represent individual samples. Circles show individuals of each population. Labels show the centroid for each population. Ellipses represent the dispersion of those points around their center. The long axis of the ellipse shows the direction of maximum dispersion and the short axis displays the direction of minimum dispersion
Extents and patterns of DNA methylation between female and male populations
To further assess differences in cytosine methylation in CCGG sites between females and males, each sex was considered as a population. Four independent analyses were done, one per developmental stage. The numbers of loci classified as MSL by the msap package for the sampling point were 411, 411, 495, and 496, respectively; and the numbers of loci classified as polymorphic MSL were 278, 269, 357, and 378, respectively. So, the frequencies of polymorphic MSL were 67.64, 65.45, 72.12, and 76.21%, accordingly. AMOVA results (March φST = 0.1719, P = 0.0062; May φST = 0.2597, P = 0.0107 and October φST = 0.1102, P = 0.0436) revealed significant differences between females and males in the months May, March, and October, respectively. In this profile, no significant differentiation was observed in July (July φST = 0.0918, P = 0.0830).
Figure 4 shows the first and second principal coordinates resulting from PCoA which indicates the epigenetic variation and the degree of differentiation during gametogenesis between female and male populations. In the late development stage, the females (Fig. 4b) were separated from males along the first coordinate (34.5% of variance explained), indicating higher differentiation than the others and in accordance with AMOVA results. In the case of early development, spawned and resorbing stages (Fig. 4a, c, d), only some differentiation could be observed, mainly along the second coordinate (14.4, 16.6 and 17.2% of variance explained). The overall clustering patterns partly disagreed with the AMOVA results of July.
Representation of principal coordinate analysis (PCoA) results for epigenetic differentiation between females and males. The first two coordinates (C1 and C2) are demonstrated with the percentage of variance explained in parentheses. Scores represent individual samples. Circles show individuals of each population. Labels show the centroid for each population. Ellipses represent the dispersion of those points around their center. The long axis of the ellipse shows the direction of maximum dispersion and the short axis displays the direction of minimum dispersion
Discussion
Our study was focused on genome-wide CCGG methylation changes with the isocaudomers, HpaII and MspI, confirming that the C. gigas genome is widely methylated in the CCGG sequence. Compared with other detection methods, F-MSAP is a cheaper and less labor-intensive approach for genome-wide DNA methylation, and its effectiveness and reliability have been verified in previous studies, including in C. gigas (Díaz-Freije et al. 2014; Jiang et al. 2016). In current study, the extent and pattern of genome-wide methylation were indicated to differ between ovaries and testes, and from the beginning of ripeness to the end of spawning. The results of this study also showed that gametogenic variability between developmental stages in C. gigas are associated with epigenetic variability as genome-wide methylation patterns were showed to differ during female and male gonad development.
There is an association displaying among the tissues jointly to promote the growth and reproduction for achieving reproductive function (Díaz-Freije et al. 2014; Trautner et al. 2017). Individuals in a family share the same or similar sets of genetic information, but some acquired factors, such as environmental change or disease interference, would make a difference to epigenetic analysis. During gametogenesis, the organism absorbs nutrients to complete its development, but also begins to supply reproductive organs in their development. During C. gigas gonadal development, the level of DNA methylation was a dynamic process (Fig. 2). This might be the result of gonadal development from early development stage converting to late development, spawned and spent/resorbing stages in this research. In the same individual gonad, the cell development of ovaries and testes is not synchronous (Guo et al. 1998), and gonadal development among different individuals does not happen at the same time, either. At all four sampling points, from both female and male individuals, the lowest degree of total methylation occurred at the early development stage, and gradually increased to the maximum until the final sampling point.
In the long-term evolutionary process, eukaryotes always have the potential for stable gene expression in response to environmental changes, that is, pattern changes of genomic methylation are essential prerequisites to these stage-specific differences (Trautner et al. 2017). Absence of methylation is supposed to facilitate transcriptional opportunities to increase phenotypic plasticity in invertebrates (Roberts and Gavery 2012), that is, a close association might exist between demethylation and gene expression. Other similar results have been confirmed in Aplysia (Rajasethupathy et al. 2012) and C. gigas (Riviere et al. 2013). Given the above, demethylation in the earlier gametogenesis of our study was supposed to facilitate the expression of reproduction related genes in the ovaries and testes; however, in the later gametogenesis, inhibition might occur based on the increasingly high frequency of methylation.
Previous studies have suggested that DNA methylation plays a critical part in sex evolution (Zhang et al. 2008, 2011; Strömqvist et al. 2010). C. gigas is an alternative and irregular protandrous hermaphrodite, most individuals mature first as males and then change sex several times (Dheilly et al. 2012). Due to lack of heteromorphic sex chromosomes (Ahmed and Sparks 1967), the sex mechanism has long been unknown in Pacific oysters. Based on genome-wide profiling of DNA methylation and RNA-seq in male gametes, methylation levels in gene regions are positively related to gene expression (Olson and Roberts 2014). In our study, marked differences were observed across gametogenesis when comparing methylation patterns between ovaries and testes (Fig. 4). Under an uncontrolled and certainly variable environment, oysters would undergo strong morphological changes mainly due to the fast growth of the first sexual maturity, so they might also changes physiologically, leading to a variety of regulatory actions in response to environmental changes. All these transformations might be mediated by DNA methylation differences in ovaries and testes during these developmental stages.
Additionally, it is suggested that there might exist two possible regulation mechanisms between the ovaries and testes. C. gigas might change and adjust the DNA methylation pattern to achieve orderly normal growth and development.
By synthesizing the PCoA results of these two profiles, gametogenesis could be divided into two groups, one including early and late development stages, the other including spawned and resorbing stages. Additionally, pairwise φST of the experimental groups showed the highest difference in ovaries was between late development and spawned stage, and the biggest disparity in testes was between early and late development stages (Table 2). The probable explanation is the turning point of methylation patterns associated with reproductive regulation of the female and male C. gigas. Díaz-Freije et al. (2014) observed some differences in MSAP patterns between early phases of development (immature and maturing) and mature stage in Octopus vulgaris. Remarkable differences in methylation patterns were found in glass, yellow and silver eels of Anguilla anguilla (Trautner et al. 2017). In addition, a significant divergence between ovaries and testes in prevalent gene expression patterns was observed in C. gigas (Dheilly et al. 2012). The selected sex-specific genes produced distinct and variable expression levels over the course of gametogenesis in males or/and females (Dheilly et al. 2012), revealing somehow analogous for female and male gonad development to our study. As corroborative evidence to support this hypothesis, there might be two possible mechanisms of DNA methylation existing in ovaries and testes and the shift between mechanisms could be regarded as a turning point in reproductive progress.
Conclusions could be obtained from our study that methylation pattern changes would play a role in the potential mechanism during gametogenesis in the C. gigas genome. There might be two possible mechanisms and a turning point in the DNA methylation regulatory mechanism during reproductive progress. Analysis of the expression of further candidate genes is needed in future studies to verify these hypotheses.
References
Ahmed M, Sparks AK (1967) A preliminary study of chromosomes of two species of oysters (Ostrea lurida and Crassostrea gigas). J Fish Res Board Can 24:2155–2159
Bonin A, Ehrich D, Manel S (2007) Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionists. Mol Ecol 16:3737–3758
Caudy M, Vässin H, Brand M, Tuma R, Jah LY, Jan YN (1988) Daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell 55:1061–1067
Deng CM (2016) Studies on reproductive biology and physiological parameters of golden shell color strain of the Pacific oyster Crassostrea gigas. MA thesis, Ocean University of China, Qingdao
Devlin RH, Nagahama Y (2002) Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208:191–364
Dheilly NM, Lelong C, Huvet A, Kellner K, Dubos MP, Riviere G, Boudry P, Favrel P (2012) Gametogenesis in the Pacific oyster Crassostrea gigas: a microarrays-based analysis identifies sex and stage specific genes. PLoS ONE 7:e36353
Díaz-Freije E, Gestal C, Castellanos-Martínez S, Morán P (2014) The role of DNA methylation on Octopus vulgaris development and their perspectives. Front Physiol 5:62
Dridi S, Romdhane MS, Elcafsi M (2007) Seasonal variation in weight and biochemical composition of the Pacific oyster, Crassostrea gigas in relation to the gametogenic cycle and environmental conditions of the Bizert lagoon, Tunisia. Aquaculture 263:238–248
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Feil R, Fraga MF (2012) Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 13:97–109
Gavery MR, Roberts SB (2014) A context dependent role for DNA methylation in bivalves. Brief Funct Genomics 13:217–222
Guo XM, Hedgecock D, Hershberger WK, Cooper K, Allen SK (1998) Genetic determinants of protandric sex in the Pacific oyster, Crassostrea gigas Thunberg. Evolution 52:394–402
Jiang Q, Li Q, Yu H, Kong LF (2016) Inheritance and variation of genomic DNA methylation in diploid and triploid Pacific oyster (Crassostrea gigas). Mar Biotechnol 18:124–132
Keyte AL, Percifield R, Liu B, Wendel JF (2006) Infraspecific DNA methylation polymorphism in cotton (Gossypium hirsutum L.). J Hered 97:444–450
Klose RJ, Bird AP (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31:89–97
La Salle S, Mertineit C, Taketo T, Moens PB, Bestor TH, Trasler JM (2004) Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev Biol 268:403–415
Li Q, Liu W, Shirasu K, Chen W, Jiang S (2006) Reproductive cycle and biochemical composition of the Zhe oyster Crassostrea plicatula Gmelin in an eastern coastal bay of China. Aquaculture 261:752–759
Naimi A, Martinez AS, Specq ML, Mrac A, Diss B, Mathieu M, Sourdaine P (2009a) Identification and expression of a factor of the DM family in the oyster Crassostrea gigas. Comp Biochem Phys A 152:189–196
Naimi A, Martinez AS, Specq ML, Diss B, Mathieu M, Sourdaine P (2009b) Molecular cloning and gene expression of Cg-Foxl2 during the development and the adult gametogenetic cycle in the oyster Crassostrea gigas. Comp Biochem Phys B 154:134–142
Nicotra AB, Segal DL, Hoyle GL, Schrey AW, Verhoeven KJ, Richards CL (2015) Adaptive plasticity and epigenetic variation in response to warming in an Alpine plant. Ecol Evol 5:634–647
Olson CE, Roberts SB (2014) Genome-wide profiling of DNA methylation and gene expression in Crassostrea gigas male gametes. Front Physiol 5:224
Pérez-Figueroa A (2013) MSAP: a tool for the statistical analysis of methylation—sensitive amplified polymorphism data. Mol Ecol Resour 13:522–527
Ponger LC, Li WH (2005) Evolutionary diversification of DNA methyltransferases in eukaryotic genomes. Mol Biol Evol 22:1119–1128
Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER (2012) A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 149:693–707
Razin A, Shemer R (1995) DNA methylation in early development. Hum Mol Genet 4:1751–1755
Reik W (2007) Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447:425–432
Reyna-López GE, Simpson J, Ruiz-Herrera J (1997) Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol Gen Genet 253:703–710
Riviere G, Wu GC, Fellous A, Goux D, Sourdaine P, Favrel P (2013) DNA methylation is crucial for the early development in the oyster C. gigas. Mar Biotechnol 15:739–753
Roberts SB, Gavery MR (2012) Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates? Front Physiol 2:116
Sanford JP, Clark HJ, Chapman VM, Rossant J (1987) Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Gene Dev 1:1039–1046
Strömqvist M, Tooke N, Brunström B (2010) DNA methylation levels in the 5′ flanking region of the vitellogenin I gene in liver and brain of adult zebrafish (Danio rerio)—sex and tissue differences and effects of 17α-ethinylestradiol exposure. Aquat Toxicol 98:275–281
Torres-Maldonado LC, Landa PA, Moreno MN, Marmolejo VA, Meza MA, Merchant LH (2002) Expression profiles of Dax1, Dmrt1, and Sox9 during temperature sex determination in gonads of the sea turtle Lepidochelys olivacea. Gen Comp Endocrinol 129:20–26
Trautner JH, Reiser S, Blancke T, Unger K, Wysujack K (2017) Metamorphosis and transition between developmental stages in European eel (Anguilla anguilla, L.) involve epigenetic changes in DNA methylation patterns. Comp Biochem Physiol D 22:139–145
Wilhelm D, Palmer S, Koopman P (2007) Sex determination and gonadal development in mammals. Physiol Rev 87:1–28
Xu ML, Li XQ, Korban SS (2000) AFLP-Based detection of DNA methylation. Plant Mol Biol Rep 18:361–368
Yi SV, Goodisman MAD (2009) Computational approaches for understanding the evolution of DNA methylation in animals. Epigenetics 4:551–556
Zhang WL, Wang XE, Yu Q, Ming R, Jiang JM (2008) DNA methylation and heterochromatinization in the male-specific region of the primitive Y chromosome of papaya. Genome Res 18:1938–1943
Acknowledgements
This study was supported by Grants from the National Natural Science Foundation of China (31772843), Shandong Seed Project, Shandong Province (2016ZDJS06A06), major project for Tianjin seed technology (15ZXZYNC00050), and Fundamental Research Funds for the Central Universities (201762014).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, X., Li, Q., Kong, L. et al. DNA methylation frequency and epigenetic variability of the Pacific oyster Crassostrea gigas in relation to the gametogenesis. Fish Sci 84, 789–797 (2018). https://doi.org/10.1007/s12562-018-1214-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12562-018-1214-5