Abstract
Norovirus (NoV) is the leading cause of acute viral gastroenteritis outbreaks in the world. These outbreaks are frequently associated with bivalve shellfish consumption, particularly because these products are often eaten raw or only slightly cooked. In Morocco, regulations concerning the acceptable levels of enteric bacteria indicator organisms in these products have been put in place. However, these regulations do not take into account the risk of viral contamination, and many gastroenteritis outbreaks have been linked to the ingestion of bivalve shellfish from areas that comply with the current food safety criteria. The aim of this study was to investigate NoV presence in shellfish samples (n = 104) collected at four sites owcff Oualidia lagoon (Moroccan Atlantic coast) from November 2015 to February 2017. Samples were analysed using real-time RT-PCR in accordance with the ISO 15216-2 method. NoVs of the genogroup II were detected in 7% of samples that were all collected during the winter months. Moreover, 71% of NoV-positive samples were harvested at sites upstream of the lagoon. These results highlight the need of regularly monitoring viral contamination in bivalve shellfish to limit the risk of viral gastroenteritis outbreaks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Norovirus (NoV) is a major cause of sporadic and epidemic foodborne gastroenteritis worldwide (Lopman et al. 2003; Glass et al. 2009; Nguyen et al. 2017). Specifically, NoVs are at the origin of 18% of all reported cases (Ahmed et al. 2014); 699 million illnesses and 219000 casualties can be attributed to NoV each year (Bartsch et al. 2016). NoV, which belongs to the Caliciviridae family, is a small, non-enveloped spherical single-stranded RNA virus that is stable in the external environment. NoV can be divided in seven genogroups based on the capsid gene sequence (Vinjé 2015). Humans are most frequently infected by the genogroups GI and GII. GII strains are implicated in 70–80% of all outbreaks and are mainly identified in clinical cases (Atmar 2010; Vega et al. 2014). NoV genetic variability does not allow building a long-term immune response, and every 2–4 years, a new pandemic strain emerges (Glass et al. 2009; Vega et al. 2011).
Contaminated food and water, person-to-person virus transmission, and infected airborne droplets represent the major source of NoV infection (Mead et al. 1999; Marks et al. 2003). NoV gastroenteritis occurs all year around with a peak in the winter season, and often affects closed communities, such as schools and hospitals (Atmar and Estes 2006; Patel et al. 2008). Acute gastroenteritis is frequently linked to the consumption of bivalve shellfish, especially those harvested in sewage-contaminated waters (Lees 2000). Indeed, bivalve molluscs satisfy their nutritional needs by filtering large quantities of water and concentrating viral particles in their digestive tract. The consumption of raw or undercooked shellfish greatly increases the human health risk.
The assessment of the sanitary quality of shellfish growing areas is based only on the enumeration of Escherichia coli, as an indicator of faecal bacterial contamination. In accordance with the Moroccan regulatory food safety criteria for shellfish (Decision 1508/12 of 15/08/2012, Moroccan Ministry of Agriculture, Rural Development and Maritime Fisheries) and European regulations, shellfish growing/harvesting areas are classified as A (100% of samples < 230 E. coli per 100 g of shellfish flesh and intravalvular fluid), B (10% of samples 4600 ≤ E. coli ≤ 46,000 per 100 g of shellfish flesh and intravalvular fluid), and C (100% of samples E. coli ≤ 46,000 per 100 g of shellfish flesh and intravalvular fluid). Conversely, the viral risk is not considered, and many gastroenteritis outbreaks linked to the ingestion of bivalve shellfish from areas that comply with the current food safety criteria have been reported (Formiga-Cruz et al. 2002; Mesquita et al. 2011). A systematic review of all shellfish-borne viral gastroenteritis outbreaks worldwide showed that NoV was the most frequently implicated viral pathogen (84% of all outbreaks) (Bellou et al. 2013).
The objective of this study was to evaluate NoV frequency in bivalve shellfish harvested at Oualidia lagoon by real-time RT-PCR analysis according to the ISO/TS 15216-2 method (ISO/TS 15216-2 2013).
Materials and Methods
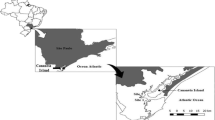
This study was conducted at Oualidia lagoon, a coastal wetland area in Morocco that is listed as Ramsar site (i.e. wetlands of international importance) since 2005. It is also included among the protected area of Morocco. This lagoon is located on the Atlantic coast, 168 km south of Casablanca and covers an area of 4 km2 with a length of 7 km (Fig. 1). It is separated from the ocean by a consolidated dune barrier and communicates with the sea through a main passage (150 m wide) throughout the year, and a secondary passage (50 m wide) that is active only during the spring tides (Carruesco 1989). A main channel with a depth that does not exceed 6 metres is connected with shallow secondary channels.
The peculiarity of this ecosystem lies in the weakness of the upstream flow velocity (Beaubrun 1976). The lagoon hydrodynamics are characterized by water renewal times that vary from 1 day (downstream near the passage entrance) to about 15 days (upstream of the lagoon), and by current velocity reduction in the upward direction according to the bathymetry and the main morphologic units (Hilmi et al. 2009; Koutitansky et al. 2007). In 2011, soil dredging was carried out in the upstream part to trap the sludge transported by the tide and reduce the rate of suspended solids (Makaoui et al. 2018).
Oualidia is a seaside resort with two distinct areas: a part located more to the North, on the national road that connects El Jadida to Safi, and a part more to the South, at the edge of the lagoon that is almost deserted, but for the summer period when it experiences a large influx of tourists. A wastewater treatment plant was put in place in 2012, but some habitations in the southern part are not connected to the sewage collection system and still have septic tanks.
Oualidia lagoon is the oldest oyster farming area in Morocco (from the 1950s). It produces about 36 tons of oysters per year (Department of Marine Fisheries 2017), and it is classified as a “B” harvesting area. To limit the bacterial contamination risk, shellfish undergo depuration before being marketed; however, this process has a limited effect on the level of viruses that can persist in the bivalve mollusc tissues for several weeks (Chalmers and McMillan 1995; Ang 1998).
Shellfish Sampling
This study was carried out at four sampling sites (S) at Oualidia lagoon (Fig. 1):
-
S1, downstream of the lagoon (sandy sedimentary),
-
S2 and S3, in the middle of the lagoon (muddy sedimentary),
-
S4, upstream of the lagoon (muddy sedimentary).
Oyster samples (n = 12 individuals/sample) were collected at three shellfish growing areas in Oualidia lagoon. The first site (S1) is downstream and is characterized by more pronounced hydrodynamics than the second site (S2) located on the main channel. The third site (S3) is the closest to the artificial dam that separates the lagoon from the salt marshes. Oyster samples were collected twice per month during the cold season (December to April) and monthly during the rest of the year, between November 2015 and February 2017. Burrowed shells [razor clams (Solen marginatus) and carpet shell clams (Ruditapes decussatus); n = 15–30 individuals/sample] were collected at S4 to evaluate NoV presence in clams, although their consumption is prohibited. Clams were sampled from November 2015 to September 2016 (twice per month from December to February and from June to August; once per month in the other months).
In total, 104 samples (n = 87 oyster samples and n = 17 clam samples) were collected.
Viral Recovery and RNA Extraction
Samples of bivalve molluscs were kept at 4 °C during the transport to the laboratory where they were processed immediately or stored at −T80 °C for 1 month at most. After thawing, samples were prepared and analysed according to the ISO/TS 15216-2/2013 method (ISO/TS 15216-2 2013), which is not validated for razor clams and carpet shells, with minor changes. For each sample, the digestive glands were removed and finely chopped. Then, 2.0 ± 0.2 g of tissue was spiked with 10 μl of Mengovirus (Mengovirus extraction control kit, Ceeramtools®) as positive control and digested with proteinase K. Samples were incubated twice (first at 37 °C for 60 min, and then at 60 °C for 15 min; always with stirring, 320 oscillations/minute). After centrifugation, the supernatant volume was measured.
Viral RNA was extracted from 500 μl of each sample using the Nucleospin® RNA Virus Extraction Kit (Macherey–Nagel, Germany) according to the manufacturer’s instructions. RNA was eluted in a final volume of 100 μl of elution buffer and stored at − 80 °C.
NoV Detection
The SuperScript®III Platinum®One-Step Quantitative RT-PCR Kit (Invitrogen) was used for NoV detection (GI and GII) by real-time RT-PCR. Briefly, 5 μl of each RNA sample was amplified in 25 μl of reaction mix that contained 1 × reaction mix, 0.5 pmol/μl of forward primer, 0.9 pmol/μl of reverse primer and 0.25 pmol/μl of probe, 1 × ROX Reference Dye and 1.25 μl of SuperScript®III RT/Platinum®Taq mix. The set of primers and probes and the respective references are listed in Table 1. Amplifications were performed in a 7500 Fast Real-time PCR System (Applied Biosystems) using the following cycling conditions: 50 °C for 60 min, 95 °C for 5 min, and then 40 cycles of 95 °C for 15 s followed by 60 °C for 1 min.
Undiluted and 1/10 diluted samples were tested. The cycle threshold (Ct), defined as the cycle number when the amplicon fluorescence is above the background signal, was considered positive when the value was < 40 with no evidence of amplification in the negative controls. The presence of PCR inhibitors was evaluated, using the mengo analysis according to the manufacturer’s instructions, by comparing the Ct values of the pure and diluted RNA samples.
A Ct value difference < 3.3 indicated the presence of inhibitors. The extraction efficiency was evaluated by comparing the Ct values of RNA samples spiked with Mengovirus to the Mengovirus standard curve. A result ≥ 1% was considered valid. NoV GI and GII RNA samples (provided by the National Institute of Hygiene, Rabat, Morocco) were used as positives controls. Wells containing only nuclease-free water or the PCR mixture without RNA were included as negative controls.
Results
For this study, only samples that provided valid results with acceptable extraction efficiencies were considered. Among the 104 samples screened by RT-PCR during the study period, two samples (clams) showed an unacceptable extraction efficiency (< 1%, although the extraction was performed a second time), 29 (28%) displayed an acceptable extraction efficiency (> 10%), and 73 (71%) an acceptable value (between 1 and 10%).
Among the 102 samples analysed, seven were NoV-positive (7%; only NoV GII). NoV GI was not detected in any sample. Considering the tested shellfish, only one clam sample (among the 15 analysed samples) and six oyster samples (among the 87 analysed samples) were NoV-positive (Table 2).
Most of the NoV-positive samples (n = 5; 71% of all positive samples) were collected in 2016. In 2017, only one oyster sample taken in February was NoV-positive (Table 3). This could be partly explained by the higher sampling frequency during the winter months.
Concerning the sampling sites, two NoV-positive oyster samples were from S1 (7% of all samples from S1), three from S2 (10% of all samples from S2), and one from S3 (3% of all samples from S3). The only NoV-positive clam sample was from S4. Moreover, five NoV-positive samples (n = 1 clam sample from S4 and n = 4 oyster samples from S2 and S3; 71% of all positive samples) were harvested at sites upstream of the lagoon.
Discussion
Several outbreaks of gastroenteritis have been linked to NoV in many countries (Nenonen et al. 2012; Bellou et al. 2013; Brake et al. 2014). This short study monitored NoV frequency in molluscs collected in a shellfish harvesting area, Oualidia lagoon (Morocco), after the construction of a wastewater treatment plant in 2012.
Oysters were collected from three sampling sites (S1 to S3) in a lagoon classified in the “B” category according to the Moroccan national regulations concerning food safety (i.e. acceptable levels of E. coli and salmonella spp in shellfish tissues). However, these indicators do not inform on NoV presence/absence (Lowther et al. 2008; Maalouf et al. 2011; Baert et al. 2011), and oyster depuration before marketing does not remove viruses (Ueki et al. 2007; Neish 2013; Polo et al. 2014).
Clams collected at S4, a natural clam deposit, were also tested although their marketing and consumption are prohibited. Previous studies found that NoV is more frequently detected in cockles/clams than oysters (Polo et al. 2014, Benabbes et al. 2013). Given the small number of samples and of positive results (n = 1), results for this species will not be discussed here.
To evaluate NoV frequency throughout the year, oysters were sampled for 16 months, with higher sampling frequency during the cold months because epidemiological data suggest higher levels of contamination during this period (Le Guyader et al. 2000; Formiga-Cruz et al. 2002). NoV seasonality is a poorly understood phenomenon, but could be linked to its ability to survive in the presence of environmental stressors (Greening 2006). In agreement, in our study, all NoV-positive oyster samples were collected in the cold season. However, a recent study (Kreidieh et al. 2017) reported that NoV detection during winter is less obvious in the Middle East and in North Africa than in the other countries of the Northern Hemisphere where NoV infections occur mainly in this season (Ahmed et al. 2013).
The rate of NoV-positive oyster samples (7%) is close to the 3% reported by a previous study carried out in Morocco between 2009 and 2010 (Benabbes et al. 2013). These values are significantly different from other geographical areas: 35% in Tunisia (Mediterranean coast) (Elamri et al. 2006), 25.6% in Galicia, Spain (Polo et al. 2014), 14.2% in Apulia, Italy (La Bella et al. 2017), and 5% in Japan (Nishida et al. 2007). At Oualidia lagoon, the presence of NoV contamination in winter time may be explained by the overflow of septic tanks at low tide, followed by the displacement of this pollution upstream of the lagoon under the effect of the water dynamics during tidal changes (Hassou et al. 2014). This might also explain why NoV-positive samples were more frequent at sites upstream of the lagoon (71%). The lower viral contamination at S1 could be linked to the sandy facies of the sediment that retains less the viruses and by the strong currents due to an important oceanic influence.
Although it has been reported that NoV GI accumulates more readily in oysters due to the presence of specific carbohydrate structures (Maalouf et al. 2010; Lowther et al. 2012), only NoV GII was detected in this survey, as previously reported in Galicia (NW Spain) where the contamination rate was much higher (53.7%) (Vilarino et al. 2009).
Molecular biology techniques reveal the presence of viral genomes, but do not provide any evidence on the presence of infectious viruses, and therefore of the potential human contamination. Despite the detection of NoV-positive samples, no outbreak was reported in relation with the tested shellfish during the study period. However, foodborne gastroenteritis cases are often under-reported by consumers. Moreover, due to the mild symptoms, cases of sporadic gastroenteritis are largely unreported (Dowell et al. 1995), especially in Africa (Kabue et al. 2016). In Morocco, an epidemiological and molecular study on children under the age of five hospitalized for acute gastroenteritis showed that NoV GII was predominant in stool samples (77.8%) (El Qazoui et al. 2014). However, no survey was conducted in Morocco to assess sporadic illness or outbreaks linked to NoV. In addition, NoV incidence in Oualidia lagoon is probably under-reported because viral particles can persist in the marine environment and represent a major risk for shellfish production.
Conclusion
Data from many countries established that NoV are a major cause of acute gastroenteritis in all age groups. This study highlights the potential risk of NoV contamination in oysters from Oualidia lagoon, Morocco. International and national seafood guidance documents have stressed the importance of implementing a national survey plan and sanitary control for the viral risk, including NoV, in bivalve molluscs.
References
Ahmed, S. M., Hall, A. J., Robinson, A. E., Verhoef, L., Premkumar, P., Parashar, U. D., et al. (2014). Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. The Lancet Infectious Diseases, 14, 725–730.
Ahmed, S. M., Lopman, B. A., & Levy, K. A. (2013). Systematic review and meta-analysis of the global seasonality of norovirus. PLoS ONE. https://doi.org/10.1371/journal.0075922.
Ang, L. H. (1998). An outbreak of viral gastroenteritis associated with eating raw oysters. Communicable Disease and Public Health, 1(1), 38–40.
Atmar, R. L. (2010). Noroviruses: State of the art. Food and Environmental Virology, 2(3), 117–126.
Atmar, R. L., & Estes, M. K. (2006). The epidemiologic and clinical importance of norovirus infection. Gastroenterology Clinics of North America, 35(2), 275–290.
Baert, L., Mattison, K., Loisy-Hamon, F., Harlow, J., Martyres, A., Lebeau, B., et al. (2011). Review: Norovirus prevalence in Belgian, Canadian and French fresh produce: A threat to human health. International Journal of Food Microbiology, 151(3), 261–269.
Bartsch, S. M., Lopman, B. A., Ozawa, S., Aron, J. H., & Bruce, Y. L. (2016). Global economic burden of norovirus gastroenteritis. PLoS ONE, 11(4), e0151219. https://doi.org/10.1371/journal.pone.0151219.
Beaubrun, P. C. (1976). Les huîtres au Maroc et l’ostréiculture dans la lagune de Oualidia. Bulletin de l’institut des Pêches Maritimes, 22, 13–143.
Bellou, M., Kokkinos, P., & Vantarakis, A. (2013). Shellfish-borne viral outbreaks: A systematic review. Food and Environmental Virology, 5(1), 13–23.
Benabbes, L., Olivier, J., Schaeffer, J., Parnaudeau, S., Rhaissi, H., Nourlil, J., et al. (2013). Norovirus and other human enteric viruses in Moroccan shellfish. Food and Environmental Virology, 5(1), 35–40.
Brake, F., Ross, T., Holds, G., Andreas Kiermeier, A., & McLeod, C. (2014). A survey of Australian oysters for the presence of human noroviruses. Food Microbiology, 44, 264–270.
Carruesco, C., (1989). Genèse et évolution de trois lagunes du littoral atlantique depuis l’holocène: Oualidia, Moulay BouSalham (Maroc) et Arcachon (France). Thèse doctorat d’état Es science université de bordeaux.
Chalmers, J. W., & McMillan, J. H. (1995). An outbreak of viral gastroenteritis associated with adequately prepared oysters. Epidemiology and Infection, 115(1), 163–167.
Da Silva, A. K., Le Saux, J. C., Parnaudeau, S., Pommepuy, M., Elimelech, M., & Le Guyader, F. S. (2007). Évaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behavior of genogroups I and II. Applied and Environmental Microbiology, 73(24), 7891–7897.
Dowell, S. F., Graves, C., & Kirkland, K. B. (1995). A multistate outbreak of oyster associated gastroenteritis: implications for interstate tracing of contaminated shellfish. Journal of Infectious Diseases, 171, 1497–1503.
El Qazoui, M., Oumzil, H., Baassi, L., El Omari, N., Sadki, K., & Amzazi, S. (2014). Rotavirus and norovirus infections among acute gastroenteritis children in Morocco. BioMed Central Infectious Diseases, 14, 300. https://doi.org/10.1186/1471-2334-14-300.
Elamri, D. E., Aouni, M., Parnaudeau, S., & Le Guyader, F. S. (2006). Detection of human enteric viruses in shellfish collected in Tunisia. Letters in Applied Microbiology, 43, 399–404.
Formiga-Cruz, M., Tofini-Quesada, G., Bofill-Mas, S., Lees, D. N., Henshilwood, K., Allard, A. K., et al. (2002). Distribution of human virus contamination in shellfish from different growing areas in Greece, Spain, Sweden and the United Kingdom. Applied and Environmental Microbiology, 68(12), 5990–5998.
Glass, R. I., Parashar, U. D., & Estes, M. K. (2009). Norovirus gastroenteritis. The New England Journal Medicine, 361, 1776–1785.
Greening, G. E. (2006). Human and animal viruses in food (including taxonomy of enteric viruses). Food Microbiology and Food Safety, 5–42.
Hassou, N., Maanan, M., Hennani, M., Zourarah, B., & Assobhei, O. (2014). Spatial and temporal variation of faecal pollution indicators (Escherichia coli and Faecal streptococci) and physico-chemical parameters at the Oualidia lagoon and its watershed (Morocco). International Journal of Current Microbiology and Applied Science, 3(3), 675–694.
Hilmi, K., Orbi, A., Lakhdar, J. L., (2009). Hydrodynamisme de la lagune de Oualidia (Maroc) Durant l’été et l’automne 2005. Bulletin de l’Institut Scientifique, Section Sciences de la Terre. Rabat, N°31.
ISO/TS 15216-2. (2013). Microbiology of food and animal feed: Horizontal method for detection of hepatitis A virus and norovirus in food using real-time RT-PCR—Part 2: Method for qualitative detection. Geneva: International Organization for Standardization.
Kabue, J. P., Meader, E., Hunter, P. R., & Potgieter, N. (2016). Human Norovirus prevalence in Africa: A review of studies from 1990 to 2013. Tropical Medicine & International Health, 21(1), 2–17.
Kageyama, T., Kojima, S., Shinohara, M., Uchida, K., Fukushi, S., Hoshino, F. B., et al. (2003). Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. Journal of Clinical Microbiology, 41(4), 1548–1557.
Koutitansky, V. G., Ouabi, M., Ibrahimi. I. (2007). L’étude du comportement hydrosédimentaire du système lagunaire Oualidia par la modélisation mathématique. Phase 2: Modélisation hydro-sédimentaire de l’état actuel et de scenarios d’aménagement. Direction des ports et du domaine public maritime. Ministère de l’Equipement et du Transport. Royaume du Maroc. 204p+Annexes.
Kreidieh, K., Charide, R., Dbaibo, G., & Melhem, N. D. (2017). The epidemiology of norovirus in the Middle East and North Africa (MENA) region: a systematic review. Virology Journal, 14, 220.
La Bella, G., Martella, V., Basanisi, M. G., Nobili, G., Terio, V., & La Salandra, G. (2017). Food-borne viruses in shellfish: Investigation on norovirus and HAV presence in Apulia (SE Italy). Food Environmental Virology, 9, 179–186.
Le Guyader, F. S., Haugarreau, L., Miossec, L., Dubois, E., & Pommepuy, M. (2000). Three-year study to assess human enteric viruses in shellfish. Applied and Environmental Microbiology, 66(8), 3241–3248.
Lees, D. N. (2000). Viruses and bivalve shellfish. International Journal Food of Microbiology., 59, 81–116.
Loisy, F., Atmar, R. L., Guillon, P., Cann, P., Pommepuy, M., & Le Guyader, F. S. (2005). Real-time RT-PCR for norovirus screening in shellfish. Journal of Virological Methods, 123, 1–7.
Lopman, B. A., Reacher, M. H., Van Duijnhoven, Y., Hanon, F. X., Brown, D., & Koopmans, M. (2003). Viral gastroenteritis outbreaks in Europe, 1995–2000. Emerging Infectious Diseases Jounal., 9(1), 90–96.
Lowther, J. A., Gustar, N. E., Powell, A. L., Hartnell, R. E., & Lees, D. N. (2012). Two-year systematic study to assess norovirus contamination in oysters from commercial harvesting areas in the United Kingdom. Applied and Environmental Microbiology, 78(16), 5812–5817. https://doi.org/10.1128/AEM.01046-12.
Lowther, J. A., Henshilwood, K., & Lees, D. N. (2008). Determination of norovirus contamination in oysters from two commercial harvesting areas over an extended period, using semi-quantitative real-time reverse transcription PCR. Journal of Food Protection, 71(7), 1427–1433.
Maalouf, H., Schaeffer, J., Parnaudeau, S., Le Pendu, J., Atmar, R. L., Crawford, S. E., et al. (2011). Strain-dependent norovirus bioaccumulation in oysters. Applied and Environmental Microbiology, 77, 3189–3196.
Maalouf, H., Zakhour, M., Le Pendu, J., Le Saux, J. C., Atmar, R. L., & Le Guyader, F. S. (2010). Distribution in tissue and seasonal variation of norovirus genogroup I and II ligands in oysters. Applied an Environmental Microbiology., 76(16), 5621–5630.
Makaoui, A., Idrissi, M., Agouzouk, A., Larissi, J., Baibai, T., El Ouehabi, Z., et al. (2018). Etat océanographique de la lagune de Oualidia, Maroc (2011–2012). European Scientific Journal, 14, 96–107.
Marks, P. J., Vipond, I. B., Regan, F. M., Wedgwood, K., Fey, R. E., & Caul, E. O. (2003). A school outbreak of Norwalk-like virus: Evidence for airborne transmission. Epidemiology and Infection, 131, 727–736.
Mead, P. S., Slutsker, L., Dietz, V., McCaig, L. F., Bresee, J. S., Shapiro, C., et al. (1999). Food-related illness and death in the United States. Emerging Infectious Diseases Journal, 5, 607–625.
Mesquita, J. R., Vaz, L., Cerqueira, S., Castilho, F., Santos, R., Monteiro, S., et al. (2011). Norovirus, hepatitis A virus and enterovirus presence in shellfish from high quality harvesting areas in Portugal. Food Microbiology, 28, 936–941.
Neish, A. (2013). (CEFAS) Investigative trials on the purification of oysters to identify ways of reducing norovirus. CEFAS contract report C5224.
Nenonen, N. P., Hannoun, C., Laesson, C. U., & Bergström, T. (2012). Marked genomic diversity of norovirus genogroup I strain in a waterborne outbreak. Applied Environmental Microbiology, 78(6), 1846–1852.
Nguyen, G. T., Phan, K., Teng, I., Pu, J., & Watanabe, T. (2017). A systematic review and meta-analysis of the prevalence of norovirus in cases of gastroenteritis in developing countries. Medicine, 96, e8139. https://doi.org/10.1097/MD.00000000008139.
Nishida, T., Nishio, O., Kato, M., Chuma, T., Kato, H., Iwata, H., et al. (2007). Genotyping and quantitation of noroviruses in oysters from two distinct sea areas in Japan. Microbiology Immunology, 51(2), 177–184.
Patel, M. M., Widdowson, M. A., Glass, R. I., Akazawa, K., Vinje, J., & Parashar, U. D. (2008). Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerging Infectious Diseases Journal, 14, 1224–1231.
Polo, D., Alvarez, C., Diez, J., Darriba, S., Longa, A., & Romalde, J. L. (2014). Viral elimination during commercial depuration of shellfish. Food Control, 43, 206–212.
Svraka, S., Duizer, E., Vennema, H., De Bruin, E., Van Der Veer, B., Dorresteijn, B., et al. (2007). Etiological role of viruses in outbreaks of acute gastroenteritis in The Netherlands from 1994 through 2005. Journal of Clinical Microbiology, 45(5), 1389–1394.
Ueki, Y., Shoji, M., & Suto, A. (2007). Persistence of caliciviruses in artificially contaminated oysters during depuration. Applied and Environmental Microbiology, 73(17), 5698–5701.
Vega, E., Barclay, L., Gregoricus, N., Hannah Shirley, S., David, L. D., & Vinjéa, J. (2014). Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. Journal of Clinical Microbiology, 52(1), 147–155.
Vega, E., Barclay, L., Gregoricus, N., Williams, K., Lee, D., & Vinjé, J. (2011). Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerging Infectious Diseases, 17, 1389–1395.
Vilariňo, M. L., Le Guyader, F. S., Polo, D., Schaeffer, J., Kröl, J., & Romalde, J. L. (2009). Assessment of human enteric viruses in cultured and wild bivalve molluscs. International Microbiology., 12(3), 145–151.
Vinjé, J. (2015). Advances in laboratory methods for detection and typing of norovirus. Journal of Clinical Microbiology, 53(2), 373–381.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
El Moqri, N., El Mellouli, F., Hassou, N. et al. Norovirus Detection at Oualidia Lagoon, a Moroccan Shellfish Harvesting Area, by Reverse Transcription PCR Analysis. Food Environ Virol 11, 268–273 (2019). https://doi.org/10.1007/s12560-019-09386-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-019-09386-0