Abstract
Modulating disease-relevant protein-protein interactions (PPIs) using small-molecule inhibitors is a quite indispensable diagnostic and therapeutic strategy in averting pathophysiological cues and disease progression. Over the years, targeting intracellular PPIs as drug design targets has been a challenging task owing to their highly dynamic and expansive interfacial areas (flat, featureless and relatively large). However, advances in PPI-focused drug discovery technology have been reported and a few drugs are already on the market, with some potential drug-like candidates already in clinical trials. In this article, we review the advances, successes and remaining challenges in the application of small molecules as valuable PPI modulators in disease diagnosis and therapeutics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Modern drug discovery is driven by molecular targets with the aim of identifying new therapeutic agents that can selectively target disease-specific molecular mechanisms or pathways (Díaz-Eufracio et al. 2018). In this context, protein-protein interactions (PPIs) are an attractive emerging class of molecular targets and are critically important in the progression of many disease states (Robertson and Spring 2018; Zhang et al. 2018). PPIs are engineered to provide a therapeutically tractable way of tweaking and manipulating the interplay in order to address the progression of many disease states (Du et al. 2018). They are involved in hubs of reversible and irreversible cellular processes, assembling and disassembling rapidly, reassembling and rearranging in order to restore normative cellular functions (Robertson and Spring 2018). There are more than 645,000 reported disease-relevant PPIs in the human interactome. However, only 2% of these had been targeted with drugs by 2011. Most of the remaining disease-relevant PPIs in protein complexes, such as transcription factors and many other signalling proteins, have been widely considered ‘undruggable’ and remain elusive, under-explored and yet to be fully understood (Gonzalez and Kann 2012; Díaz-Eufracio et al. 2018; Robertson and Spring 2018; Zhang et al. 2018).
Inhibiting PPIs using small molecules is a tremendously important diagnostic and therapeutic strategy that may lead to greatly protracted remissions and even curative therapies for a number of diseases (Stevers et al. 2018). The emergence of new technologies has unveiled the potential of PPIs in drug discovery and has enabled regular discovery of small-molecule PPI modulators as significant smart-drug targets (Grossmann et al. 2015; Jana et al. 2017). Over the years, PPI-focused drug technology has been regarded as prototypically intractable because of the highly dynamic and expansive PPIs interfacial areas (Taylor et al. 2018). However, recent advances have resulted in a few drugs being placed on the market, with some potential drug-like candidates already in clinical trials. In this study, we review the advances, successes and remaining challenges in the application of small molecules as valuable PPI modulators in disease diagnosis and therapeutics.
Strategies for targeting protein-protein interactions
Over the years, technological progress has played an imperative role in the identification of small-molecule modulators of PPIs that have to date reached clinical production (Stevers et al. 2018). The use of structural biology to determine ‘hotspots’ in PPIs’ binding interfaces has been an important strategy in discovering small-molecule modulators (Robertson and Spring 2018; Zhang et al. 2018). Despite the large sizes of PPIs’ interfaces, only a small subset of amino acid residues that comprise the hotspot contributes most of the binding free energy. These ‘hotspot’ regions are potential targets for drug discovery (Zhang et al. 2018). A classic way of identifying and defining hotspots in PPIs has been the combination of alanine-scanning mutagenesis and X-ray crystallography (Moreira et al. 2007; Wells and McClendon 2007). The initial application of this strategy was used to identify a hotspot in the binding interface between the extracellular domain of human growth hormone and its receptor (Clackson and Wells 1995).
Using alanine-scanning mutagenesis, other classic PPI hotspots of high-fidelity protein regions, such as the Fc fragment hinge region-binding domain, have been identified (Wells and McClendon 2007). Mutagenesis and structural studies of the binding events of interleukin-2 (IL-2) and the IL-2 receptor alpha chain (IL-2Rα) provide more classical insight (Wilson and Arkin 2011). The first small molecule (Ro26-4550) capable of inhibiting the IL-2/IL-2Rα interaction was discovered in 1997 (Wilson and Arkin 2011). Despite the compound not qualifying as a drug, it provided proof-of-principle that small-molecule PPI inhibitor drug discovery or design might be feasible. Moreover, structural studies of the Ro26-4550/IL-2 complex helped to characterise the IL-2 binding site, and served as the starting point for the design of higher affinity small-molecule IL-2/IL-2Rα PPI inhibitors (Wilson and Arkin 2011).
Fragment-based drug discovery (FBDD) (often referred to as fragment-based lead discovery, FBLD) is a key strategy for the discovery and design of small-molecule modulators of PPIs (Everts 2008; Erlanson et al. 2016; Robson-Tull 2018). It involves first identifying small chemical fragments (~ 200 Da), which may only bind at millimolar affinity to their targets (shown in Fig. 1). The fragments are then expanded or linked to other fragments that bind to nearby regions on the target in order to design a ‘lead’ with stronger affinity (Everts 2008; Erlanson et al. 2016). These ‘leads’ are then optimised via medicinal chemistry and may then be entered into preclinical and eventually clinical studies. Tethering and structure-activity relationship by nuclear magnetic resonance (SAR by NMR) are the two methods for FBDD/FBLD of potential modulators of PPIs (Erlanson et al. 2016; Robson-Tull 2018). Tethering involves constructing mutant forms of the target protein that contain the amino acid cysteine near a domain involved in PPIs (shown in Fig. 2), and then exposing it to a fragment library of disulphide moiety-linked organic compounds of less than 200 Da in molecular weight (Wilson and Arkin 2011). The goal is to select for compounds that bind weakly to the PPIs’ binding site near a native or engineered cysteine residue. The tethering technology was used to explore the IL-2 binding site for Ro26-4550 and to discover the IL-2Rα receptor antagonist (SP4206), which is an inhibitor of the IL-2/IL-2Rα PPIs (Arkin et al. 2003; Wilson and Arkin 2011).
Fragment-based drug discovery strategy. a Selection of a target compatible with the biophysical screening technique. b Production and purification of the target protein. c The fragment library design. d Biophysical screening of the fragment library. e Validation of hits to identify the fragment binding mode. f Development of the fragment(s) into a lead molecule (figure taken from Robson-Tull 2018)
The application of tethering in identifying leads in fragment-based drug design. Protein mutant forms with the cysteine mutation near a domain involved in PPIs are constructed. The mutant is exposed to a fragment library of disulphide moiety-linked organic compounds of less than 200 Da and compounds that bind weakly to the PPIs’ binding site near a native or engineered cysteine residue are selected (figure taken from Haberman 2012)
SAR by NMR involves the use of a high-throughput NMR technique to screen chemical libraries for fragment-sized compounds that bind to a protein sub-site with micromolar binding constants (Ma et al. 2016; Rüdisser et al. 2016). Using structural information from NMR to locate the binding sites for the compounds within a sub-site, ligands that bind to distinct but nearby sites within the sub-site can be identified. The two ligands can then be linked together to produce a new compound that binds to the domain with high affinity. This compound can thereafter be further optimised via medicinal chemistry to yield ‘lead’ compounds and ultimately, drug candidates (Ma et al. 2016; Robson-Tull 2018). SAR by NMR has been used to discover inhibitors of Bcl-2 family members that inhibit apoptosis (Oltersdorf et al. 2005). A very good example is ABT-263 (also known as Navitoclax, shown in Fig. 9a), which works by stimulating apoptosis in tumours (Duan et al. 2018).
Computational identification of hotspots for fragment-based drug design is another useful strategy in the discovery and design of lead compounds that address PPIs (Kozakov et al. 2011; Pan et al. 2018). It predicts the ability to bind fragment-sized small molecules and the side-chain flexibility necessary for the expansion of ‘pockets’. This strategy is known as computational solvent (CS) mapping (Landon et al. 2007; Hall et al. 2012). The emergence of CS mapping has enabled the virtual identification of druggable binding sites within PPIs and the subsequent discovery or design of small-molecule PPI modulators (Cencic et al. 2011; Kozakov et al. 2011). Druggable hotspots for well-studied PPI targets that were identified via CS mapping include IL-2/IL-2Ra, Bcl-XL/BAK, MDM2/p53, HPV-11 E2/HPV-11 E1, ZipA/FtsZ and TNF-α/TNFR1. CS mapping constitutes a powerful enabling technology that can help move the field of small-molecule PPI inhibitors up the technology curve and enable the discovery and development of numerous drugs that target PPIs (Kozakov et al. 2011; Pan et al. 2018).
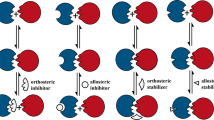
Another indirect strategy for discovering small-molecule PPI modulators involves searching for allosteric modulators. An allosteric site is a region of a protein that lies outside the binding site for the protein natural ligand, but when modulated, changes the conformation of the protein in such a way that it affects the activity of the binding site (Hansen et al. 2018; Trinh et al. 2018). For example, the anti-HIV/AIDS drug maraviroc, an allosteric PPI inhibitor of the CCR5 chemokine receptor, was discovered via high-throughput screening (HTS) (Dorr et al. 2005). Another serendipitous discovery of small-molecule allosteric modulators of PPIs is the discovery of compounds that act at an allosteric site to inhibit the sub-units of the heterodimeric transcription factor core binding factor (CBF) CBFβ and Runx1 (also known as CBFα) (Gorczynski et al. 2007). Developing methods for identification of allosteric sites in proteins and discovering drugs that modulate the activity of these proteins by binding to these allosteric sites is now the focus of pharmaceutical studies (Hansen et al. 2005; Hansen et al. 2018; Trinh et al. 2018).
The design of improved chemical libraries for targeting PPIs using methods such as diversity-oriented synthesis (DOS) has also been of benefit to PPI-focused drug technology (Hajduk et al. 2011; Basso et al. 2019; Luise and Wyatt 2019). In DOS, chemical libraries are developed to cover larger portions of chemical space than the standard libraries derived from combinatorial chemistry. Synthetic schemes are developed to maximise the number of structures and scaffolds produced in as few steps as possible to fill the largest amount of chemical space (Nielsen and Schreiber 2008; Schreiber 2009; Luise and Wyatt 2019). DOS is the best way to create drug-size and structurally diverse molecules efficiently. It is applicable in cases where a drug for a specific disease has to be developed without knowledge of the specific targets involved in the pathophysiological cues (Basso et al. 2019; Luise and Wyatt 2019). Library screening of structurally diverse compounds using the chemical genetics approach augments the ideal strategy to identify the targets. DOS compounds that inhibit key biological targets such as PPIs involved in signal transduction pathways are ideal probes and such compounds are very useful in exploring biological pathways in vitro and in small, easily permeated organisms, such as zebrafish embryos (Nielsen and Schreiber 2008; Schreiber 2009).
In recent years, the synthesis of diverse libraries in as small a number of steps as possible has been developed through a modular strategy known as ‘Build/Couple/Pair’ (B/C/P) (Galloway et al. 2010). Compounds that have been discovered via screening of DOS libraries using such a modular strategy have been found to modulate PPIs, transcription factor/DNA interactions and multidrug resistance in pathogens (Galloway et al. 2010). One example is robotnikinin. It inhibits the interaction between the 12-pass transmembrane receptor Patched 1 (Ptc1) and the extracellular protein Sonic Hedgehog (Shh) (Hitzenberger et al. 2017; Carballo et al. 2018). The Shh/Ptc1 interaction activates the cancer-implicated Hedgehog pathway, which is necessary for embryonic development (Stanton et al. 2009; Carballo et al. 2018). Robotnikinin is a useful probe, not a drug, and like many macrocycles (robotnikinin is itself a macrocycle) has drug-like physicochemical properties (Stanton et al. 2009; Hitzenberger et al. 2017).
With the rise in DOS and the B/P/C strategy, macrocycles have also become an emerging and promising strategy in targeting PPIs (Driggers et al. 2008; Song et al. 2017). Macrocyclic natural products have provided many drugs, including macrolide antibiotics (e.g. erythromycin and azithromycin), other antibiotics (rifampin and vancomycin), immunosuppressors (e.g. cyclosporine, rapamycin and sirolimus) and cancer chemotherapy drugs (temsirolimus, everolimus and epothilone) (Song et al. 2017). Some of these compounds are modulators of PPIs, e.g. paclitaxel, epothilone B, dictyostatin and halichondrin B (Driggers et al. 2008; Miller et al. 2018). These compounds modulate (stabilise or disrupt) the interaction between α and β sub-units of the tubulin heterodimer, thus disturbing microtubule dynamics and acting as antimitotics. Other PPI modulators include the mammalian target of rapamycin (mTOR) inhibitor. Rapamycin (also known as sirolimus) forms a complex with FK-binding protein 12 (FKBP12). This complex forms a PPI with the mTOR complex 1, thus inhibiting its activity (Driggers et al. 2008). Rapamycin/sirolimus’ anticancer derivatives temsirolimus and everolimus work via the same mechanism as well. Similarly, the macrocyclic natural product cyclosporine A, an immunosuppressant, forms a complex with cyclophilin A, which forms a PPI with calcineurin. This complex inhibits the action of calcineurin, which when not inhibited activates the expression of IL-2 (Driggers et al. 2008).
Macrocycles demonstrate drug-like physicochemical properties with respect to factors such as solubility and lipophilicity. They display oral bioavailability, metabolic stability and good pharmacokinetic and pharmacodynamic properties (Driggers et al. 2008; Alihodzić et al. 2018). However, macrocyclic natural products have not received much attention in addressing challenging PPI targets and in solving such pressing problems as the need for new drugs to combat microbial antibiotic resistance (Alihodzić et al. 2018). The emergence of strategies for the synthesis of macrocycle-rich chemical libraries such as B/C/P and the recent breakthrough in respect of the synthesis of organic compounds, such as olefin metathesis, have put macrocycles within easier reach of medicinal chemists (Lee and Grubbs 2001; Yu et al. 2011). The synthesis of macrocyclic compounds via DNA-programmed chemistry (DPC) technology has also beneficiated the PPI-focused drug technology (Franzini and Randolph 2016; Zhou et al. 2018). DPC allows the control of high-fidelity chemical reactions needed for synthesis of the desired libraries through tagging specific reactants with hybridised DNA molecules. Once libraries are synthesised, screening for biological activity via affinity selection follows. The DPC strategy has led to the discovery of macrocycles, such as E-32712, that disrupt the interaction of tumour necrosis factor (TNF) with its receptor, TNFR (Drahl 2009; Franzini and Randolph 2016; Zhou et al. 2018).
These new technologies, combined with cellular screening assays (including high-content screening), help to discover and design PPI modulators. Cellular assays help to assess the activity of whole intracellular pathways or portions of pathways to identify compounds that inhibit these pathways via disruption of key PPIs (Mella et al. 2018; Booij et al. 2019). Two examples of cellular screening assays are ligand signal transducers and activators of transcription (STATs) technology and BioImage Redistribution technology. Ligand STATs technology takes advantage of the activation of the intracellular signal transduction pathway through STATs and high-throughput fluorescent reporter assay (Pándy-Szekeres et al. 2018), whereas BioImage redistribution technology focuses on pathways that involve intracellular translocation of a signalling protein from one intracellular compartment to another, such as from the cytoplasm to the nucleus or vice versa (Lage et al. 2018). Taken together, these strategies help to discover and design PPI modulators. Potent PPI modulators have already been developed and some potential drug-like candidates have already reached clinical production.
The challenges of targeting protein-protein interactions
Undruggability of PPIs
PPIs represent challenging targets for small-molecule drugs that have the potential to become available orally (Zhang et al. 2018). They have highly dynamic and expansive interfacial areas of approximately 1500–3000 Å, compared with those involved in interactions between proteins and small molecules, which are approximately 300–1000 Å (Gonzalez and Kann 2012; Robertson and Spring 2018). PPIs’ contact surfaces are usually flat, featureless and relatively large. They are deficient in the kind of cavities present in the surfaces of proteins that bind to small-molecule ligands. Previously, PPIs were regarded as prototypically ‘intractable’ and ‘undruggable’ (Robertson and Spring 2018; Zhang et al. 2018). PPIs do not have natural small-molecule ligands; hence, these ligands cannot be used to initiate design of drug molecules. The contact surface area in PPIs often involves ramified amino acid residues whose sequences in the polypeptide chain are not juxtaposed. The amino acid residues are only augmented through the three-dimensional folding state of the native protein (Jochim and Arora 2010). This makes it impossible to use short peptide chains derived from the protein structure as starting points for the design of peptidomimetic drugs. Furthermore, high-throughput screening using combinatorial libraries rarely identifies compounds that address PPIs (Haberman 2012).
Target validation and druggability
In PPI-focused drug technology, target validation and druggability have been the focal points in selecting targets for drug discovery (Feng et al. 2017). Target validation refers to a process of determining that a target is critically involved in a disease pathway and that modulating the target with a drug is likely to have a positive therapeutic effect (Modell et al. 2016; Feng et al. 2017). However, druggable targets refer to biomolecules that can be modulated with drugs, usually using well-proven drug discovery science and technology aimed at developing both large-molecule and small-molecule drugs (Modell et al. 2016). Over the years, large-molecule drugs or biologics have been the fastest-growing and most successful class of biologics. Examples include monoclonal antibody (mAb) drugs and recombinant proteins (Sinha et al. 2012). Most of these drugs are involved in PPIs and are indicated for oncology and inflammatory diseases. They address appropriate targets, such as cell-surface receptors (e.g. HER2 in breast cancer and CD20 in non-Hodgkin’s lymphoma) and cytokines (e.g. TNF-alpha (TNF-α) in inflammatory diseases such as rheumatoid arthritis) (Allison 2009; Feng et al. 2017).
Large-molecule biologics circumvent the druggability problems encountered when developing small-molecule drugs in PPI-focused drug technology (Wan 2016). However, they have limitations and setbacks. Large-molecule biologics do not address intracellular targets, are usually not orally available and are administered parenterally (Debouck and Metcalf 2000). Thus, small-molecule drugs have been synthesised to curb these challenges. Small-molecule drugs are orally available, less expensive, easy to administer and convenient for patients. The discovery of small-molecule drugs typically entails the use of medicinal and combinatorial chemistry, in some cases augmented by structure-based drug design (Debouck and Metcalf 2000; Wan 2016). The central goal of medicinal chemistry was to design and select small-molecule compounds that have ‘drug-like properties’ with good absorption, distribution, metabolism and elimination properties needed for orally deliverable drugs. Hence, the ‘Rule of Five’, a set of parameters that predicts a compound’s solubility and permeability, was often used (Lipinski et al. 2012). However, natural products and some small-molecule drugs did not fit the Rule of Five criteria. This led to the ascendancy of two waves of technology-driven drug discovery techniques: combinatorial chemistry combined with HTS and genomics-driven drug discovery (Debouck and Metcalf 2000; Wan 2016).
Combinatorial chemistry produces libraries of small organic compounds, which are subjected to HTS to discover compounds that address targets derived from genomics (Taylor et al. 2018). Once an active skeleton has been identified, combinatorial chemistry is a superb technology for optimising the structures of lead compounds. The use of combinatorial chemistry and HTS permits the examination of large numbers of compounds in a short time, implying that a few drugs could reach the market and achieve blockbuster status (Li and Vederas 2009; Taylor et al. 2018). Moreover, emphasis have been on organic compounds that can be synthesised using combinatorial chemistry because they are more amenable to use in HTS and have relatively simpler structures than natural products. However, in the majority of cases, combinatorial libraries of these synthetic organic compounds are based on structural modification of existing drugs, thus rendering other classes of drug targets such as PPIs ‘hard targets,’ ‘challenging targets,’ or simply ‘undruggable’ (Bauer et al. 2010). Hence, approximately 10–14% of human proteins are druggable according to the current libraries of drug-like molecules. The use of current combinatorial libraries of synthetic organic molecules for de novo drug discovery has been sorely lacking (Newman and Cragg 2007).
In contrast to combinatorial libraries of synthetic compounds, natural products play a crucial role in the discovery of drug leads. Current synthetic libraries are indirectly based on natural products (Bauer et al. 2010; Wan 2016). Thus, the envisioned governing paradigm of medicinal and combinatorial chemistry is to develop new libraries based on natural product scaffolds that are underrepresented in current libraries. Approximately 83% of small natural product scaffolds and 20% of small metabolite scaffolds are not represented in commercially available libraries. These underrepresented scaffolds may enable to address what are now considered ‘undruggable,’ ‘hard’ or ‘challenging’ targets (Haberman 2012). Hence, drugs or drug candidates that modulate PPIs (for example natural products or natural product-like compounds), screening natural product libraries and synthesising natural product-like compounds remain one approach to discovering drugs that address PPIs (Bauer et al. 2010; Haberman 2012; Wan 2016).
Targeted protein-protein interactions
Cell-surface receptors
Many cell-surface receptors modulate their physiological functions through PPIs and these include receptors for cytokines, chemokines, growth factors and integrins. Integrins bind to extracellular matrix proteins or to members of the immunoglobulin superfamily, such as intercellular adhesion molecule-1 (ICAM-1) and Fc receptors (which bind to the Fc regions of antibodies) (Guidolin et al. 2019; Husain et al. 2019). Large-molecule drugs such as recombinant proteins and mAbs target many of the cell-surface receptors. These include biologic inhibitors of VEGF and TNF, and recombinant versions of proteins that are ligands for cell-surface receptors such as erythropoietin and granulocyte colony-stimulating factor (G-CSF) (Parveen et al. 2019). Erythropoietin is used for cancer chemotherapy and the treatment of anaemia associated with dialysis undertaken in the treatment of chronic renal failure (Fecková et al. 2019). G-CSF is used for the treatment of neutropenia associated with bone marrow transplantation, cancer chemotherapy and increasing the number of blood haematopoietic stem cells in haematopoietic stem cell transplantation (Zhao et al. 2019). In cases where developing biologic drugs to modulate such cell-surface receptors has not been possible, small-molecule receptor modulators are orally available, less expensive and safer than the corresponding biologics (Parveen et al. 2019). The development of small-molecule drugs that modulate these receptors is challenging; however, some potential drugs have already reached clinical production (Fecková et al. 2019; Zhao et al. 2019). The modulators of cell-surface receptors that interact with proteins or peptides other than chemokine receptors are listed in Table 1.
Cytokine receptors
Most small-molecule PPI modulators of cytokine receptors are PPI inhibitors or antagonists. Using the proprietary cellular screening assay, STATs technology, small-molecule cytokine mimetics that are agonists have been developed (Silva et al. 2019). These include SB-247464, a small-molecule non-peptide mimetic of murine G-CSF (Tian et al. 1998). G-CSF (like many other cytokines and growth factors) binds to the extracellular domains of its receptors and dimerises them, resulting in the activation of intracellular signalling through a pathway that leads to the phosphorylation of members of the STAT class of signalling proteins. The phosphorylated STATs form homodimers, which are translocated into the nucleus and bind to a synthetic STAT-responsive promoter that drives expression of a fluorescent reporter gene (Tian et al. 1998; Zhao et al. 2019). Screened small organic compounds selected SB-247464 (shown in Fig. 3a). In support of this model, SB-250017, a related non-symmetrical compound was developed. However, it can bind to the extracellular domain of the receptor but cannot dimerise it. Hence, SB-250017 acts as an antagonist of SB-247464 but has no effect on the activation of the receptor by G-CSF (Haberman 2012).
Chemical structure of a SB-247464—a small-molecule cytokine non-peptide mimetic agonist of murine G-CSF developed using the proprietary cellular screening assay, STATs technology (figure taken from Grosdidier et al. 2009); and b eltrombopag—a synthetic small-molecule TPO agonist approved for the treatment of idiopathic thrombocytopenic purpura (figure taken from Susanto 2015)
More so, STATs technology has been used to discover small-molecule non-peptide agonists of the human thrombopoietin (TPO) receptor (Erickson-Miller et al. 2005). Optimisation of their initial compound, SB-394725, led to the development of eltrombopag (SB-497115-GR). The small-molecule TPO agonist eltrombopag (shown in Fig. 3b) provides an alternative to recombinant human TPO, which can elicit anti-endogenous TPO antibodies in some patients, resulting in profound thrombocytopenia (Li et al. 2001). Eltrombopag is the only synthetic small-molecule direct (i.e. not allosteric) PPI modulator to reach the market. It is approved for the treatment of the rare disease, idiopathic thrombocytopenic purpura. Eltrombopag is also being tested in phase III clinical trials to treat low platelet count in patients with liver cirrhosis due to hepatitis C and in phase II trials in oncology. GSK2285921 (follow-on compound to eltrombopag) is being tested in phase II clinical trials in oncology. With respect to oncology, SB-559457 (a related compound to eltrombopag) was found to be specifically toxic to primary human myeloid leukaemia cells in culture (Kalota and Gewirtz 2010).
More recently, small-molecule agonists of the human receptors for erythropoietin (EPO) and G-CSF were discovered using STATs technology (Miller et al. 2015). STATs cellular assay technology is good for discovering such compounds, provided the chemical libraries screened in these assays contain viable hits. Research has discovered a lead series of small-molecule, selective EPO receptor agonists (including LG5640) that display partial efficacy compared with recombinant human EPO in several models of EPO-induced erythropoiesis (Haberman 2012; Miller et al. 2015; Zhao et al. 2019).
Integrins
The integrin superfamily of proteins consists of cell-surface receptors that mediate attachment between cells and either the extracellular matrix or other cells (Takada et al. 2007). Among these proteins is the leukocyte integrin lymphocyte function-associated antigen 1 (LFA-1). LFA-1 is found on such leukocytes as T cells, B cells, macrophages and neutrophils, and is involved in recruitment to sites of infection or inflammation. LFA-1 on T cells binds to the immunoglobulin superfamily member ICAM-1 on, for example, antigen-presenting cells and endothelial cells (Dustin et al. 2004). The LFA-1/ICAM-1 PPI plays an important role in processes such as T cell activation, T cell homing to peripheral lymphoid organs and sites of inflammation (Dustin et al. 2004; Graff et al. 2008). This PPI is thus a target for the discovery of drugs to treat inflammatory conditions. Using a tethering-based FBDD platform, compounds that potently inhibit both human T cell migration and T cell activation by disrupting LFA-1/ICAM-1 PPIs have so far been discovered (Haberman 2012; Zhong et al. 2012).
One of the compounds (SAR1118) showed good pharmacokinetic properties and oral availability in rodents and inhibited neutrophil migration in a murine peritonitis model (Zhong et al. 2012). A phase I clinical trial (conducted in 2008) and phase II clinical trial (conducted in 2011) of SAR1118 observed that it was safe, well tolerated and demonstrated statistically significant improvements in tear production and visual function. A phase III study of SAR1118 observed positive ophthalmic solution in the treatment of dry eye syndrome. In addition to dry eye, SAR1118 will be tested in a broad range of ocular inflammatory conditions, including diabetic macular oedema (Haberman 2012; Zhong et al. 2012).
Chemokine receptor
Chemokines and their receptors are attractive drug targets because of their role in inflammatory diseases. Chemokines induce chemotaxis. They are members of a family of small cytokines. Chemokine receptors are members of the G protein-coupled receptor (GPCR) superfamily. They bind to natural small-molecule ligands (Guidolin et al. 2019; Husain et al. 2019). Numerous small-molecule drugs that are competitive inhibitors of these ligands already exist. GPCR antagonists represent the largest class of drugs produced (Miszta et al. 2018; Husain et al. 2019). Chemokine receptors bind to chemokines (small proteins) and thus represent a class of PPIs. Discovering small molecules that directly inhibit chemokine receptors at their chemokine binding sites has been an insurmountable task, presenting difficulties in discovering PPI modulators. However, discovering small-molecule chemokine receptor antagonists that act via an allosteric mechanism is easier, because GPCRs exert their signalling activities via complex ligand-mediated conformational changes, which may be a particularly ‘allosteric’ class of proteins (Miszta et al. 2018). Natural ligands and drugs that bind to ‘orthosteric’ sites on GPCRs induce unique GPCR conformational states that activate a discrete subset of signalling pathways and cellular behaviours. Since allosteric modulators of GPCRs work by causing conformational changes in the structures of these proteins, some of them may also give rise to functional selectivity in the actions of orthosteric natural ligands that co-bind to the GPCR (Conn et al. 2009; Husain et al. 2019; Miszta et al. 2018).
In the case of GPCRs—including chemokine receptors—whose natural ligands are peptides or proteins, allosteric sites (defined as binding sites for known allosteric modulators of these receptors) are located in distinct sites on the receptor proteins from the orthosteric peptide binding sites (Conn et al. 2009; Miszta et al. 2018). Chemokines bind specifically to orthosteric sites that are located in the extracellular domains of their receptors. Allosteric sites on chemokine receptors are located in transmembrane domains that are distant from the chemokine binding sites (Conn et al. 2009). Small-molecule allosteric modulators that bind to these sites were developed via fairly standard medicinal chemistry and high-throughput screening, augmented with structure-based drug design. Thus, although orthosteric binding sites on chemokine receptors (and on other GPCRs that have peptide ligands) have proven so far to be intractable for the discovery of small-molecule modulators, the discovery of drug-like small-molecule allosteric modulators of chemokine receptors is much more feasible (Conn et al. 2009; Haberman 2012; Miszta et al. 2018; Husain et al. 2019).
However, the discovery of small-molecule chemokine receptor antagonists has been a challenging task. Most of the agents that have been entered into clinical trials have failed, purely because the diseases addressed by these compounds have complex biology and, poor predictive animal models and target redundancy was used (Horuk 2009; Haberman 2012; Miszta et al. 2018). So far, only two small-molecule chemokine antagonists have entered the market. One example is the allosteric CCR5 antagonist maraviroc for the treatment of HIV/AIDS (shown in Fig. 4a). The other is the CXCR4 inhibitor plerixafor (shown in Fig. 4b). Plerixafor is a partial antagonist of the chemokine receptor CXCR4 and an allosteric agonist of CXCR7 (Kalatskaya et al. 2009). It is used together with G-CSF to mobilise haematopoietic stem cells to the peripheral blood for autologous transplantation in patients with non-Hodgkin lymphoma and multiple myeloma (Davies et al. 2007) (Table 2).
Chemical structure of a maraviroc—a small-molecule allosteric CCR5 chemokine antagonist used for the treatment of HIV/AIDS (figure taken from Xu et al. 2014); and b plerixafor—a partial antagonist of the chemokine receptor CXCR4 and an allosteric agonist of CXCR7. It is used for autologous transplantation in patients with non-Hodgkin lymphoma and multiple myeloma (figure taken from Venkata Narasimha Rao et al. 2017)
TNF/TNFR PPI
TNF/TNFR PPIs were previously regarded as intractable. However, macrocycles that block the interaction of TNF with its receptor, TNFR, were discovered (Drahl 2009; Parveen et al. 2019). The TNF/TNFR PPIs are involved in numerous inflammatory diseases such as rheumatoid arthritis, and TNF is the target of several large-selling biologic TNF inhibitors. E-32712 is one example of an orally active TNF/TNFR inhibitory macrocycle discovered so far (Drahl 2009; Parveen et al. 2019).
Intracellular signalling pathways
Signal transduction is a process by which extracellular signals mediate changes within a cell via intracellular signalling pathways (Miszta et al. 2018; Husain et al. 2019). It begins when an extracellular signalling molecule activates a cell-surface receptor or an intracellular receptor. Once activated, receptors mediate changes in intracellular target molecules and subsequently initiate cascades of molecular changes through pathways. The end result is a physiological response such as cellular differentiation, cell growth, cell proliferation, secretion of signalling molecules (such as growth factors or cytokines), cellular motility, cellular adhesion or apoptosis. Signal transduction and intracellular signalling pathways are also fundamental in pathophysiological cues and disease progression. They often become dysregulated in metabolic diseases, immune diseases, cancer and many other diseases (Conn et al. 2009; Miszta et al. 2018; Husain et al. 2019).
Studies have targeted tractable signalling receptors such as GPCRs, nuclear receptors, growth factor receptors and cytokine receptors as drug targets (Miszta et al. 2018). The emergence of kinase inhibitors has been another very significant breakthrough. It began with the discovery of imatinib and led to the discovery of other inhibitors aimed at treating different types of cancer (Bhullar et al. 2018; Ferguson and Gray 2018). However, many intracellular signal transduction pathways remain inaccessible because they are driven by key components that have so far been intractable. Most important are the ‘undruggable’ PPIs, which are key components of all signalling pathways, for example PPIs between transcription factors and multicomponent protein complexes that are key mediators of intracellular signalling. Small-molecule modulators of PPIs that target signal transduction pathways are yet to be developed and there have been a keen interest in targeting these ‘undruggable’ targets (Ferguson and Gray 2018; Miszta et al. 2018; Husain et al. 2019).
Tcf/β-catenin transcription factor complex
Small-molecule inhibitors of the oncogenic Tcf/β-catenin transcription factor complex that is central to the Wnt pathway have been discovered (Lepourcelet et al. 2004; Yan et al. 2017; Jeong et al. 2018). The Wnt pathway is dysregulated in several types of cancer, which include hepatocellular carcinoma, multiple myeloma (MM), B cell chronic lymphocytic leukaemia (B-CLL) and colorectal cancer. Due to the central role of the Wnt pathway in these cancers, PPIs that are critically involved in the pathway have been targeted. One example is the ‘destruction complex’, a multicomponent cytoplasmic protein complex that includes among others the proteins adenomatous polyposis coli (APC) and glycogen synthase kinase 3 (GSK-3). Under normative cellular functions (when the ‘destruction complex’ is intact), GSK-3 phosphorylates β-catenin, a multifunctional protein that is involved both in signal transduction and in intercellular adhesion. This phosphorylation targets β-catenin for degradation in the cytoplasm (Yan et al. 2017; Jeong et al. 2018).
However, when the ‘destruction complex’ is disrupted via signalling from Wnt family ligands bound to their cell-surface receptor, β-catenin accumulates in the cytoplasm and moves into the nucleus. There, it binds to transcription factors of the T cell factor (Tcf) family, including Tcf4, the major Tcf expressed in stem cells of the gut and in colorectal cancer. In the absence of β-catenin, Tcf proteins are transcriptional repressors (Lepourcelet et al. 2004; Yan et al. 2017). β-Catenin binding changes Tcf proteins from repressors into transcriptional activators that activate a set of downstream genes, including the oncogene c-Myc and the cell cycle protein cyclin D1. In precancerous colonic adenomas or colorectal cancers, APC is often mutated and no destruction complex forms. This results in constitutive stabilisation of β-catenin, which can freely move into the nucleus and bind to Tcf4. In the case of other cancers caused by dysregulation of the Wnt pathway, β-catenin also becomes stabilised, via other genetic changes that do not involve ACP (Yan et al. 2017; Jeong et al. 2018).
Several small-molecule inhibitors of the human Tcf/β-catenin PPI have been developed through structural and mutagenesis studies of the PPI (which identified a hotspot), followed by assay development and screening of natural product libraries (Lepourcelet et al. 2004). Of the tested compounds, two fungal derivatives, PKF115-584 and CGP049090, gave the best results in all the assays and were tested in preclinical studies. However, the molecular mechanisms by which the compounds act on this PPI remain elusive and are not yet clear. Hence, studies have been focussed on other means to target the Wnt pathway by performing chemical genetic studies to identify novel targets that modulate the pathway (Huang et al. 2009).
BCL6/SMRT PPI in B cell lymphoma
Diffuse large B cell lymphoma (DLBCL) is the most common type of non-Hodgkin’s lymphoma and accounts for about 30% of all lymphomas (Friedberg 2011). B cell lymphoma 6 (BCL6) acts as an oncogene in the majority of cases of DLBCLs (Compton and Hiebert 2010). The BTB domains of BCL6 interact with a co-repressor known as SMRT (silencing mediator for retinoid or thyroid-hormone receptors). SMRT in turn facilitates the recruitment of histone deacetylase 3 (HDAC3) to the DNA promoters bound by BCL6. This results in the repression of the genes controlled by the promoters, via removal of acetyl groups from histones of chromatin (Compton and Hiebert 2010; Friedberg 2011).
In normal lymphoid germinal centre B cell development, immunoglobulin genes undergo recombinations and somatic mutations to generate antibody diversity. Despite this genomic instability, germinal centre B cells are able to undergo rapid proliferation because BCL6 represses a set of genes that regulate the DNA damage response and cell cycle checkpoints (Compton and Hiebert 2010). Among these genes are CHK1 (checkpoint kinase 1), cyclin-dependent kinase inhibitor 1 (CDKN1A), ATR (ataxia telangiectasia and Rad3-related protein) and TP53 (which codes for p53). Once B cell clonal diversity has been achieved, BCL6 expression is downregulated. This allows restoration of cell cycle checkpoints, normal DNA damage control and B cell differentiation and maturation. Oncogenic overexpression of BCL6 through chromosomal translocation, gene amplification or promoter mutation results in continued B cell progenitor proliferation and acquisition of additional mutations. This results in an aggressive B cell lymphoma (Compton and Hebert 2010; Friedberg 2011; Haberman 2012).
Cerchietti and co-workers (Cerchietti et al. 2010) discovered small-molecule antagonists of the PPI between BCL6 and SMRT, and demonstrated that these compounds could kill DLBCL cells in vitro. Using computer-aided drug design (CADD) (to first identify putative small-molecule binding sites), screening by virtual docking of the compounds into the putative binding site and selection based on maximising chemical diversity and Lipinski’s Rule of Five, a lead compound whose reproducibly inhibited BCL6/SMRT PPI in DLBCL was designed (Cerchietti et al. 2010). Furthermore, they performed structural and mutagenesis studies (which identified a hotspot), performed CADD, and used their models for virtual screening of 1,000,000 commercially available compounds. Compound selection was based on chemical diversity, drug-likeness, immediate commercial availability and the ability to block BCL6-mediated transcriptional repression in a cellular assay (Haberman 2012).
A lead compound designated ‘79-6’ (PubChem CID5721353, shown in Fig. 5a), which specifically killed BCL6-positive lymphoma cell lines and BCL6-positive tumour cells in xenograft models was identified. However, the five-membered ring of 79-6 contains sulphur, which is prone to oxidation and may consequently result in loss of efficacy, hence the need to optimise 79-6 in order to develop a clinical candidate for BCL6-targeted therapy (Cerchietti et al. 2010). Furthermore, epigenetic regulation was identified as a potentially important area of opportunity for drug discovery (Haberman 2012). So far, two inhibitors of class I HDACs have been approved for the treatment of cutaneous T cell lymphoma—vorinostat (Zolinza) and romidepsin (Istodax). Other agents are being tested in clinical trials for various types of cancer. However, the mechanisms by which these compounds work, and why they appear to be active against certain cancers but not others and not normal cells, remain elusive (Cerchietti et al. 2010; Haberman 2012).
Chemical structure of a ‘79-6’ (PubChem CID5721353)—a target-specific lead compound known to kill BCL6-positive lymphoma cell lines and BCL6-positive tumour cells in xenograft models (figure taken from Yasui et al. 2017) and b FMP-API-1—a 3′,3-diamino-4,4′-dihydroxydiphenylmethane ‘drug-like’ compound identified to disrupt the AKAP18δ/PKA PPIs via an allosteric mechanism with a micromolar dissociation constant (figure taken from Christian et al. 2011)
HDAC inhibitors have been studied in clinical trials against DLBCL as single agents, but the results have been disappointing (Compton and Hiebert 2010). BCL6/SMRT PPI antagonist in combination with rituximab (which targets the B cell-specific cell-surface protein CD20) is a potential agent to treat DLBCL (Haberman 2012). Currently, rituximab is used in combination with chemotherapy to treat DLBCL. However, there is still a large proportion of patients with unfavourable prognosis. Hence, a drug that specifically targets the BCL6/SMRT PPI, perhaps used in combination with rituximab, may address the unmet medical needs and reduce the need for toxic chemotherapy in DLBCL patients. The discovery and development of inhibitors of the BCL6/SMRT PPI has gained wider interest in the treatment of DLBCL (Compton and Hiebert 2010; Haberman 2012).
AKAP-protein kinase A interaction
Scaffolding proteins are a tremendously important means by which the cell organises signal transduction pathways. They form PPIs with the signalling proteins (Conn et al. 2009). Thus, discovering small-molecule inhibitors of these PPIs is a potential strategy for targeting a wide array of signalling pathways (Miszta et al. 2018; Husain et al. 2019). A-kinase anchoring proteins (AKAPs) (also known as cAMP-dependent protein kinases) are scaffolding proteins that tether protein kinase A (PKA) and other signalling proteins to specific intracellular sites. They are a family of serine/threonine kinases whose activity is dependent on cellular levels of cyclic AMP (cAMP). Thus, the tethering of PKA via PPIs with an AKAP results in the compartmentalisation of cAMP signalling within the cell (Christian et al. 2011).
Small-molecule inhibitors of the AKAP/PKA interaction have been developed for the potential treatment of chronic heart failure. AKAP18δ is an AKAP isoform that serves as a scaffold for organising the adrenaline-beta-adrenoreceptor-cAMP-PKA signalling pathway in cardiac muscle (Lygren and Taskén 2008). Targeting AKAP18δ and its PPIs with PKA is a potential approach to modulate the cardiac myocyte cAMP-PKA system (Christian et al. 2011). Hence, research efforts have been centred on developing a screening assay for the disruption of the AKAP18δ/PKA PPIs by screening a library of over 20,000 ‘drug-like’ compounds. This led to the identification of 3′,3-diamino-4,4′-dihydroxydiphenylmethane, designated FMP-API-1 (shown in Fig. 5b), which disrupt the AKAP18δ/PKA PPIs via an allosteric mechanism with micromolar dissociation constant (Christian et al. 2011). In the meantime, FMP-API-1 might be used as a tool compound to investigate the function of AKAP-PKA PPIs in cells and in animal models. However, higher affinity drug-like small-molecule AKAP/PKA PPI antagonists are still sought. Thus, scaffolding proteins provide numerous opportunities for targeting by small-molecule PPI modulators (Miszta et al. 2018; Husain et al. 2019).
The ubiquitin system
The ubiquitin (Ub) system is a vital regulatory system based on covalently linking the small (8.5 kDa) regulatory protein, Ub, to numerous specific protein targets in all eukaryotic cells (Cohen and Tcherpakov 2010; Varshavsky 2017; Wertz and Wang 2019). This system is involved in functions such as regulation of intracellular protein turnover, mitosis, innate immunity and certain protein kinases and other enzymes (Cohen and Tcherpakov 2010). The ubiquitinylation cascade is a potential target for the development of specific smart drugs. In protein degradation the Ub system works together with the proteasome in a pathway known as the Ub proteasome system (UPS) where Ub is used to tag proteins for degradation by the proteasome (Cohen and Tcherpakov 2010; Varshavsky 2017). The ubiquitinylation pathway is complex and involves several levels of mediators, which include Ub activators (E1), Ub-conjugating enzymes (E2) and Ub ligases (E3) (as shown in Fig. 6). In this pathway, Ub moves from E1s to E2s. E3s interact with ubiquitinylated E2s and substrate proteins via PPIs where Ub is transferred from E2 to the substrate. This Ub moiety is recycled when the cycle is complete, resulting in tagging of substrates with polyubiquitin chains. In humans, there are about 10 E1s, 40 E2s and over 600 E3s (Varshavsky 2017; Wertz and Wang 2019).
A schematic illustration of the Ubiquitin-proteasome pathway (UPP). Ub is conjugated to proteins that are destined for degradation by an ATP-dependent process that involves three enzymes. A chain of five Ub molecules attached to the protein substrate is sufficient for the complex to be recognised by the 26S proteasome. In addition to ATP-dependent reactions, Ub is removed and the protein is linearised and fed into the central core of the proteasome, where it is digested to peptides. The peptides are degraded to amino acids by peptidases in the cytoplasm or used in antigen presentation (figure taken from Haq and Ramakrishna, 2017)
The Ub system is a virtually untapped area of opportunity for drug discovery and development. Drugs that target the UPS and the ubiquitinylation pathway itself are in clinical development (Wertz and Wang 2019). Examples include bortezomib, MLN4924 (which inhibits a pathway that activates one class of E3s), the Cullin RING E3 ligases (CRLs) and CC0651 (an allosteric modulator that inhibits the ubiquitinylation activity of an E2 that interacts with CRLs) (Appel 2011; Varshavsky 2017; Wertz and Wang 2019). Bortezomib targets protease activity of the proteasome. It blocks proteasomal disposal of all ubiquitinylated proteins in the cell. However, bortezomib remains a useful drug in MM, where blocking of the proteasome results in an overload of damaged proteins, which subsequently destroys the cell via apoptosis (Appel 2011). Based on bortezomib mechanism of action, it is highly nonspecific and has severe adverse effects. As a result, developing second-generation proteasome inhibitors, which include MLN9708 and MLN4924 became a necessity. MLN9708 is an oral drug, more specific and with fewer side effects than bortezomib. MLN9708 is being tested in phase I and phase II trials in MM patients (Appel 2011).
MLN4924 is another drug in clinical trials that targets an arm of the ubiquitinylation cascade itself (Deshaies 2009; Soucy et al. 2009). It is an AMP analog that inhibits NEDD8-activating enzyme (NAE). NEDD8 is a Ub-like protein and NAE is an E1 for neddylation. Inhibition of neddylation by MLN4924 at the E1 (NEDD8-activating) step thus indirectly inhibits one class of E3s, the CRLs, without the need to inhibit the PPIs between members of this class of E3s and the substrate proteins that bind to them, or the need to inhibit the catalytic activity of E2s or the PPIs between E2s and the CRL E3s. MLN4924 is thus much more specific than bortezomib in inhibiting the Ub system. However, it is not very specific with respect to targeting proteins for degradation. MLN4924 has potentially important anti-tumour effects and induces apoptosis. MLN4924 is now being tested in phase I clinical trials in patients with solid and haematological tumours (Deshaies 2009; Soucy et al. 2009).
Another strategy to inhibit CRLs indirectly, this time at the E2 level, was developed (Ceccarelli et al. 2011). This led to the discovery of CC0651, an allosteric inhibitor of the E2 Ub-conjugating enzyme Cdc34. CC0651 analogs inhibited proliferation of human cancer cell lines. They also caused accumulation of the CDK inhibitor p27Kip, which led to uncontrolled DNA synthesis in the S-phase of the cell cycle, leading to DNA damage and induction of apoptosis. The E2 inhibitor CC0651 and its analogs appear to be more specific than MLN4924, which inhibits the activity of all CRLs. However, the target of CC0651 still ubiquitinylates hundreds of substrate proteins, so it is relatively nonspecific (Ceccarelli et al. 2011; Haberman 2012).
The best agents that target the Ub system are the PPI modulators that target E3s, which interact with their substrates via PPIs (Lecker et al. 2006; Haberman 2012). However, because of the intractability of PPIs, the development of specific agents that target the Ub system has been a major bottleneck. Research efforts have led to the development of an agent that targets one E3, the human homolog of mouse double minute 2 (MDM2) protein. HDM2 interacts with p53 via a PPI (as shown in Fig. 7) (Moll and Petrenko 2003). The p53 is often referred to as the ‘guardian of the genome’, controls pathways that respond to DNA damage or other stress signals by blocking cell proliferation by inducing either DNA repair or apoptosis. The p53 is mutated or inactivated in nearly all human cancers, which allows the uncontrolled proliferation of cancer cells, rendering them resistant to cytotoxic chemotherapy (Moll and Petrenko 2003). In approximately half of human cancers, p53 is inactivated via mutation. In the other half, p53 remains unmutated but is inactivated. The main means of inactivation is via HDM2, which is overexpressed in the majority of cancers with wild-type p53. HDM2 regulates p53 in three ways: inhibition of p53-induced transcription, promotion of export of p53 out of the nucleus and inducing p53 degradation by the proteasome (Moll and Petrenko 2003). The last two activities mentioned both involve HDM2’s E3 Ub ligase activity, thus presenting a potential target for PPI-focused drug development (Lecker et al. 2006; Haberman 2012; Varshavsky 2017; Wertz and Wang 2019).
A schematic diagram showing HDM2/p53 PPI. MDM2 and p53 form an auto-regulatory feedback channel. p53 stimulates the expression of MDM2; MDM2 in turn inhibits p53 activity because it stimulates its degradation in the nucleus and the cytoplasm, blocks its transcriptional activity and promotes its nuclear export. A broad range of DNA-damaging agents or deregulated oncogenes induces p53 activation (figure taken from Carry and Garcia-Echeverria 2013)
Currently, there are two leading drug candidates that specifically disrupt the HDM2/p53 PPI (Shangary and Wang 2009). The most advanced compound, now being tested in phase I clinical trials, is RG7112, is an analog of nutlin-3a (Vassilev et al. 2004; Shangary and Wang 2009; Cheok et al. 2011). Nutlin-3a is the active enantiomer isolated from racemic nutlin-3, which have been studied in various cell culture and preclinical animal models, as a monotherapy and in combination therapies (Shangary and Wang 2009; Cheok et al. 2011; Crane et al. 2015). These studies showed that nutlin-3 potently induced apoptosis in cell lines derived from such haematologic cancers as acute myeloid leukaemia, acute lymphoblastoid leukaemia, MM and B-CLL. These haematologic tumours and other HDM2/p53 PPI-disrupting agents are potential targets for treatment with nutlin-3, since they exhibit a high percentage of unmutated TP53 at diagnosis. These studies concur with the development of nutlins in cancer, either as single agents or in combination therapies. They led to the entry of the nutlin-3 analog RG7112 into phase I clinical trials in haematologic malignancies and advanced solid tumours (Haberman 2012).
The other compound is MI-219, analogs of which are currently being tested in advanced preclinical studies (Shangary et al. 2008; Shangary and Wang 2009). MI-219 binds to HDM2 with an inhibition constant (Ki) of 5 nM. It is designed to mimic not only phenylalanine 19, tryptophan 23 and leucine 26 in the p53 binding site for HDM2 but also a fourth residue, leucine 22, which appears to play an important role in the HDM2/p53 PPI as well (Shangary and Wang 2009). MI-219 has good pharmacological properties, a high degree of specificity for MDM2 and induced accumulation of p53, as well as inhibiting the growth of cancer cell lines with wild-type p53 with submicromolar IC50 values (Shangary and Wang 2009; Cheok et al. 2011). MI-219 analogs have also been tested and have demonstrated activity in combination therapies with etoposide, doxorubicin and cisplatin in mouse models of lung cancer, rhabdomyosarcoma and pancreatic cancer. These agents were found to have minimal toxic effects in normal cells. Given the large number and specificity of E3 Ub ligases and their important role in intracellular pathways, there is a large field of possibility for discovery of novel PPI modulators that target these biomolecules and their interactions with their substrates (Shangary and Wang 2009; Cheok et al. 2011).
JNJ-26854165, a novel tryptamine derivative is another compound in phase I clinical trials in advanced or refractory solid tumours. JNJ-26854165 is thought to be a possible HDM2/p53 inhibitor; however, it appears to work via a different mechanism of action that does not involve HDM2 (Kojima et al. 2010). It appears to work via accelerating the proteasomal degradation of p21 and to antagonise the p53-mediated transcriptional induction of p21. JNJ-26854165 also induces apoptosis in tumour cells with mutant p53 via inducing delay in the S-phase of mitosis and upregulation of expression of the transcription factor E2F1 (a key mediator of an important pathway that controls cellular proliferation). This results in apoptosis, preferentially of S-phase cells. JNJ-26854165 has similar effects on tumours to HDM2/p53 PPIs like the nutlins and MI-219 (Cheok et al. 2011; Haberman 2012).
Apoptosis regulators
Apoptosis is an ATP-dependent pathway of programmed cell death in all-multicellular animals (Danial and Korsmeyer 2004; Wyllie 2010). It is essential for normal embryonic development and for maintaining normal cellular homeostasis in adults, as well as for response to infectious agents. Apoptosis is dysregulated in several major diseases. Cancer is the major focus of studies seeking to develop drugs that modulate apoptotic pathways, since apoptosis is blocked in perhaps all cancers. This is a significant target in uncontrolled cellular proliferation in cancer (Danial and Korsmeyer 2004; Wyllie 2010). An illustration of apoptosis in ovarian cancer and some of the targeted therapeutic approaches is shown in Fig. 8.
A schematic mechanism of apoptosis in ovarian cancer and some current targeted molecular therapeutic approaches. Two main pathways of apoptosis have been elucidated: the death receptor (extrinsic) pathway and the mitochondrial (intrinsic) pathway. Targeted molecular therapeutic approaches include angiogenesis inhibitors, inhibitors of the epidermal growth factor receptor (EGFR), aurora kinase inhibitors, poly ADP Ribose Polymerase (PARP) inhibitors, platelet-derived growth factor (PDGF) receptor inhibitors, MTOR inhibitors, targeting Bcl-2 family in ovarian cancer and apoptosis, minimising expression of inhibitors of apoptosis (IAP) as target for ovarian cancer, therapeutic potential of TNF family members, wild-type p53: the genomic guardian target, interferons (IFN), integrins and insulin-like growth factor (IGF) (figure taken from Ubanako et al. 2015)
Central to apoptotic pathways are two families of proteins, the caspases and the B cell lymphoma-2 (Bcl-2) family (Danial and Korsmeyer 2004; Wyllie 2010). Caspases are a class of serine proteases that function in apoptosis. They form a cascade that ultimately results in cell death. Bcl-2 family proteins control this process, either halting the processes that result in apoptotic cell death or allowing these processes to go forward. However, the central pathways of apoptosis are controlled by a complex system of pro-apoptotic and anti-apoptotic Bcl-2 family members, which act to ensure that apoptosis is only triggered when it is appropriate. Bcl-2 family member interactions that control apoptosis are PPIs. Thus, it has been difficult to discover agents that affect the central pathways of apoptosis and that are capable of being taken into the clinic (Danial and Korsmeyer 2004; Wyllie 2010; Haberman 2012).
The central pathways of apoptosis include an intrinsic and an extrinsic pathway (Ubanako et al. 2015). The intrinsic pathway is triggered by cellular stress (which includes among others the deprivation of growth factors needed for survival, drug treatments or ionising radiation), p53-mediated apoptotic signals triggered by DNA damage, virus infection, hypoxia and energy deprivation. The intrinsic pathway is also modulated by other signal transduction pathways, such as ‘oncogene overdrive’ in which the Myc oncogene may trigger apoptosis instead of hyperproliferation and malignant transformation, the Akt/PTEN pathway (which when dysregulated is a factor in several types of cancer) and the UPS pathway for degradation of unwanted or defective intracellular proteins (Boone et al. 2011; Ubanako et al. 2015). Triggers of the intrinsic pathway operate mainly via modulating members of the Bcl-2 family. Bcl-2 family proteins possess homologous domains that can enter into PPIs among the family members. Anti-apoptotic members of the Bcl-2 family, such as Bcl-2, Bcl-xL and Mcl-1, possess four conserved domains called BH1, BH2, BH3 and BH4. BH 1, 2 and 4 define a hydrophobic groove within the molecule, and BH3 is an 8-to-12 amino acid domain that binds within that groove. These anti-apoptotic proteins localise to the mitochondria, where they specifically bind and sequester pro-apoptotic multi-domain Bcl-2 family members, such as Bak and Bax (Danial and Korsmeyer 2004; Wyllie 2010; Haberman 2012; Ubanako et al. 2015).
The extrinsic apoptotic pathway is triggered by a class of cell-surface receptors of the TNFR family and their corresponding TNF family ligands (Danial and Korsmeyer 2004; Wyllie 2010; Ubanako et al. 2015). These receptor/ligand pairs include TNFR/TNF-α (tumour necrosis factor-alpha), Fas/FasL (Fas ligand) and TRAIL receptor/TRAIL (TRAIL = TNF-related apoptosis-inducing ligand). TRAIL is of great interest to cancer biologists and oncology drug developers, since it has been found to induce apoptosis selectively in cancer cells, independent of p53, which is usually inactivated in human cancers. However, the physiological role of TRAIL is not well understood (Zaba et al. 2010; Haberman 2012).
The binding of a specific ligand to TNFR family receptors on the cell surface leads to clustering of the receptors. The intracellular domains of the receptor complexes then bind to death adaptor proteins, such as Fas-associated death domain protein (Danial and Korsmeyer 2004; Ubanako et al. 2015). These in turn bind to an initiator caspase, caspase 8, which is autocatalytically activated. Caspase 8 activates effector caspases (for example, caspase 3 and 7) leading to apoptosis. Caspase 8 also initiates the intrinsic programme of apoptosis via cleavage of the inactive p22 form of Bid, resulting in the formation of an active form of Bid. This in turn triggers the intrinsic pathway of apoptosis via mitochondria membrane pore formation by complexing of pro-apoptotic Bcl-2 family proteins (Danial and Korsmeyer 2004; Wyllie 2010; Ubanako et al. 2015).
The intention of cancer research is to develop PPI-focused small-molecule drugs specifically targeting the relevant PPIs of Bcl-2 family proteins, since all the interactions between the Bcl-2 family members are PPIs (Adams and Cory 2007). Currently, three Bcl-2 family PPI-disrupting agents (BH3 mimetics) are being tested in clinical trials, namely navitoclax, obatoclax and ABT-199 (Venetoclax). Navitoclax (ABT-263) (shown in Fig. 9a) is the result of the fragment-based drug discovery methodology known as SAR by NMR, which led to a Bcl-2 inhibitor designated as ABT-737 (shown in Fig. 9b). ABT-737 might be useful for the treatment of lymphoma and small cell lung cancer (SCLC) as a monotherapy. It can also be used for a wide variety of cancers in combination with cytotoxic agents or radiation. However, it has poor physiochemical and pharmaceutical properties (Keller et al. 2006; Tse et al. 2008).
Chemical structure of a ABT-263 (navitoclax)—an orally active anticancer drug which does not have off-target effects (figure taken from Tse et al. 2008). b ABT-737—an orally bioavailable, selective small molecule B cell lymphoma 2 (Bcl-2) homology 3 (BH3) mimetic, with potential pro-apoptotic and antineoplastic activities (figure taken from Tse et al. 2008). c Obatoclax mesylate (GX15-070)—an experimental drug for the treatment of various types of cancer which include leukaemia, myelofibrosis, Hodgkin’s lymphoma and mantle-cell lymphoma among others (figure taken from Goard and Schimmer 2013). d ABT-199 (venetoclax)—a BH3-mimetic Bcl-2 inhibitor, does not cause Ca2+-signalling dysregulation or toxicity in pancreatic acinar cells (figure taken from Souers et al. 2013)
ABT-737 is deficient in oral bioavailability properties and its poor solubility makes formulation for intravenous delivery exigent. Hence, these issue rendered ABT-737 a poor prospect for clinical development (Tse et al. 2008). However, ABT-737 was later optimised to produce a second-generation compound, navitoclax, which has improved physicochemical and pharmacological properties and is orally available. Navitoclax is being tested in phase I and phase II clinical trials in various cancers, including combination therapies with drugs such as Rituxan (rituximab) and Tarceva (erlotinib), as well as with cytotoxic chemotherapies and as a single agent (Roberts et al. 2012).
Obatoclax (GX15-070) (shown in Fig. 9c) is a pan-Bcl-2 inhibitor that inhibits Mcl-1. Mcl-1 is overexpressed in several types of cancer, which include prostate cancers (Dash et al. 2010). The interest of cancer research is in the development of small-molecule PPI inhibitor drugs that target Mcl-1. One such drug, obatoclax, was discovered by using a high-throughput protein-protein interaction assay to screen natural product libraries (Shore and Viallet 2005; Nguyen et al. 2007). Studies indicated that obatoclax inhibited the Mcl-1 apoptotic pathway by disrupting the Mcl-1/BAK and the Mcl-1/BAX PPIs. It overcame apoptosis Mcl-1-mediated resistance in lymphoma cells and cultured melanoma cells (Nguyen et al. 2007). However, because of its poor solubility, obatoclax cannot be an oral drug. Nevertheless, intravenous administration of obatoclax showed single-agent anti-tumour activity in mouse xenograft models bearing several different types of human carcinomas. This suggests that obatoclax is an important Mcl-1-inhibitor in cases where Mcl-1 is involved in blocking apoptosis (Nguyen et al. 2007; Haberman 2012).
ABT-199 (Venetoclax) (shown in Fig. 9d) is a BH3 mimetic that selectively inhibits Bcl-2 (Jakubowska et al. 2018). It was initially used for the treatment of relapsed chronic lymphocytic leukaemia. Early generations of Bcl-2 inhibitors induced sustained Ca2+ retaliations in pancreatic acinar cells (PACs), inducing cell death. Therefore, BH3 mimetics are assumably toxic to the pancreas when used to treat cancer. Although ABT-199 was shown to kill Bcl-2-dependent cancer cells without affecting intracellular Ca2+ signalling, its effects on PACs remain elusive (Jakubowska et al. 2018). Hence, it is of paramount importance to assess whether this recently approved anti-leukaemic drug might potentially have pancreatotoxic effects. Inhibition of Bcl-2 via ABT-199 did not elicit intracellular Ca2+ signalling on its own. It did not potentiate Ca2+ signalling induced by physiological or pathophysiological stimuli in PACs, although ABT-199 did not affect cell death in PACs under conditions that killed ABT-199-sensitive cancer cells. Cytosolic Ca2+ extrusion was slightly enhanced in the presence of ABT-199. In contrast, inhibition of Bcl-xL potentiated pathophysiological Ca2+ responses in PACs without exacerbating cell death (Jakubowska et al. 2018).
Stapled peptides
In parallel with second-generation technologies and the discovery and development of small-molecule PPI modulators, peptides have been designed that mimic the amino acid sequence and secondary structures of protein domains that are involved in PPIs. This has led to the emergence of ‘stapled peptides’ comprising a small loop rigidifying the peptide conformation (Ali et al. 2019; Verhoork et al. 2019). These mimetics are resistant to degradation by proteolytic enzymes have favourable pharmacological properties and are able to penetrate cells (Schafmeister et al. 2000; Walensky et al. 2010; Kim et al. 2011). The initial application of stapled peptide technology was to the Bcl-2 family PPI system, which controls apoptosis (Verhoork et al. 2019). As discussed earlier, Bid is one of the BH3-only pro-apoptotic members of the Bcl-2 family. It appears to exert its pro-apoptotic activity by binding to the hydrophobic groove of anti-apoptotic BH-1/2/3/4 proteins such as Bcl-2 and Bcl-xL. This releases pro-apoptotic BH-1/2/3 proteins from their complexes with the anti-apoptotic proteins, enabling them to initiate the intrinsic pathway of apoptosis. Beginning in the mid-1990s, studies to develop a Bid mimetic peptide have been ongoing until now (Kim et al. 2011; Ali et al. 2019; Verhoork et al. 2019).
The BH3 domain of Bid that triggers apoptosis possesses an α-helical structure, which was lost upon synthesis of a peptide with the amino acid sequence of the Bid BH3 domain. Additionally, this protein was ineffective in disrupting PPIs between Bcl-2 or Bcl-xL and BH-1/2/3 pro-apoptotic proteins, and was subject to degradation by serum and cellular proteases (Kim et al. 2011; Ali et al. 2019). Moreover, such peptides could not penetrate cells; hence, the stapled peptide technology was applied to produce a stable α-helical form of the peptide. To construct stapled peptides, synthesised peptides with the amino acid sequence of the domain of interest (in this case, the Bid BH3 domain) incorporate two appropriately spaced non-natural amino acids bearing olefin side chains (Walensky et al. 2004; Kim et al. 2011; Ali et al. 2019; Verhoork et al. 2019). The peptides were designed using the three-dimensional structures of the target protein and the interacting partner domain to provide the sequence of the stapled peptide. Using molecular visualisation software, they analysed the binding interface between these two biomolecules and confirmed that the partner-binding domain was α-helical. Thereafter, they selected amino acid sequences that were not directly involved in target recognition as candidates for substitution with non-natural amino acids. The synthesised peptides each contain two olefin-bearing amino acid residues 3, 4 or 7 amino acid residues apart. These modified peptides were designed to project the reactive olefin residues on the same face of the α-helix (Haberman 2012; Ali et al. 2019; Verhoork et al. 2019).
The bridge between the two non-natural amino acids (staple) was formed by using ruthenium-mediated ring-closing olefin metathesis (shown in Fig. 10). The ‘staple’ in a stapled peptide forms a macrocyclic ring with the amino acid residues it encompasses. The range of bridge lengths of 3, 4 or 7 amino acid residues is used to identify the optimal stapled peptide with respect to affinity for its target and other properties relevant to serving as a drug candidate (Kim et al. 2011; Haberman 2012).
Construction of a stapled peptide. A ‘staple’ is formed between two non-natural amino acids by using Grubb’s ruthenium-mediated ring-closing olefin metathesis. The staple forms a macrocyclic ring with the amino acid residues (figure taken from Haberman 2012)
Peptide stapling results in stabilisation of the α-helical form of peptides such as the Bid BH3 domain and other peptides derived from domains that are α-helical in their natural proteins. Such peptides are resistant to proteases and can also be cell-penetrant (Kim et al. 2011; Haberman 2012). Hydrocarbon staples promote effective cellular uptake via endocytic vesicle trafficking. Cellular penetration by a stapled peptide is also dependent on the net charge of the molecule; those with net positive charges show better cellular uptake (Kim et al. 2011; Haberman 2012).
In the original application of the stapled peptide technology, the constructed stapled peptide Bid BH3 mimetics was called ‘stabilized alpha-helix of Bcl-2 domains’ (SAHBs: Walensky et al. 2004). SAHBs were α-helical, protease-resistant and penetrated cells. One example of the SAHBs that were studied further is SAHBA, which was found to cause the same structural changes in Bcl-xL as did Bid. Intravenous administration of SAHBA consistently caused tumour regression in mouse xenograft models of human leukaemia, as well as improved survival (median survival 5 days for control animals, and 11 days for SAHBA-treated animals). SAHBA showed no overt toxicity in normal tissues. The studies designed to create a Bid mimetic using stapled peptide technology provided proof of principle for the technology and served as a starting point to build a pipeline of preclinical candidates based on stapled peptides (Walensky et al. 2004; Kim et al. 2011; Haberman 2012).
The notch pathway
The notch pathway regulates various aspects of cell proliferation, cellular differentiation, cell-cell communication and cellular survival or death. It is essential for the development of the nervous and haematopoietic systems (Huang et al. 2019). Deregulation of the notch pathway is involved in various cancers, including lung cancer, pancreatic cancer, cancer of the ovary and cancer of immature T cells (T cell acute lymphoblastic leukaemia (T-ALL)). Notch is a cell-membrane receptor just like its ligands (members of the Delta, Serrate and Lag-2 family). The notch pathway becomes activated when the extracellular domain of notch binds to a notch ligand on the surface of an adjacent cell, leading to sequential proteolytic cleavage of the notch intracellular domain, first by an A disintegrin and metalloproteinase (ADAM) family metalloprotease such as TNF-α-converting enzyme (also known as ADAM17) and then by a γ-secretase complex (Moellering et al. 2009; Haberman 2012; Huang et al. 2019).
The free intracellular domain of notch (also known as the intracellular domain of NOTCH1 (ICN1)) translocates to the nucleus and docks with the DNA-bound transcription factor CSL (CBF1, Suppressor of Hairless and Lag-1) (Moellering et al. 2009). The CSL/ICN1 interaction creates a long shallow groove along the interface of the two proteins, which serves as a docking site for coactivator proteins of the mastermind-like (MAML) family, such as MAML1. The resulting trimolecular complex initiates specific transcription of notch-dependent target genes, which uses a stapled peptide to target the notch signal transcription pathway (Moellering et al. 2009; Haberman 2012).
As with other key signal transduction pathways, the notch pathway is centred on PPIs, especially the crucial ternary transcription factor complex CSL/ICN1/MAML1. The discovery of small-molecule drugs that modulate this pathway had thus been considered unfeasible (Moellering et al. 2009; Haberman 2012) until the application of stapled peptide technology to discover agents that modulate this pathway, in particular the key transcription factor complex (Huang et al. 2019). Previous research found that a dominant-negative fragment of MAML1, designated as dnMAML1 (consisting of amino acid residues 13–74) antagonised notch signalling and cell proliferation when expressed in T-ALL cells (Moellering et al. 2009). X-ray diffraction studies showed that the dnMAML1 polypeptide formed an α-helix, which docks with the elongated groove formed by the ICN1/CSL complex. This suggested that it might be possible to design a stapled peptide based on a portion of the sequence of dnMAML1. Such a stapled peptide might inhibit binding of MAML1 to the ICN1/CSL complex, thus blocking transcription of downstream notch-dependent target genes (Moellering et al. 2009; Haberman 2012; Huang et al. 2019).
Moellering and colleagues therefore synthesised a set of six short candidate peptides, which together encompassed the entire contact surface of dnMAML1 with the ICN1/CSL complex (Moellering et al. 2009). Functional studies with these peptides led them to select the 15-amino-acid stapled peptide SAHM1, which showed that stapling conferred a marked helical character (94% helical) to SAMH1, compared with its non-stapled counterpart. SAMH1 was found to be cell-penetrant. Biochemical studies showed that SAMH1 bound to the ICN1/CSL complex competitively with MAML1. Cell culture studies in the KOPT-K1 human T-ALL cell line with a notch-regulated fluorescent reporter gene showed that SAMH1 specifically repressed transcription of notch target genes, in a dose-dependent manner. Gene expression analysis in KOPT-K1 and HPB-ALL human T cell leukaemia cells showed that SAMH1 specifically repressed transcription of notch pathway downstream target genes (Moellering et al. 2009; Haberman 2012; Huang et al. 2019).
More so, SAMH1 markedly reduced proliferation of several T-ALL cell lines in vitro, but was ineffective against leukaemia cell lines that did not depend on the notch pathway for cell proliferation (Al-Shehabi et al. 2019; Majer et al. 2019). In sensitive T-ALL cell lines, SAMH1 treatment also triggered apoptosis. In a mouse model of T-ALL, twice-daily intraperitoneal injection of SAMH1 resulted in significant dose-dependent regression of leukaemia, compared with vehicle-treated mice. Studies of mononuclear cells from SAMH1- and vehicle-treated mice confirmed that SAMH1 treatment resulted in a significant decrease in notch target gene transcription (Al-Shehabi et al. 2019; Majer et al. 2019). These studies showed that SAMH1 is a direct transcriptional antagonist of the notch pathway. This suggests that SAHM1 can be used to determine the role of this pathway in normal physiology and development, and in disease processes. SAMH1 also provides a starting point for drug development, especially in notch-driven cancers such as T-ALL (Haberman 2012; Al-Shehabi et al. 2019; Majer et al. 2019).
Conclusions and future perspectives
The discovery and development of protein-protein interaction modulators has been difficult, because of the structure of protein-interacting interfaces, the lack of natural ligands for PPIs to serve as starting points for drug design and the unsuitability of proprietary chemical libraries for use in HTS campaigns to identify compounds that modulate PPIs (Jana et al. 2017; Stevers et al. 2018; Albert et al. 2019). Despite the difficulty of discovering and developing PPI modulatory drugs, developing PPI modulators is becoming of increasing strategic importance. However, most of the known small-molecule drugs discovered to date address only 2% of human proteins. Most of the remaining proteins are critically involved in disease pathways, lie within the category of intractable targets and are PPIs (Feng et al. 2017; Lage et al. 2018; Guidolin et al. 2019; Husain et al. 2019). Despite the difficulties in discovering small-molecule PPI modulators that are capable of being taken into clinical trials, drugs are already on the market and some drug-like candidates have reached clinical production. Central to these successes has been the determination of ‘hotspots’ in protein-protein interfaces. By targeting hotspots, several compounds that directly modulate PPIs have been discovered (Robertson and Spring 2018; Zhang et al. 2018). However, this has been on a sporadic ‘one compound at a time’ basis (Haberman 2012). Examples include eltrombopag, as well as several others currently being tested in clinical trials. In addition to these direct PPI modulators, there are allosteric chemokine receptor modulators, which include maraviroc, plerixafor and several others now undergoing clinical trials (Kalota and Gewirtz 2010).
Following failure to meet the strategic needs to expand the number of targets that can be addressed by developable drugs using the sporadic, ‘one compound at a time’ approach (Haberman 2012), developing second-generation technologies designed to enable the fabrication of small-molecule and peptide PPI modulators on a more consistent basis becomes imperative. These include CS mapping, diversity-oriented synthesis of chemical libraries, DPC technology for synthesis of libraries of macrocyclic compounds and stapled-peptide technology (Drahl 2009; Franzini and Randolph 2016; Zhou et al. 2018). Responding to these new technologies, research vistas are moving back into the PPI modulator field. Whether the new suite of enabling technologies for PPI modulator discovery and development will enable this area to be successful and to meet the strategic needs for discovery and development remains an open question. Most of the compounds that have been discovered using these technologies are in clinical and preclinical stages (Taylor et al. 2018; Stevers et al. 2018; Albert et al. 2019; Fecková et al. 2019; Zhao et al. 2019). It will be necessary for some of these compounds to enter clinical production, achieve proof of concept and thereafter reach the market. Nevertheless, the PPI modulator field is an exciting area that is gaining increasing interest and investment in the PPI-focused technology development curve.
Abbreviations
- PPI:
-
Protein-protein interactions
- IL-2:
-
Interleukin-2
- IL-2Rα:
-
Interleukin-2 receptor alpha chain
- FBDD:
-
Fragment-based drug discovery
- FBLD:
-
Fragment-based lead discovery
- SAR:
-
Structure-activity relationship
- NMR:
-
Nuclear magnetic resonance
- Bcl-2:
-
B cell lymphoma 2
- Bcl-XL:
-
B cell lymphoma-extra large
- CS:
-
Computational solvent
- MDM2:
-
Mouse double minute 2 homolog
- HPV:
-
Human papillomavirus
- TNF:
-
Tumour necrosis factor
- TNF-α:
-
Tumour necrosis factor alpha
- TNFR1:
-
Tumour necrosis factor receptor 1
- FtsZ:
-
Filamenting temperature-sensitive mutant Z
- ZipA:
-
Z interacting protein A
- HTS:
-
High-throughput screening
- CBF:
-
Core binding factor
- DOS:
-
Diversity-oriented synthesis
- Shh:
-
Sonic Hedgehog
- Ptc1:
-
Protein phosphatase 2C homolog 1
- mTOR:
-
Mammalian target of rapamycin
- FKBP12:
-
FK-binding protein 12
- DPC:
-
DNA-programmed chemistry
- STATs:
-
Signal transducers and activators of transcription
- mAb:
-
Monoclonal antibody
- HER2:
-
Human epidermal growth factor receptor 2
- ICAM-1:
-
Intercellular adhesion molecule-1
- VEGF:
-
Vascular endothelial growth factor
- G-CSF:
-
Granulocyte colony-stimulating factor
- TPO:
-
Thrombopoietin
- LFA-1:
-
Leukocyte integrin lymphocyte function-associated antigen 1
- GPCR:
-
G protein-coupled receptor
- CCR5:
-
C-C chemokine receptor type 5
- CXCR4:
-
C-X-C chemokine receptor type 4
- CXCR7:
-
C-X-C chemokine receptor type 7
- MM:
-
Multiple myeloma
- B-CLL:
-
B cell chronic lymphocytic leukaemia
- APC:
-
Adenomatous polyposis coli
- GSK-3:
-
Glycogen synthase kinase 3
- Tcf:
-
T cell factor
- DLBCL:
-
Diffuse large B cell lymphoma
- BCL6:
-
B cell lymphoma 6
- SMRT:
-
Silencing mediator for retinoid or thyroid-hormone receptors
- HDAC3:
-
Histone deacetylase 3
- CHK1:
-
Checkpoint kinase 1
- CDKN1A:
-
Cyclin-dependent kinase inhibitor 1
- ATR:
-
Ataxia telangiectasia and Rad3-related protein
- CADD:
-
Computer-aided drug design
- AKAP:
-
A-kinase anchoring protein
- cAMP:
-
Cyclic adenosine monophosphate
- Ub:
-
Ubiquitin
- UPS:
-
Ubiquitin proteasome system
- UPP:
-
Ubiquitin-proteasome pathway
- CRL:
-
Cullin RING E3 ligase
- Ki :
-
Inhibition constant
- TRAIL:
-
TNF-related apoptosis-inducing ligand
- SCLC:
-
Small cell lung cancer
- PACs:
-
Pancreatic acinar cells
- SAHBs:
-
Stabilised alpha-helix of Bcl-2 domains
- T-ALL:
-
T cell acute lymphoblastic leukaemia
- ADAM:
-
A disintegrin and metalloproteinase
- ICN1:
-
Intracellular domain of NOTCH1
- MAML:
-
Mastermind-like
- dnMAML1:
-
Dominant-negative fragment of MAML1
References
Adams JM, Cory S (2007) The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 26:1324–1337. https://doi.org/10.1038/sj.onc.1210220
Albert L, Peñalver A, Djokovic N (2019) Modulating protein-protein interactions with visible-light responsive peptide backbone photoswitches. ChemBioChem 20:1–14. https://doi.org/10.1002/cbic.201800737
Ali AM, Atmaj J, Oosterwijk NV (2019) Stapled peptides inhibitors: a new window for target drug discovery. Comput Struct Biotechnol J 17:263–281. https://doi.org/10.1016/j.csbj.2019.01.012
Alihodzić S, Bukvić M, Elenkov I et al (2018) Current trends in macrocyclic drug discovery and beyond -Ro5. Prog Med Chem 57:113–233. https://doi.org/10.1016/bs.pmch.2018.01.002
Allison M (2009) Bristol-Myers Squibb swallows last of antibody pioneers. Nat Biotechnol 27:781–783. https://doi.org/10.1038/nbt0909-781
Al-Shehabi H, Fiebig U, Kutzner J et al (2019) Human SAMHD1 restricts the xenotransplantation relevant porcine endogenous retrovirus (PERV) in non-dividing cells. J Gen Virol 100:656–661. https://doi.org/10.1099/jgv.0.001232
Appel A (2011) Drugs: more shots on target. Nature 480:S40–S42. https://doi.org/10.1038/480S40a
Arkin MR, Randal M, DeLano WL et al (2003) Binding of small molecules to an adaptive protein–protein interface. Proc Natl Acad Sci USA 100:1603–1608. https://doi.org/10.1073/pnas.252756299
Bauer RA, Wurst JM, Tan DS (2010) Expanding the range of “druggable” targets with natural product-based libraries: an academic perspective. Curr Opin Chem Biol 14:308–314. https://doi.org/10.1016/j.cbpa.2010.02.001
Basso A, Park SB, Moni L (2019) Editorial: diversity oriented synthesis. Front Chem 6:668. https://doi.org/10.3389/fchem.2018.00668
Booij TH, Price LS, Danen EHJ (2019) 3D cell-based assays for drug screens: challenges in imaging, image analysis, and high-content analysis. SLAS discovery 1–13. https://doi.org/10.1177/2472555219830087
Boone DN, Qi Y, Li Z et al (2011) Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Pro Nat Acad Sci USA 108:632–637. https://doi.org/10.1073/pnas.1008848108
Bhullar KS, Lagarón NO, McGowan EM et al (2018) Kinase-targeted cancer therapies: progress, challenges and future directions. Mol Cancer 17:48. https://doi.org/10.1186/s12943-018-0804-2
Carballo GB, Honorato JR, Farias de Lopes GP (2018) A highlight on Sonic Hedgehog pathway. Cell Commun Signal 16:11. https://doi.org/10.1186/s12964-018-0220-7
Carry JC, Garcia-Echeverria C (2013) Inhibitors of the p53/hdm2 protein–protein interaction—path to the clinic. Bioorg Med Chem Lett 23:2480–2485. https://doi.org/10.1016/j.bmcl.2013.03.034
Ceccarelli DF, Tang X, Pelletier B et al (2011) An allosteric inhibitor of the human Cdc34 ubiquitin conjugating enzyme. Cell 145:1075–1087. https://doi.org/10.1016/j.cell.2011.05.039
Cencic R, Hall DR, Robert F et al (2011) Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci USA 108:1046–1051. https://doi.org/10.1073/pnas.1011477108
Cerchietti LC, Ghetu AF, Zhu X et al (2010) A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell 17:400–411. https://doi.org/10.1016/j.ccr.2009.12.050
Cheok CF, Verma CS, Baselga J et al (2011) Translating p53 into the clinic. Nat Rev Clin Oncol 8:25–37. https://doi.org/10.1038/nrclinonc.2010.174
Christian F, Szaszák M, Friedl S et al (2011) Small molecule AKAP-protein kinase a (PKA) interaction disruptors that activate PKA interfere with compartmentalized cAMP signaling in cardiac myocytes. J Biol Chem 286:9079–9096. https://doi.org/10.1074/jbc.M110.160614
Clackson T, Wells JA (1995) A hot spot of binding energy in a hormone-receptor interface. Science 267:383–386. https://doi.org/10.1126/science.7529940
Cohen P, Tcherpakov M (2010) Will the ubiquitin system furnish as many drug targets as protein kinases? Cell 143:686–693. https://doi.org/10.1016/j.cell.2010.11.016
Compton LA, Hiebert SW (2010) Anticancer therapy SMRT-ens up: targeting the BCL6-SMRT interaction in B cell lymphoma. Cancer Cell 17:315–316. https://doi.org/10.1016/j.ccr.2010.03.012
Conn PJ, Christopoulos A, Lindsley CW (2009) Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov 8:41–54. https://doi.org/10.1038/nrd2760
Crane EK, Kwan SY, Izaguirre DI (2015) Nutlin-3a: a potential therapeutic opportunity for TP53 wild-type ovarian carcinomas. PLoS One 10(8):e0135101. https://doi.org/10.1371/journal.pone.0135101
Danial NN, Korsmeyer SJ (2004) Cell death: critical control points. Cell 116:205–219. https://doi.org/10.1016/S0092-8674(04)00046-7
Dash R, Richards JE, Su ZZ et al (2010) Mechanism by which Mcl-1 regulates cancer-specific apoptosis triggered by mda-7/IL-24, an IL-10-related cytokine. Cancer Res 70:5034–5045. https://doi.org/10.1158/0008-5472.CAN-10-0563
Davies SL, Serradell N, Bolos J et al (2007) Plerixafor hydrochloride. Drug Today 32:123–136
Debouck C, Metcalf B (2000) The impact of genomics on drug discovery. Annu Rev Pharmacol Toxicol 40:193–208. https://doi.org/10.1146/annurev.pharmtox.40.1.193
Deshaies RJ (2009) Drug discovery: fresh target for cancer therapy. Nature 458:709–710. https://doi.org/10.1038/458709a
Díaz-Eufracio BI, JesúsNaveja J, Medina-Franco JL (2018) Protein-protein interaction modulators for epigenetic therapies. In: Donev R (ed) Advances in protein chemistry and structural biology, 1st edn. Swansea University, UK, pp 65–84. https://doi.org/10.1016/bs.apcsb.2017.06.002
Dorr P, Westby M, Dobbs S et al (2005) Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother 49:4721–4732. https://doi.org/10.1128/AAC.49.11.4721-4732.2005
Drahl C (2009) Big hopes ride on big rings. ACS Meeting News: constraining molecules in macrocyclic rings could help address challenges in drug discovery. Chem Eng News 87:54–57. https://doi.org/10.1021/cen-v087n036.p054
Driggers EM, Hale SP, Lee J et al (2008) The exploration of macrocycles for drug discovery – an underexploited structural class. Nat Rev Drug Discov 7:608–624. https://doi.org/10.1038/nrd2590
Du L, Grigsby SM, Yao A et al (2018) Peptidomimetics for targeting protein–protein interactions between DOT1L and MLL oncofusion proteins AF9 and ENL. ACS Med Chem Lett 9:895–900. https://doi.org/10.1021/acsmedchemlett.8b00175
Duan Z, Tu M, Zhang Q et al (2018) Novel therapeutic strategy to inhibit growth of pancreatic cancer organoids using a rational combination of drugs to induce mitotic arrest and apoptosis. J Clin Oncol 36:322–322. https://doi.org/10.1200/JCO.2018.36.4_suppl.322
Dustin ML, Bivona TG, Philips MR (2004) Membranes as messengers in T cell adhesion signaling. Nat Immunol 5:363–372. https://doi.org/10.1038/ni1057
Erlanson DA, Fesik SW, Hubbard RE et al (2016) Twenty years on: the impact of fragments on drug discovery. Nat Rev Drug Discov 15:605–619. https://doi.org/10.1038/nrd.2016.109
Erickson-Miller CL, DeLorme E, Tian SS et al (2005) Discovery and characterization of a selective, nonpeptidyl thrombopoietin receptor agonist. Exp Hematol 33:85–93. https://doi.org/10.1016/j.exphem.2004.09.006
Everts S (2008) Piece by Piece. Chem Eng News 86:15–23
Fecková B, Kimáková P, Ilkovičová L et al (2019) Methylation of the first exon in the erythropoietin receptor gene does not correlate with its mRNA and protein level in cancer cells. BMC Genet 20:1. https://doi.org/10.1186/s12863-018-0706-8
Feng Y, Wang Q, Wang T et al (2017) Drug target protein-protein interaction networks: a systematic perspective. Biomed Res Int 2017:1–13. https://doi.org/10.1155/2017/1289259
Ferguson FM, Gray NS (2018) Kinase inhibitors: the road ahead. Nat Rev Drug Discov 17:353–377. https://doi.org/10.1038/nrd.2018.21
Franzini R, Randolph C (2016) Chemical space of DNA-encoded libraries. J Med Chem 59:6629–6644. https://doi.org/10.1021/acs.jmedchem.5b01874
Friedberg JW (2011) New strategies in diffuse large B-cell lymphoma: Translating findings from gene expression analyses into clinical practice. Clin Cancer Res 108:1046–1051. https://doi.org/10.1158/1078-0432.CCR-11-1073
Galloway WR, Isidro-Llobet A, Spring DR (2010) Diversity-oriented synthesis as a tool for the discovery of novel biologically active small molecules. Nat Commun 1:80. https://doi.org/10.1038/ncomms1081
Goard CA, Schimmer AD (2013) An evidence-based review of obatoclax mesylate in the treatment of hematological malignancies. Core Evidence 8:15–26. https://doi.org/10.2147/CE.S42568
Gonzalez MW, Kann MG (2012) Protein interactions and disease. PLoS Comput Biol 8:e1002819. https://doi.org/10.1371/journal.pcbi.1002819
Gorczynski MJ, Grembecka J, Zhou Y et al (2007) Allosteric inhibition of the protein-protein interaction between the leukemia-associated proteins Runx1 and CBFbeta. Chem Biol 14:1186–1197. https://doi.org/10.1016/j.chembiol.2007.09.006
Graff JR, Konicek BW, Carter JH et al (2008) Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res 68:631–634. https://doi.org/10.1158/0008-5472.CAN-07-5635
Grosdidier S, Totrov M, Fernández-Recio J (2009) Computer applications for prediction of protein–protein interactions and rational drug design. Adv Appl Bioinforma Chem 2:101–123. https://doi.org/10.2147/AABC.S6272
Grossmann TN, Pelay-Gimeno M, Glas A et al (2015) Structure-based design of inhibitors of protein–protein interactions: mimicking peptide binding epitopes. Angew Chem Int Ed 54:8896–8927. https://doi.org/10.1002/anie.201412070
Guidolin D, Marcoli M, Tortorella C et al (2019) Receptor-receptor interactions as a widespread phenomenon: novel targets for drug development? Front Endocrinol 10:53. https://doi.org/10.3389/fendo.2019.00053
Haberman AB (2012) Advances in the discovery of protein-protein interaction modulators. SCRIP Insights Informa 2012. https://biopharmconsortium.com/2012/04/25/advances-in-the-discovery-of-protein-protein-interaction-modulators-published-by-informas-scrip-insights. Accessed 21 April 2019
Hajduk PJ, Galloway WR, Spring DR (2011) Drug discovery: a question of library design. Nature 470:42–43. https://doi.org/10.1038/470042a
Hall DR, Kozakov D, Vajda S (2012) Analysis of protein binding sites by computational solvent mapping. Methods Mol Biol 819:13–27. https://doi.org/10.1007/978-1-61779-465-02
Hansen SK, Cancilla MT, Shiau TP et al (2005) Allosteric inhibition of PTP1B activity by selective modification of a non-active site cysteine residue. Biochemistry 44:7704–7712. https://doi.org/10.1021/bi047417s
Hansen KB, Yi F, Perszyk RE et al (2018) Structure, function, and allosteric modulation of NMDA receptors. J Gen Physiol 150:1081–1105. https://doi.org/10.1085/jgp.201812032
Haq S, Ramakrishna S (2017) Deubiquitylation of deubiquitylases. Open Biol 7:170016. https://doi.org/10.1098/rsob.170016
Hitzenberger M, Schuster D, Hofer TS (2017) The binding mode of the Sonic Hedgehog inhibitor robotnikinin, a combined docking and QM/MM MD study. Front Chem 5:76. https://doi.org/10.3389/fchem.2017.00076
Horuk R (2009) Chemokine receptor antagonists: overcoming developmental hurdles. Nat Rev Drug Discov 8:23–33. https://doi.org/10.1038/nrd2734
Huang SM, Mishina YM, Liu S et al (2009) Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461:614–620. https://doi.org/10.1038/nature08356
Huang O, Li J, Zheng J (2019) The carcinogenic role of the notch signaling pathway in the development of hepatocellular carcinoma. J Cancer 10:1570–1579. https://doi.org/10.7150/jca.26847
Husain B, Paduchuri S, Ramani SR et al (2019) Extracellular protein microarray technology for high throughput detection of low affinity receptor-ligand interactions. J Vis Exp 143:e58451. https://doi.org/10.3791/58451
Jana T, Ghosh A, Mandal SD et al (2017) PPIMpred: a web server for high-throughput screening of small molecules targeting protein–protein interaction. R Soc Open Sci 4:160501. https://doi.org/10.1098/rsos.160501
Jakubowska MA, Kerkhofs M, Martines C (2018) ABT-199 (Venetoclax), a BH3-mimetic Bcl-2 inhibitor, does not cause Ca2+ -signalling dysregulation or toxicity in pancreatic acinar cells. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1864:968–976. https://doi.org/10.1111/bph.14505
Jeong WJ, Ro EJ, Choi KY (2018) Interaction between Wnt/β-catenin and RAS-ERK pathways and an anti-cancer strategy via degradations of β-catenin and RAS by targeting the Wnt/β-catenin pathway. Precision Oncology 2:5. https://doi.org/10.1038/s41698-018-0049-y
Jochim AL, Arora PS (2010) Systematic analysis of helical protein interfaces reveals targets for synthetic inhibitors. ACS Chem Biol 5:919–923. https://doi.org/10.1021/cb1001747
Kalatskaya I, Berchiche YA, Gravel S et al (2009) AMD3100 is a CXCR7 ligand with allosteric agonist properties. Mol Pharmacol 75:1240–1247. https://doi.org/10.1124/mol.108.053389
Kalota A, Gewirtz AM (2010) A prototype nonpeptidyl, hydrazone class, thrombopoietin receptor agonist, SB-559457, is toxic to primary human myeloid leukemia cells. Blood 115:89–93. https://doi.org/10.1182/blood-2009-06-227751
Keller TH, Pichota A, Yin Z (2006) A practical view of “druggability”. Curr Opin Chem Biol 10:357–361. https://doi.org/10.1016/j.cbpa.2006.06.014
Kim YW, Grossmann TN, Verdine GL (2011) Synthesis of all-hydrocarbon stapled α-helical peptides by ring-closing olefin metathesis. Nat Protoc 6:761–771. https://doi.org/10.1038/nprot.2011.324
Kojima K, Burks JK, Arts J et al (2010) The novel tryptamine derivative JNJ-26854165 induces wild-type p53- and E2F1-mediated apoptosis in acute myeloid and lymphoid leukemias. Mol Cancer Ther 9:2545–2557. https://doi.org/10.1158/1535-7163.MCT-10-0337
Kozakov D, Hall DR, Chuang GY et al (2011) Structural conservation of druggable hot spots in protein-protein interfaces. Proceedings of the National Academy of Sciences USA 108:13528–13533. https://doi.org/10.1073/pnas.1101835108
Landon M, Lancia DR, Yu J et al (2007) Identification of hot spots within druggable binding regions by computational solvent mapping of proteins. J Med Chem 50:1231–1240. https://doi.org/10.1021/jm061134b
Lage OM, Ramos MC, Calisto R et al (2018) Current screening methodologies in drug discovery for selected human diseases. Mar Drugs 16:279. https://doi.org/10.3390/md16080279
Lecker SH, Goldberg AL, Mitch WE (2006) Protein degradation by the ubiquitin–proteasome pathway in normal and disease states. J Am Soc Nephrol 17:1807–1819. https://doi.org/10.1681/ASN.2006010083
Lee CW, Grubbs RH (2001) Formation of macrocycles via Ring-closing olefin metathesis. J Org Chem 66:7155–7158. https://doi.org/10.1021/jo0158480
Lepourcelet M, Chen YN, France DS et al (2004) Small-molecule antagonists of the oncogenic Tcf/beta-catenin protein complex. Cancer Cell 5:91–102. https://doi.org/10.1016/S1535-6108(03)00334-9
Li J, Yang C, Xia Y et al (2001) Thrombocytopenia caused by the development of antibodies to thrombopoietin. Blood 98:3241–3248. https://doi.org/10.1182/blood.V98.12.3241
Li JW, Vederas JC (2009) Drug discovery and natural products: end of an era or an endless frontier? Science 325:161–165. https://doi.org/10.1126/science.1168243
Lipinski CA, Lombardo F, Dominiv BW et al (2012) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. https://doi.org/10.1016/j.addr.2012.09.019
Luise N, Wyatt PG (2019) Diversity-oriented synthesis of bicyclic fragments containing privileged azines. Bioorg Med Chem Lett 29:248–251. https://doi.org/10.1016/j.bmcl.2018.11.046
Lygren B, Taskén K (2008) The potential use of AKAP18delta as a drug target in heart failure patients. Expert Opin Biol Ther 8:1099–1108. https://doi.org/10.1517/14712598.8.8.1099
Ma R, Wang P, Wu J et al (2016) Process of fragment-based lead discovery - a perspective from NMR. Molecules 21:E854. https://doi.org/10.3390/molecules21070854
Majer C, Schüssler JM, König R (2019) Intertwined: SAMHD1 cellular functions, restriction, and viral evasion strategies. Med Microbiol Immunol. 1–17. https://doi.org/10.1007/s00430-019-00593-x
Mella RM, Kortazar D, Roura-Ferrer M (2018) Nomad biosensors: a new multiplexed technology for the screening of GPCR ligands. SLAS Technol 23:207–216. https://doi.org/10.1177/2472630318754828
Miller JL, Church TJ, Leonoudakis D et al (2015) Discovery and characterization of nonpeptidyl agonists of the tissue-protective erythropoietin receptor. Mol Pharmacol 88:357–367. https://doi.org/10.1124/mol.115.098400
Miller JH, Field JJ, Kanakkanthara A et al (2018) Marine invertebrate natural products that target microtubules. J Nat Prod 81:691–702. https://doi.org/10.1021/acs.jnatprod.7b00964
Miszta P, Jakowiecki J, Rutkowska E (2018) Approaches for differentiation and interconverting GPCR agonists and antagonists. Methods Mol Biol 1705:265–296. https://doi.org/10.1007/978-1-4939-7465-8_12
Moellering RE, Cornejo M, Davis TN et al (2009) Direct inhibition of the NOTCH transcription factor complex. Nature 462:182–188. https://doi.org/10.1038/nature08543
Modell AE, Blosser SL, Arora PS (2016) Systematic targeting of protein-protein interactions. Trends Pharmacol Sci 37:702–713. https://doi.org/10.1016/j.tips.2016.05.008
Moll UM, Petrenko O (2003) The MDM2-p53 interaction. Mol Cancer Res 1:1001–1008
Moreira IS, Fernandes PA, Ramos MJ (2007) Hot spots - a review of the protein-protein interface determinant amino-acid residues. Proteins 6:803–812. https://doi.org/10.1002/prot.21396
Newman DJ, Cragg GM (2007) Natural products as sources of new drugs over the last 25 years. J Nat Prod 70:461–477. https://doi.org/10.1021/np068054v
Nguyen M, Marcellus RC, Roulston A et al (2007) Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proceedings of the National Academy of Sciences USA 104:19512–19517. https://doi.org/10.1073/pnas.0709443104
Nielsen TE, Schreiber SL (2008) Towards the optimal screening collection: a synthesis strategy. Angew Chem Int Ed Engl 47:48–56. https://doi.org/10.1002/anie.200703073
Oltersdorf T, Elmore SW, Shoemaker AR et al (2005) An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435:677–681. https://doi.org/10.1038/nature03579
Pan Y, Wang Z, Zhan W et al (2018) Computational identification of binding energy hot spots in protein-RNA complexes using an ensemble approach. Bioinformatics 34:1473–1480. https://doi.org/10.1093/bioinformatics/btx822
Pándy-Szekeres G, Munk C, Tsonkov TM et al (2018) GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res 46:D440–D446. https://doi.org/10.1093/nar/gkx1109
Parveen A, Subedi L, Kim HW et al (2019) Phytochemicals targeting VEGF and VEGF-related multifactors as anticancer therapy. J Clin Med 8:350. https://doi.org/10.3390/jcm8030350
Roberts AW, Seymour JF, Brown JR et al (2012) Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of Navitoclax in patients with relapsed or refractory disease. J Clin Oncol 30:488–496. https://doi.org/10.1200/JCO.2011.34.7898
Robertson NS, Spring DR (2018) Using peptidomimetics and constrained peptides as valuable tools for inhibiting protein–protein interactions. Molecules 23:959. https://doi.org/10.3390/molecules23040959
Robson-Tull J (2018) Biophysical screening in fragment-based drug design: a brief overview. Bioscience Horizons: The International Journal of Student Research 11:hzy01512. https://doi.org/10.1093/biohorizons/hzy015
Rüdisser S, Vangrevelinghe E, Maibaum J (2016) An integrated approach for fragment-based lead discovery: virtual, NMR, and high-throughput screening combined with structure-guided design. Application to the aspartyl protease renin. In: Erlanson DA, Jahnke W (eds) Fragment-based drug discovery lessons and outlook, 1st edn. Wiley, New York, pp 447–480. https://doi.org/10.1002/9783527683604
Schafmeister CE, Po J, Verdine GL (2000) An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J Am Chem Soc 122:5891–5892. https://doi.org/10.1021/ja000563a
Schreiber SL (2009) Organic chemistry: molecular diversity by design. Nature 457:153–154. https://doi.org/10.1038/457153a
Shangary S, Qin D, McEachern D et al (2008) Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A 105:3933–3938. https://doi.org/10.1073/pnas.0708917105
Shangary S, Wang S (2009) Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol.49:223-241. https://doi.org/10.1146/annurev.pharmtox.48.113006.094723
Shore GC, Viallet J (2005) Modulating the bcl-2 family of apoptosis suppressors for potential therapeutic benefit in cancer. Hematology Am Soc Hematol Educ Program 2005:226–230. https://doi.org/10.1182/asheducation-2005.1.226
Silva D, Yu S, Ulge UY et al (2019) De novo design of potent and selective mimics of IL-2 and IL-15. Nature 565:186–191. https://doi.org/10.1038/s41586-018-0830-7
Sinha D, Chowdhury D, Vino S (2012) Monoclonal antibodies (mAbs): the latest dimension of modern therapeutics. Int J Curr Sci 2:9–23
Song X, Lu L, Passioura T (2017) Macrocyclic peptide inhibitors for the protein–protein interaction of Zaire Ebola virus protein 24 and karyopherin alpha 5. Org Biomol Chem 15:5155–5160. https://doi.org/10.1039/c7ob00012j
Soucy TA, Smith PG, Milhollen MA et al (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458:732–736. https://doi.org/10.1038/nature07884
Souers AJ, Leverson JD, Boghaert ER et al (2013) ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 19:202–208. https://doi.org/10.1038/nm.3048
Stanton BZ, Peng LF, Maloof N et al (2009) A small molecule that binds Hedgehog and blocks its signaling in human cells. Nat Chem Biol 5:154–156. https://doi.org/10.1038/nchembio.142
Stevers LM, Sijbesma E, Botta M et al (2018) Modulators of 14-3-3 protein-protein interactions. J Med Chem 61:3755–3778. https://doi.org/10.1021/acs.jmedchem.7b00574
Susanto JP (2015) The role of Eltrombopag and Romiplostim as the thrombopoietin receptor agonist (TPO-RA) in treatment of idiopathic thrombocytopenic purpura (ITP): what is TPO-RA, when TPO-RA is used and how to take TPO-RA? Folia Medica Indonesiana 51:203–207. https://doi.org/10.20473/fmi.v51i3.2840
Takada Y, Ye X, Simon S (2007) The integrins. Genome Biol 8:215. https://doi.org/10.1186/gb-2007-8-5-215
Taylor IR, Dunyak BM, Komiyama T et al (2018) High throughput screen for inhibitors of protein-protein interactions in a reconstituted heat shock protein 70 (Hsp70) complex. J Biol Chem 293:4014–4025. https://doi.org/10.1074/jbc.RA117.001575jbc.RA117.001575
Tian SS, Lamb P, King AG et al (1998) A small, nonpeptidyl mimic of granulocyte-colony-stimulating factor. Science 281:257–259. https://doi.org/10.1126/science.281.5374.257
Tse C, Shoemaker AR, Adickes J et al (2008) ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res 68:3421–3428. https://doi.org/10.1158/0008-5472.CAN-07-5836
Trinh PNH, May LT, Leach K et al (2018) Biased agonism and allosteric modulation of metabotropic glutamate receptor 5. Clin Sci 132:2323–2338. https://doi.org/10.1042/CS20180374
Ubanako PN, Choene M, Motadi L (2015) Mechanisms of apoptosis in ovarian cancer: the small molecule targeting. Int J Med Med Sci 7:46–60. https://doi.org/10.5897/IJMMS2014.1081
Varshavsky A (2017) The ubiquitin system, autophagy, and regulated protein degradation. Annu Rev Biochem 86:123–128. https://doi.org/10.1146/annurev-biochem-061516-044859
Vassilev LT, Vu BT, Graves B et al (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848. https://doi.org/10.1126/science.1092472
Venkata Narasimha Rao G, Ravi B, Sunil Kumar M et al (2017) Ultra performance liquid chromatographic method for simultaneous quantification of plerixafor and related substances in an injection formulation. Cogent Chemistry 3:1275955. https://doi.org/10.1080/23312009.2016.1275955
Verhoork SJM, Jennings CE, Rozatian N (2019) Tuning the binding affinity and selectivity of perfluoroaryl-stapled peptides by cysteine-editing. Chemistry 25:177–182. https://doi.org/10.1002/chem.201804163
Walensky LD, Kung AL, Escher I et al (2004) Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science 305:1466–1470. https://doi.org/10.1126/science.1099191
Walensky LD, Korsmeyer SJ, Verdine G (2010) Stabilized alpha helical peptides and uses thereof. United States Patent Number 7:469 https://patents.google.com/patent/US7723469B2/en. Accessed 24 Mar 2019
Wan H (2016) An overall comparison of small molecules and large biologics in ADME testing. ADMET & DMPK 4:1–22. https://doi.org/10.5599/admet.4.1.276
Wells JA, McClendon CL (2007) Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature 450:1001–1009. https://doi.org/10.1038/nature06526
Wertz IE, Wang X (2019) From discovery to bedside: targeting the ubiquitin system. Cell Chem Biol 26(2):156–177. https://doi.org/10.1016/j.chembiol.2018.10.022
Wilson CG, Arkin MR (2011) Small-molecule inhibitors of IL-2/IL-2R: lessons learned and applied. Curr Top Microbiol Immunol 348:25–59. https://doi.org/10.1007/82_2010_93
Wyllie AH (2010) “Where, O death, is thy sting?” A brief review of apoptosis biology Mol Neurobiol 42:4–9. https://doi.org/10.1007/s12035-010-8125-5
Xu GG, Guo J, Wu Y (2014) Chemokine receptor CCR5 antagonist maraviroc: medicinal chemistry and clinical applications. Curr Top Med Chem 13:1504–1514. https://doi.org/10.2174/1568026614666140827143745
Yan M, Li G, An J (2017) Discovery of small molecule inhibitors of the Wnt/β-catenin signaling pathway by targeting β-catenin/Tcf4 interactions. Exp Biol Med 242:1185–1197. https://doi.org/10.1177/1535370217708198
Yasui T, Yamamoto T, Sakai N et al (2017) Discovery of a novel B-cell lymphoma 6 (BCL6)–corepressor interaction inhibitor by utilizing structure-based drug design. Bioorg Med Chem 25:4876–4886. https://doi.org/10.1016/j.bmc.2017.07.037
Yu M, Wang C, Kyle AF et al (2011) Synthesis of macrocyclic natural products by catalysts-controlled stereoselective ring-closing metathesis. Nature 479:88–93. https://doi.org/10.1038/nature10563
Zaba LC, Fuentes-Duculan J, Eungdamrong NJ et al (2010) Identification of TNF-related apoptosis inducing ligand and other molecules that distinguish inflammatory from resident dendritic cells in patients with psoriasis. J Allergy Clin Immunol 125:1261–1268. https://doi.org/10.1016/j.jaci.2010.03.018
Zhang G, Andersen J, Gerona-Navarro G (2018) Peptidomimetics targeting protein-protein interactions for therapeutic development. Protein Pept Lett 25:1076–1089. https://doi.org/10.2174/0929866525666181101100842
Zhao F, Liu W, Yue S et al (2019) Pre-treatment with G-CSF could enhance the antifibrotic effect of BM-MSCs on pulmonary fibrosis. Stem Cells Int 2019:1726743. https://doi.org/10.1155/2019/1726743
Zhong M, Gadek TR, Bui M (2012) Discovery and development of potent LFA-1/ICAM-1 antagonist SAR 1118 as an ophthalmic solution for treating dry eye. ACS Med Chem Lett 3:203–206. https://doi.org/10.1021/ml2002482
Zhou X, Pathak P, Jayawickramarajah J (2018) Design, synthesis, and applications of DNA–macrocyclic host conjugates. Chem Commun 54:11668–11680. https://doi.org/10.1039/c8cc06716c
Acknowledgements
Abidemi Paul Kappo is thankful to the National Research Foundation (NRF), South Africa for a Thuthuka Grant (Grant No: 107262) award and University of Zululand Research Committee for their support. Llyod Mabonga is thankful to the Department of Science and Technology (DST) and the NRF for a Doctoral Research Bursary. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the DST, NRF and the University of Zululand.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Lloyd Mabonga declares that he has no conflict of interest. Abidemi Paul Kappo declares that he has no conflict of interest.
Ethical approval
The article does not contain any studies with human participants or animals hence do not require ethics approval by the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of a Special Issue on ‘Biophysics & Structural Biology at Synchrotrons’ edited by Trevor Sewell.
Rights and permissions
About this article
Cite this article
Mabonga, L., Kappo, A.P. Protein-protein interaction modulators: advances, successes and remaining challenges. Biophys Rev 11, 559–581 (2019). https://doi.org/10.1007/s12551-019-00570-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-019-00570-x