Abstract
Arabidopsis Cytochrome P450 85A2 (AtCYP85A2) was introduced to Arabidopsis thaliana seeds using a seed-specific promoter, pAt5g54000. GUS (β-Glucuronidase) activity and RT-PCR analysis demonstrated that AtCYP85A2 overexpression only occurred in seeds of a transgenic plant, pAt5g54000-AtCYP85A2::Col-0. A crude enzyme solution prepared pAt5g54000-AtCYP85A2::Col-0 seeds successfully catalyzed the conversion of castasterone (CS) to brassinolide (BL), which was not detected in wild-type seeds. Furthermore, a higher level of CS and BL was detected in pAt5g54000-AtCYP85A2::Col-0 seeds compared to untransformed seeds, thus demonstrating that seed-specific overexpression of AtCYP85A2 efficiently encoded a bi-functional enzyme for brassinosteroids 6-oxidase/brassinolide synthase to generate CS and BL in seeds of pAt5g54000-AtCYP85A2::Col-0. Compared to the wild type, pAt5g54000-AtCYP85A2::Col-0 produced substantially larger seeds with a high concentration of nutrients due to an enhancement in brassinosteroids signaling. Additionally, pAt5g54000-AtCYP85A2::Col-0 exhibited superior seed germination, seedling and rosette plant growth, and flower and silique formation, indicating that seed-specific AtCYP85A2 expression activates overall vegetative and reproductive growth and development in A. thaliana.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brassinosteroids (BRs) are steroidal plant hormones that control various plant processes including growth, development, and differentiation. More than 50 naturally occurring BRs have been identified, among which brassinolide (BL) and its direct biosynthetic precursor castasterone (CS) have shown stronger activity than other natural BRs, suggesting that CS and BL are physiologically the most important BRs in plants (Grove et al. 1979; Bajguz 2007). In Arabidopsis thaliana, CS has been identified as the most abundant BR in every plant organ, suggesting that CS is a biologically active BR that mediates the overall growth and development of Arabidopsis plants (Yokota et al. 1982; Fujioka et al. 1998; Bajguz and Tretyn 2003; Roh et al. 2017; Kim et al. 2018). Vegetative organs such as the root, shoot/stem, and leaf in Arabidopsis exhibit little to no BL, whereas the endogenous levels of BL are much higher in reproductive organs such as flowers, siliques, and seeds, suggesting that BL plays essential roles in the growth and development of Arabidopsis reproductive organs (Kim et al. 2005; Montoya et al. 2005; Nomura et al. 2005).
Both CS and BL in Arabidopsis are sensed by the heterodimeric membrane-localized receptor kinases brassinosteroid insensitive 1 (AtBRI1) and AtBRI1-associated receptor kinase 1 (AtBAK1) (Sun et al. 2013). The signal is transferred to the BR signaling kinases (AtBSKs) and AtBRI1 suppressor 1 (AtBSU1), which suppress the kinase activity of brassinosteroid insensitive 2 (AtBIN2), a negative regulator in BR signaling (He et al. 2002; Tang et al. 2008; Kim et al. 2011). The inactivation of AtBIN2 leads to the accumulation of BR transcription factors, such as brassinazole resistant 1 (AtBZR1) and AtBRI1 EMS suppressor 1 (AtBES1) in the nucleus, which regulates the transcription of BR-responsive genes, thus enabling BRs to mediate plant growth and development (Kim and Wang 2010; Wang et al. 2012).
CS and BL are known to be biosynthesized by AtCYP85A1 and/or AtCYP85A2 in Arabidopsis (Kim et al. 2005; Nomura et al. 2005). Both AtCYP85A1 and AtCYP85A2 consist of 465 amino acids and share 83% identity and 92% similarity. However, AtCYP85A1 and AtCYP85A2 show different enzyme activities in the biosynthesis of CS and/or BL in Arabidopsis (Kim et al. 2005). The conversion of 6-deoxocastasterone (6-deoxoCS) to CS is catalyzed by both AtCYP85A1 and AtCYP85A2; however, the conversion of CS to BL is only mediated by AtCYP85A2, indicating that AtCYP85A1 possesses only BR 6-oxidase activity, whereas AtCYP85A2 acts as a bifunctional enzyme with both BR 6-oxidase and BL synthase (Fig. 1). The enzyme kinetics indicated that the BR 6-oxidase activity in AtCYP85A2 is more powerful than that in AtCYP85A1, suggesting that endogenous CS and BL seem to be predominantly maintained by AtCYP85A2 in the plant. Therefore, the manipulation of AtCYP85A2 is a promising approach for the regulation of BR activity in plant organs.
Compared to wild-type plants, Arabidopsis plants with universally overexpressed AtCYP85A2 (35S-AtCYP85A2) synthesized higher levels of endogenous CS and BL, resulting in the promotion of growth and development in 35S-AtCYP85A2 (Kim et al. 2005). Specifically, 35S-AtCYP85A2 yielded larger siliques that contained higher numbers of seeds than the wild type, indicating that AtCYP85A2 plays important roles in the growth and development of reproductive organs in Arabidopsis. However, the mechanisms by which AtCYP85A2 regulates the growth and development of plant reproductive organs remain largely uncharacterized. Therefore, to elucidate these mechanisms, our study sought to introduce AtCYP85A2 into Arabidopsis seeds using a seed-specific promoter, after which the biochemical and physiological effects of seed-specific AtCYP85A2 overexpression on transgenic Arabidopsis seed development were investigated. In addition to enhancing the growth and development of reproductive organs such as flower, silique and seeds, AtCYP85A2 overexpression also promoted the vegetative growth of roots, shoots/stems, and leaves in the transformed plant, demonstrating that seed-specific BR upregulation not only promotes seed yields but also increases crop biomass.
Materials and Methods
Plant Materials and Growth Conditions
A. thaliana Columbia-0 (Col-0) was used as a background ecotype in this study. Col-0 seeds were washed in 70% ethanol (EtOH) (v/v) and rinsed with autoclaved sterile water for 3 times. After cold treatment for 3 days at 4 °C, the seeds were planted in a 0.5 × Murashige and Skoog (MS) medium containing 1% sucrose and 0.8% plant agar. Plants were grown in an environmental growth chamber (Vision Scientific, Seoul, Korea) at 22 °C, with a 16 h light (120 µmol photons m−2 s−1)/8 h dark photoperiod at 20 °C.
Preparation of pAt5g54000-AtCYP85A2::Col-0
A seed-specific expression construct was produced to transform Arabidopsis seeds. A promoter of the At5g54000 (pAt5g54000) construct, pAt5g54000/pBI101, was provided by Jeong Sheop Shin at Korea University. To obtain AtCYP85A2 CDS, the leaves of two-week-old plants were harvested to extract total RNA. The total RNAs of Arabidopsis seedlings were extracted with the TRI Reagent (Invitrogen, Carlsbad, CA, USA). Using an MMLV-reverse transcription system (Promega, Madison, WI, USA), cDNAs were synthesized from 1 μg of the total RNAs according to the manufacturer’s instructions. For amplification of AtCYP85A2, Ex-Taq polymerase (Takara Bio, Shiga, Japan) was used and cloned into the pTA-Topo (MGmed) vector using the BamHI restriction enzyme site.
After sequencing the AtCYP8A2/pTA-topo DNA, pAt5g54000/pBI101 and AtCYP85A2/pTA-topo were digested by BamHI (New England Biolab). T4 DNA ligase (Takara Bio) was then used to connect the cut vector and the AtCYP85A2 CDS fragment. This construct, pAt5g54000-AtCYP85A2/pBI101, was confirmed through DNA sequencing and was transformed into Agrobacterium strain GV3101 via electroporation (Bio-Rad).
Using the floral-dip method (Clough and Bent 1998), the GV3101 Agrobacterium strain containing the seed-specific expression of AtCYP85A2 construct (pAt5g54000-AtCYP85A2/pBI101) was introduced to Col-0 to produce a transgenic Arabidopsis mutant. Transformed seeds were then selected using kanamycin.
Semi-qPCR and qRT-PCR Analysis
Total RNA and cDNA were prepared from Col-0 and the transgenic mutant using the TRI reagent and the MMLV-reverse transcription system as described above. Semi-qPCR was then conducted to confirm the expression level of AtCYP85A2 in the transgenic mutant. For the amplification of AtCYP85A2 and reference gene UBQ5, cDNAs extracted from the same amount of each tissue as the template were used. AtCYP85A2 and UBQ5 were amplified for 20 thermal cycles (95 °C for 10 s, 58 °C for 10 s, and 72 °C for 10 s) using rTaq polymerase (ELPIS, Seoul, South Korea).
Next, the expression of AtCYP85A1, AtCYP85A2, GA/ABA biosynthetic genes, seed size-determining genes and BR signaling genes was quantified via qRT-PCR using a CFX96TM Real-Time PCR Detection system and iQ SYBR Green Supermix (Bio-Rad). The thermal cycling program included an initial denaturation step at 95 °C for 3 min, followed by 45 cycles of 95 °C for 10 s, 50 °C for 15 s, and 75 °C 15 s. PP2A was used to normalize the gene expression level. The primers used for the semi-qPCR and qRT-PCR are described in Supplementary Table 1.
Histochemical GUS Staining
Histochemical GUS staining was performed as described in a previous method (Glazebrook and Weigel 2002). Samples were fixed in cold 90% acetone overnight, then transferred to a staining solution containing 2 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (Duchefa, Haarlem, Netherlands) in 50 mM Na2HPO4 buffer (pH 7.2), 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, and 0.2% triton X-100. The samples in the staining solution were infiltrated under a vacuum on ice for 30 min. Afterward, the samples were incubated at 37 °C overnight. All samples were transferred to 20, 35, and 50% ethanol and sequentially fixed in 50% ethanol, 10% acetic acid, and 5% formaldehyde for 20 min. The samples were then washed with 70% ethanol and observed under a dissecting microscope (Olympus SZ-PT).
Endogenous BR Analysis and Crude Enzyme Assay
Crude enzyme preparation and purification of BL as an enzyme product and endogenous BRs were conducted as described in a previous study (Roh et al. 2020). The seeds of Col-0 and pAt5g54000-AtCYP85A2::Col-0 were harvested, and the remaining portion (shoot and root) from the plants was harvested to be used as a control. Fifty grams of samples for each part were used to purify endogenous BRs. Reverse-phase HPLC (SenshuPak, C18) was then performed after purification by chromatography. The sample was eluted using a MeCN-water gradient 45% MeCN for 0–20 min, for 20–40 min, 45–100% MeCN gradient, for 40–70 min, 100% MeCN. Fractions were collected every minute, and the flow rate was 2.5 mL min−1. BL and CS were detected at 13/14 and 22/23 min in this HPLC condition, respectively. For both CS and BL, these fractions were dried and combined before being analyzed via capillary GC-selected ion monitoring (SIM)/MS after bismethaneboronation.
Twenty grams of samples for each part were used to prepare the cell-free crude enzyme solution. The crude enzyme solution was quantified using the Bradford solution (Bio-Rad) coupled with bovine serum albumin (BSA) as a standard. Concretely, 5 μg of CS was added to 1 mg of cell-free enzyme solution with a cofactor (NADPH). Enzyme mixtures were incubated at 37 °C for 3 h with gentle shaking. The products for the enzyme reactions were extracted using water-saturated ethyl acetate and then purified. Reverse-phase HPLC (SenshuPak, C18) was conducted as described above. The fractions for BL were dried, combined, and analyzed via capillary GC-SIM/MS after bismethaneboronation.
A Hewlett-Packard 5973 mass spectrometer (electron impact ionization, 70 electron voltage; Agilent, Santa Clara, CA, USA) connected to a 6890 gas–chromatography fitted with a fused silica capillary column (HP-5, 0.25 mm × 15 m, 0.25 μm film thickness; Agilent) was used for GC-SIM/MS analysis. Helium was used as a carrier gas at a 1 mL min−1 flow rate. The samples were processed using an on-column injection mode. The initial oven temperature was maintained for 2 min at 175 °C, after which it was elevated to 280 °C at a 40 °C min−1 rate and finally maintained at 280 °C for 15 min. Methaneboronation was performed by dissolving the samples in pyridine containing of 2 mg mL−1 methaneboronic acid (Sigma) and incubating them at 80 °C for 20 min.
Phenotype and Statistical Analysis
With an optical microscope, a total of 200 seeds of Col-0, 35S-AtCYP85A2, and pAt5g54000-AtCYP85A2::Col-0 were examined to measure length and width. Phenotype changes were determined by measuring the width, length, and mass of the seed.
Pair-wise comparisons between groups were conducted via Student's t-test and Shapiro–Wilk test was performed to test for normality. Multiple comparisons were conducted via one-way analysis of variance (ANOVA) followed by the Holm-Sidak test.
Quantification of Soluble Starch, Proteins, and Lipids in Arabidopsis Seeds
One-hundred seeds for Col-0 and pAt5g54000-AtCYP85A2::Col-0 were used for the quantification of each substance. The Starch Assay Kit (Abcam, Cambridge, United Kingdom) was used to measure endogenous starch in Arabidopsis seeds according to the manufacturer’s instructions. After color development for each sample, their absorbances were measured at 570 nm. The standard curve was constructed using the starch standard included in the kit.
The modified version of the Focks and Benning method was used to quantify the total soluble proteins (Focks and Benning 1998). Briefly, Arabidopsis seeds were ground in liquid nitrogen and mixed with 250 µL of cold acetone. This mixture was incubated at − 20 °C for 10 min to precipitate the proteins. The samples were then centrifuged at 16,000×g, and 4 °C for 20 min, after which the supernatant was discarded. The pellets were resuspended into 250 µL of total protein extraction buffer (50 mM Tris–HCl, 250 mM NaCl, 1 mM EDTA, 1% SDS [w/v]). Afterward, the samples were incubated at room temperature (RT) for 2 h with gentle shaking, then centrifuged at 16,000×g and 4 °C for 5 min. The protein concentration of the supernatant was measured using the Bradford solution (Bio-Rad) at 595 nm, and BSA was used as a standard.
Next, a modified version of the method employed by Mishra was used to quantify the total lipids in the seeds (Mishra et al. 2014). Arabidopsis seeds were ground in liquid nitrogen and mixed with 600 μL of CHCl3/MeOH at a 2:1 ratio. The samples were then vortexed for 2 min and centrifuged at 13,000 rpm for 15 min. The supernatants were collected and dried in a vacuum at 50 °C. The dried supernatants were resuspended in sterile water and incubated at 100 °C for 10 min. The samples were then cooled for 5 min in ice, and 5 mL of a vanillin-phosphoric acid reagent (SPV reagent, 0.2 mg mL−1 vanillin, 17% phosphoric acid) was added to each sample. Next, the samples were incubated with shaking at 200 rpm and 37 °C. Upon color development, the absorbance was measured at 530 nm. Cholesterol (CHR) was used to produce the standard curve.
Germination Assay
Germination assays were performed five times. Briefly, 150 Col-0 and pAt5g54000-AtCYP85A2::Col-0 seeds were used for each assay. Arabidopsis seeds were washed twice with 70% EtOH (v/v) and rinsed with sterile water for four times. After cold treatment (4 °C incubation) for 3 days, the seeds were planted in 0.5 × MS medium. Seed germination was monitore3d 24 h after the seeds were planted.
Results
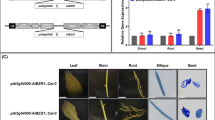
pAt5g54000, an Arabidopsis seed-specific promoter, was cloned into a binary vector. The AtCYP85A2 gene was then cloned into the vector containing pAt5g54000 (Fig. 2A). The resulting construct was transferred into Agrobacterium tumefaciens and introduced into the wild type (Col-0), after which antibiotic-resistant seeds were collected. Semi-qRT-PCR analysis for the expression of AtCYP85A2 in transgenic Arabidopsis (pAt5g54000-AtCYP85A2::Col-0) indicated that AtCYP85A2 expression was slightly enhanced in the shoot and root (approximately 1.1 times higher) but significantly increased in seeds (3.75 times higher) compared to those in the wild-type seeds (Fig. 2B, C). In the GUS report system, GUS activity was not detected in the root, stem, leaf, and flower, whereas strong GUS activity was identified in the seeds from the siliques of transgenic Arabidopsis (Fig. 2D). These findings thus demonstrated the seed-specific introduction of AtCYP85A2 into pAt5g54000-AtCYP85A2::Col-0.
Seed-specific expression of AtCYP85A2 in the constructed pAt5g54000-AtCYP85A2::Col-0 mutant. A Schematic diagram of the seed-specific expression of AtCYP85A2 constructs. The seed-specific promoter and AtCYP85A2 were ligated in the pBI101 vector. NOS promoter, nopaline synthase promoter; NPT II (KanR), neomycin phosphotransferase II (kanamycin resistance); NOS terminator, nopaline synthase terminator; GUS, β-glucuronidase gene (uidA); LB, left border; RB, right border. B Expression levels of AtCYP85A1 and AtCYP85A2 of the shoots, roots, and seeds of Arabidopsis measured via semi-qPCR. UBQ5 was used as the reference gene in A. thaliana. C Expression levels of AtCYP85A1 and AtCYP85A2 in leaves, roots, and seeds of Arabidopsis measured via qPCR. PP2A was used for normalization. The numbers on each bar represent the relative gene expression levels of pAt5g54000-AtCYP85A2::Col-0 compared to that of Col-0. D GUS staining in the roots, stems, leaves, flowers, siliques, and seeds of pAt5g54000-AtCYP85A2::Col-0. GUS activity was concentrated on the siliques and seeds of transgenic plants
The seed-specific expression of AtCYP85A2 can alter the endogenous levels of CS and BL in pAt5g54000-AtCYP85A2::Col-0. To verify this, a quantitative analysis of CS and BL in wild type and pAt5g54000-AtCYP85A2::Col-0 was performed. After adding [26. 28-2H3-CS] and [26. 28-2H3-BL] as internal standards, ethyl acetate soluble fractions obtained from wild type and pAt5g54000-AtCYP85A2::Col-0 were purified using silica gel and C18 column chromatography followed by reverse-phase HPLC. The active HPLC fractions corresponding to CS (fraction 22/23), and BL (fraction 13/14) were collected and analyzed via capillary GC–MS or GC- SIM after methaneboronation.
As summarized in Table 1, our full GC–MS analyses indicated that the bismethaneboronate (BMB) of the active compound in the 22/23 HPLC fraction obtained from plants without seeds and seeds of the wild type and pAt5g54000-AtCYP85A2::Col-0 exhibited the same MS spectrum at m/z 512 [M+], 357, 327, 287, and 155 at the same retention time on GC as those of authentic CS BMB. This demonstrated that CS was present in every organ in the wild type and transgenic plants. Endogenous level of CS calculated by determining the ratio of endogenous CS/[2H3]-labeled CS in plants without seeds were almost equal to that in the wild type. However, the CS level in the seeds of pAt5g54000-AtCYP85A2::Col-0 was higher (approximately 2.2 times) than that in the seeds of the wild type, indicating that seed-specific overexpression of AtCYP85A2 enhanced BR 6-oxidase activity to generate CS from 6-deoxoCS only in seeds of pAt5g54000-AtCYP85A2::Col-0.
Furthermore, a crude enzyme solution prepared from plants without seeds and seeds of the wild type did not mediate the conversion of CS to BL (Fig. 3A, B). Additionally, even in the GC-SIM analyses that showed a higher sensitivity for BR detection than a full GC–MS analysis, no BL was identified from plants without seeds and seeds of the wild type (Table 1), indicating that the wild type plants exhibited little to no BL synthase activity for the conversion of CS to BL. Similar to the wild type, no BL synthase activity nor endogenous BL were found in pAt5g54000-AtCYP85A2::Col-0 plants without seeds. However, a crude enzyme from the seeds of pAt5g54000-AtCYP85A2::Col-0 successfully catalyzed the conversion of CS to BL. Additionally, the BMB of the active compound in the HPLC fraction 13/14 obtained from the seeds exhibited the same MS spectrum (at m/z 528[M+], 374, 332, 177, and 155) and GC retention time as those of authentic BL BMB, thus demonstrating the occurrence of endogenous BL in the transgenic seeds. Quantitative analysis revealed that the pAt5g54000-AtCYP85A2::Col-0 seeds contained a fair amount of BL (approximately 6.4 ng g−1 fresh weight), which demonstrated that BL synthase activity was strongly concentrated in the transgenic seeds. Taken together, our findings demonstrated that AtCYP85A2 acted as a bifunctional enzyme possessing both BR 6-oxidase and BL synthase activity. Further, this newly created enzyme was synthesized via the seed-specific introduction of AtCYP85A2 to synthesize CS and BL in seeds of pAt5g54000-AtCYP85A2::Col-0 seeds.
BL synthase activity and BR signaling were specifically increased in pAt5g54000-AtCYP85A2::Col-0 seeds. A GC–MS results for the conversion of CS to BL with the crude enzyme solution prepared from tissues of Col-0 and pAt5g54000-AtCYP85A2::Col-0. [2H3]-BL was used as an internal standard. N.D. indicates not detected. B Specific enzyme activity of the crude enzyme prepared from tissues of Col-0 and pAt5g54000-pAtCYP85A2::Col-0. N.D. indicates not detected. C Gene expression level of BR signaling component genes in Col-0 and pAt5g54000-AtCYP85A2::Col-0. qRT-PCR was performed with total RNA. PP2A was used to normalize the gene expression level. Two biological replicates and three technical replicates were performed for the quantification of gene expression levels. Bar graphs were generated using Prism 8.0 software. The asterisks indicate the statistical significance of Student's t-test: *(P < 0.05), **(P < 0.01), and ***(P < 0.001)
To investigate whether BR signaling was activated in pAt5g54000-AtCYP85A2::Col-0 seeds, the expression level of genes involved in BR signaling were compared to those in untransformed seeds. As shown in Fig. 3C, the expression of genes for positive regulators such as AtBRI1, AtBES1, and AtBZR1 was activated, whereas the expression of AtBIN2, a negative regulator, was inhibited in the seeds. These findings suggested that AtCYP85A2 overexpression activated the biosynthesis of CS, and BL enriched BR signaling in pAt5g54000-AtCYP85A2::Col-0 seeds.
The physiological effects of the seed-specific overexpression of AtCYP85A2 on the growth and development of pAt5g54000-AtCYP85A2::Col-0 were investigated at several developmental stages. During seed germination, the rate of germination was activated by pAt5g54000-AtCYP85A2::Col-0, resulting in faster seedling growth compared to the wild type (Fig. 4A). In germinating pAt5g54000-AtCYP85A2::Col-0 seeds where AtCYP85A2 expression gradually increased, the expression of GA biosynthetic genes such as GA20 oxidase (GA20ox) and GA3 oxidase (GA3ox) was enhanced, whereas the expression of an ABA biosynthetic gene such as Abscisic acid deficient2 (ABA2) was suppressed (Fig. 4B). In seedlings grown under light conditions, the length of primary roots, length of hypocotyls, and size of cotyledons in pAt5g54000-AtCYP85A2::Col-0 were longer than those of wild type seedlings (Fig. 5A). In the rosette plant stage, pAt5g54000-AtCYP85A2::Col-0 formed larger expanded leaves with longer petioles than wild-type leaves, which accelerated the bolting of the inflorescent stem in the transgenic plants (Fig. 5B). In adult plants, the growth and branching of inflorescent stems were activated in transgenic plants (Fig. 5C). In the reproductive growth stage, flowers and siliques developed more quickly in pAt5g54000-AtCYP85A2::Col-0, resulting in an increased number of flowers and siliques in the transgenic plants (Fig. 6A). Compared to the wild type, pAt5g54000-AtCYP85A2::Col-0 produced larger siliques containing a greater number of seeds. Further, pAt5g54000-AtCYP85A2::Col-0 produced seeds that were fairly larger in length, width, and weight than untransformed seeds (Fig. 6B). Moreover, the endogenous amounts of starch, proteins, and lipids in the seeds of pAt5g54000-AtCYP85A2::Col-0 were increased by as much as approximately two-to-threefold compared to the wild type (Fig. 6C), which indicated that both the size and quality of seeds were significantly improved in pAt5g54000-AtCYP85A2::Col-0. Collectively, our findings demonstrated that seed-specific introduction of AtCYP85A2 enhanced both the vegetative and reproductive growth and development of pAt5g54000-AtCYP85A2::Col-0 plants.
Enhanced germination rates in the pAt5g54000-AtCYP85A2::Col-0 mutant. A Germination rate of seeds in Col-0 and pAt5g54000-AtCYP85A2::Col-0. The germination assay was performed three times, and 150 seeds per line were used for each germination assay. B Relative gene expression levels of AtCYP85A2, AtGA20OX, AtGA3OX, and AtABA2 at several time points compared to the gene expression level at 1DAG (days after germination) in pAt5g54000-AtCYP85A2::Col-0. Relative gene expression levels calculated by dividing the gene expression levels of each time point by 1DAG. PP2A was used to normalize the gene expression level. The graphs were generated using Prism 8.0
Increases in vegetative growth in the pAt5g54000-AtCYP85A2::Col-0 mutant. Phenotypes of Col-0 and pAt5g54000-AtCYP85A2::Col-0 at various times. All scale bars in each figure indicate 1 cm. A Phenotypes of seedlings in Col-0 and pAt5g54000-AtCYP85A2::Col-0. The photographs were taken in 10 days after planting. B Phenotypes of the rosette plant in Col-0 and pAt5g54000-AtCYP85A2::Col-0. The photographs depict 4-week-old plants. C Phenotypes of adult Col-0 and pAt5g54000-AtCYP85A2::Col-0 plants. Photographs of 6-week-old plants were taken. The asterisks indicate the statistical significance of Student's t-test: ns (P > 0.05), *(P < 0.05), **(P < 0.01), and ***(P < 0.001). The dot plots were generated using Prism 8.0
The pAt5g54000-AtCYP85A2::Col-0 mutants exhibited superior reproductive growth and seed yields both in terms of quality and quantity. All graphs in this figure were drawn using Prism 8.0. The asterisks indicate the statistical significance of Student's t-test: *(P < 0.05), **(P < 0.01), and ***(P < 0.001). A Phenotypes of siliques in Col-0 and pAt5g54000-AtCYP85A2::Col-0. The siliques were obtained from 6-week-old plants. The scale bar indicates 1 cm. Thirty siliques were measured for each line. B Seed phenotypes in Col-0, 35S-AtCYP85A2, and pAt5g54000-AtCYP85A2::Col-0. The scale bar indicates 500 µm. A total of 200 seeds were measured for each line. C Quantitative analysis of total starch, proteins, and lipids in seeds of Col-0 and pAt5g54000-AtCYP85A2::Col-0. Each quantitative analysis was performed in triplicate. D Gene expression levels of seed size-determinizing genes (SHB1, IKU2, and MINI3) in seeds of Col-0 and pAt5g54000-AtCYP85A2::Col-0. PP2A was used for normalization. Two biological replicates and three technical replicates were evaluated for the quantification of gene expression levels
Discussion
CS and BL directly bind to receptors on the plasma membrane of cells to trigger the activation of BR signaling for the regulation of growth and development in plants. Therefore, the regulation of the endogenous level of CS and BL directly modulates BR activity in plant tissues and organs. Our study demonstrated that AtCYP85A2 overexpression significantly increased the endogenous levels of CS in pAt5g54000-AtCYP85A2::Col-0 seeds. Arabidopsis AtCYP85A1, a homolog of AtCYP85A2, encodes AtCYP85A1, which exerts BR 6-oxidase activity and synthesizes CS from 6-deoxoCS. However, the expression of AtCYP85A1 in pAt5g54000-AtCYP85A2::Col-0 and wild-type seeds was largely equal (Fig. 2B, C), suggesting that the increased levels of CS in the transgenic seeds were caused by the AtCYP85A2 overexpression. Considerable amounts of BL, which were absent in wild-type seeds, were synthesized in the pAt5g54000-AtCYP85A2::Col-0 seeds. Assuming that the biological activity of BL was 5 to 10 times higher than that of CS, our findings indicated that AtCYP85A2 overexpression resulted in BR activity levels tens of times higher in the seeds of transgenic Arabidopsis compared to the controls.
AtSHB1, AtIKU2, and AtMINI3 are known as positive regulatory genes for the determination of seed size/mass and are direct target genes of AtBZR1 in Arabidopsis (Jiang et al. 2013; Li et al. 2019). Coupled with the enhanced expression of AtBZR1, the expression of AtSHB1, AtIKU2, and AtMINI3 increased in the pAt5g54000-AtCYP85A2::Col-0 seeds (Figs. 3C, 6D), suggesting that a signaling pathway of AtBZR1 → AtSHB1 → AtIKU2 → AtMINI3 plays a role in increasing the size of seeds in transgenic plants.
Interactions between plant hormones such as gibberellins (GAs), abscisic acid (ABA), and BRs, are known to be important for regulating seed germination in plants. When endogenous levels of GAs and BRs are increased, the germination of seeds is activated. In contrast, when the ABA level is increased, seed germination is inhibited. Recent findings have reported that BR transcription factors BZR1 and BES1 directly bind to promoters of GA20ox/GA3ox and ABA2 to increase and decrease the synthesis of GAs and ABA, respectively. In our study, the endogenous levels of GA and ABA appeared to be respectively, increased and decreased by high BR activity in the transgenic seeds (Fig. 4B). Therefore, seed-specific AtCYP85A2 overexpression appeared to alter the hormonal balance of GA and ABA in pAt5g54000-AtCYP85A2::Col-0 seeds, which promoted the germination of seeds in transgenic Arabidopsis.
Compared to 35S-AtCYP85A2, pAt5g54000-AtCYP85A2::Col-0 grew larger siliques. Furthermore, the siliques of the pAt5g54000-AtCYP85A2::Col-0 plants produced seeds that were substantially larger than those produced by the 35S-AtCYP85A2 mutant (Fig. 6B). This demonstrated that the overexpression of AtCYP85A2 using a seed-specific promoter such as pAt5g54000 was more effective at inducing biologically active BRs and enhance the development of reproductive organs such as siliques and seeds compared to AtCYP85A2 overexpression using the 35S universal promoter in Arabidopsis.
Interestingly, the vegetative growth of pAt5g54000-AtCYP85A2::Col-0 was also activated by AtCYP85A2 overexpression in the seeds (Fig. 5), suggesting that BR activity may also have been increased in the vegetative organs of pAt5g54000-AtCYP85A2::Col-0. However, the expression of AtCYP85A1 and AtCYP85A2 to generate CS and BL in the shoots and roots of pAt5g54000-AtCYP85A2::Col-0 were almost identical to those in the wild type. Additionally, the endogenous levels of CS and BL in the vegetative organs of pAt5g54000-AtCYP85A2::Col-0 were almost the same as those in the wild type (Table 1), suggesting that the seed-specific expression of AtCYP85A2 did not affect the biosynthesis of active BRs to increase BR activity in vegetative organs. Instead, we speculate that the resulting abundance of nutrients such as starch, lipids, and proteins, in the seeds supplied enough energy to promote the growth of seedlings. As a result, signals other than BR appeared to activate processes involved in the vegetative growth of pAt5g54000-AtCYP85A2::Col-0. Additional studies are thus being conducted to identify the molecular mechanisms that drive the vegetative growth in pAt5g54000-AtCYP85A2::Col-0 mutants without altering BR activity.
BRs are considered ideal crop enhancers in agriculture research. However, the high cost of BR synthesis discourages their direct application to plants (Tong and Chu 2012). For this reason, the genetic modulation of BR biosynthesis genes is a good approach to increase BR activity in plants. We previously demonstrated that heterologously expressed AtCYP85A2 in seeds of Brachypodium distachyon, a model for monocotyledonous crops where no BL synthase activity is present, improved grain yields and quality (Roh et al. 2021). In this study, we found that seed-specific AtCYP85A2 overexpression in Arabidopsis increased the overall vegetative and reproductive growth and seed formation of the mutant plants. This suggests that seed-specific activation of BR activity, as well as the seed-specific introduction of AtCYP85A2, are promising approaches to improve both seed yields and biomass, thus improving the productivity of commercially relevant monocotyledonous and dicotyledonous plants.
References
Bajguz A (2007) Metabolism of brassinosteroids in plants. Plant Physiol Biochem 45(2):95–107
Bajguz A, Tretyn A (2003) The chemical characteristic and distribution of brassinosteroids in plants. Phytochemistry 62(7):1027–1046
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Focks N, Benning C (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118(1):91–101
Fujioka S, Noguchi T, Yokota T, Takatsuto S, Yoshida S (1998) Brassinosteroids in Arabidopsis thaliana. Phytochemistry 48(4):595–599
Glazebrook J, Weigel D (2002) Arabidopsis: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Grove MD, Spencer GF, Rohwedder WK, Mandava N, Worley JF, Warthen JD, Steffens GL, Flippen-Anderson JL, Cook JC (1979) Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281(5728):216–217
He J-X, Gendron JM, Yang Y, Li J, Wang Z-Y (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci 99(15):10185–10190
Jiang W-B, Huang H-Y, Hu Y-W, Zhu S-W, Wang Z-Y, Lin W-H (2013) Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol 162(4):1965–1977
Kim T-W, Wang Z-Y (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol 61:681–704
Kim T-W, Hwang J-Y, Kim Y-S, Joo S-H, Chang SC, Lee JS, Takatsuto S, Kim S-K (2005) Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17(8):2397–2412
Kim T-W, Guan S, Burlingame AL, Wang Z-Y (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43(4):561–571
Kim S, Moon J, Roh J, Kim S-K (2018) Castasterone can be biosynthesized from 28-homodolichosterone in Arabidopsis thaliana. J Plant Biol 61(5):330–335
Li N, Xu R, Li Y (2019) Molecular networks of seed size control in plants. Annu Rev Plant Biol 70:435–463
Mishra SK, Suh WI, Farooq W, Moon M, Shrivastav A, Park MS, Yang J-W (2014) Rapid quantification of microalgal lipids in aqueous medium by a simple colorimetric method. Bioresour Technol 155:330–333
Montoya T, Nomura T, Yokota T, Farrar K, Harrison K, Jones JG, Kaneta T, Kamiya Y, Szekeres M, Bishop GJ (2005) Patterns of Dwarf expression and brassinosteroid accumulation in tomato reveal the importance of brassinosteroid synthesis during fruit development. Plant J 42(2):262–269
Nomura T, Kushiro T, Yokota T, Kamiya Y, Bishop GJ, Yamaguchi S (2005) The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J Biol Chem 280(18):17873–17879
Roh J, Yeom HS, Jang H, Kim S, Youn JH, Kim S-K (2017) Identification and biosynthesis of C-24 ethylidene brassinosteroids in Arabidopsis thaliana. J Plant Biol 60(5):533–538
Roh J, Moon J, Youn J-H, Seo C, Park YJ, Kim S-K (2020) Establishment of biosynthetic pathways to generate castasterone as the biologically active brassinosteroid in Brachypodium distachyon. J Agric Food Chem 68(13):3912–3923
Roh J, Moon J, Lee YE, Park CH, Kim S-K (2021) Seed-specific expression of Arabidopsis AtCYP85A2 produces biologically active brassinosteroids such as castasterone and brassinolide to improve grain yield and quality in seeds of brachypodium distachyon. Front Plant Sci. https://doi.org/10.3389/fpls.2021.639508
Sun Y, Han Z, Tang J, Hu Z, Chai C, Zhou B, Chai J (2013) Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res 23(11):1326–1329
Tang W, Kim T-W, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang Z-Y (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321(5888):557–560
Tong H, Chu C (2012) Brassinosteroid signaling and application in rice. J Genet Genomics 39(1):3–9
Wang Z-Y, Bai M-Y, Oh E, Zhu J-Y (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46:701–724
Yokota T, Arima M, Takahashi N (1982) Castasterone, a new phytosterol with plant-hormone potency, from chestnut insect gall. Tetrahedron Lett 23(12):1275–1278
Acknowledgements
We would like to thank Prof. Jeong Sheop Shin (Korea University, Seoul, Republic of Korea) for providing the seed-specific promoter construct pAt5g54000/pBI101. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government’s Ministry of Science, ICT, and Future Planning (2021R1A2C1007516).
Author information
Authors and Affiliations
Contributions
S-KK, JR, MHY, and C-HP planned the experiments; MHY, C-HP, and YL carried out the experiments; S-KK, JR, MHY, and C-HP analyzed the data; S-KK, JR, MHY, and C-HP wrote and revised the manuscript for publication. All authors agreed on the contents of this manuscript and declare no conflicts of interest.
Corresponding authors
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yeon, M.H., Park, CH., Lee, Y.E. et al. Seed-Specifically Overexpressed Arabidopsis Cytochrome P450 85A2 Promotes Vegetative and Reproductive Growth and Development of Arabidopsis thaliana. J. Plant Biol. 65, 75–86 (2022). https://doi.org/10.1007/s12374-021-09340-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-021-09340-3