Abstract
Phosphorus is an important macronutrient for plant growth and is acquired by plants mainly as phosphate. Phosphate fertilizer is usually used to reduce inorganic phosphate (Pi) deficiency in the soil, improve the low phosphorus and drought tolerance of plants, and promote plant growth. Phosphate transporters (PHTs) play an important role in absorbing phosphorus from the soil. MdPHT1;7 was induced by Pi deficiency and drought in roots in our previous research. In this study, we cloned MdPHT1;7 and showed its heterologous expression can complement a high-affinity Pi transporter gene in the Pi uptake-defective yeast mutant BY4743. MdPHT1;7 is a phosphorus transporter located on the cell membrane. Overexpression of MdPHT1;7 in ‘Orin’ apple and ‘Micro-Tom’ tomato enhanced Pi accumulation, low Pi tolerance and drought tolerance. We hypothesized that MdPHT1;7 can enhance Pi absorption and play an important role in improving plant resistance to low phosphorus and drought stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phosphate is a major macronutrient for plant growth and development (Raghothama 1999). In plants, inorganic phosphate (Pi) is an important biomolecule, and in the form of phosphate, pyrophosphate, adenosine triphosphate, adenosine diphosphate, or adenosine monophosphate, Pi has a crucial role in energy transfer and metabolic regulation. Drought is the most significant environmental stress in agriculture worldwide, and improving yield under drought is a major goal of plant breeders. Water and nutrients are two of the most important factors controlling the growth of plants. Drought can prevent Pi absorption by reducing the distribution of Pi in the root system (Ackerson 1985). Pi can also increase tissue osmotic pressure by adjusting the water content in the cell structure and the soluble sugar content in the cell and enhance tolerance to drought stress (Liu et al. 2015a). Plants fertilized with Pi showed a moderate growth increase under drought compared to that of nonfertilized plants (Graciano et al. 2005). Moreover, phosphate fertilizer is also usually used to reduce Pi deficiency in the soil, improve the drought tolerance of plants, and promote plant growth (Faustino et al. 2013). In Alnus cremastogyne, phosphorous fertilization alleviates drought stress by regulating its antioxidant and osmotic potential (Tariq et al. 2018). A number of recent studies have highlighted the role of phosphorus in mitigating drought stress in plants by promoting growth, especially in seedlings (Tng et al. 2014). Plants mainly take up Pi from the soil using phosphate transporter (PHT) located on the cell membranes of plant roots, and Pi is then transported to the plant (Mimura et al. 2003). Increased phosphorus uptake enhance tolerance to low phosphorus (Kakar et al. 2001; Bucher et al. 2015) and drought stresses (Begum et al. 2020; Faustino et al. 2013).
Genome sequence analyses and experimental evidence have indicated that plants contain numerous PHT families, including PHT1, PHT2, PHT3, PHT4, and PHT5, which are distinguished by their protein sequences, structures, locations, and functions (Rausch and Bucher 2002; Knappe et al. 2003; Guo et al. 2008; Liu et al. 2011; Liu et al. 2016; Zhang et al. 2016a). The PHT1 family is the most widely studied in plants, and PHT1 proteins represent a group of phosphate carriers usually found in the plasma membrane (Pao et al. 1998). The first high-H+/Pi phosphate transporter identified in higher plants was AtPT1 from Arabidopsis thaliana (Muchhal et al. 1996). This gene has significant roles in the uptake of phosphorus from the soil (Lopez-Arredondo et al. 2014).
Most members of the PHT1 family are induced by low phosphorus stress and are mainly expressed in the roots. AtPht1;1 and AtPht1;4 are capable of Pi transport and are localized mainly at the plasma membrane of epidermis and stele cells of Arabidopsis roots, confirming their important roles in Pi acquisition and root-to-shoot translocation (Schnable 2012). AtPht1;8 and AtPht1;9 also show high expression levels in roots, suggesting their potential in root Pi uptake and translocation (Misson et al. 2004). In contrast to those four genes, AtPht1;5 is expressed in shoots and actively involved in the mobilization of phosphorous from the Pi source to sink organs in accordance with Pi status (Paszkowski et al. 2002). In rice, the PHT1 family has 13 genes, and eight members have been functionally characterized (Glassop et al. 2005). OsPT1, OsPT6, OsPT9, and OsPT10 are highly expressed in roots and responsible for absorbing Pi in rice (Nagy et al. 2006; Sekhon et al. 2013; Tian et al. 2013). OsPT1 is constitutively expressed in roots, indicating that this gene plays an important role in Pi acquisition under various Pi conditions (Nagy et al. 2006). In contrast to OsPT1, the other three genes are increasingly expressed by low Pi stress and function under low Pi conditions. After being taken up by rice roots, most Pi should be transported into shoots and then distributed into various organs. This process is complex and requires the cooperation of many Pi transporters. OsPT2 and OsPT8 are both highly expressed in stele cells of rice roots and leaves of shoots, and they play essential roles in Pi root-to-shoot translocation and Pi homeostasis in shoots (Rubio et al. 2001; Ye et al. 2015). Moreover, OsPT8 also contributes to Pi translocation from the panicle axis to the rice hull. Additionally, OsPT11 and OsPT13 appear to be involved in arbuscular mycorrhizal fungal symbiosis (Lynch and Conery 2000).
Some reports have indicated that PHT can respond to not only low phosphorus stress but also drought stress. In poplar, PtPHT1.2 and PtPHO9 expression was increased under drought conditions, irrespective of the phosphate levels. In particular, PtPHT1.2 expression was significantly induced by approximately 90-fold in poplar (Zhang et al. 2016a). Under drought stress, 18 PHT genes were upregulated in the leaves, and 11 PHT genes were downregulated in Brassica napus roots (Li et al. 2019). The expression of 15 MdPHT genes was upregulated in apple under drought stress (Sun et al. 2017). Cao et al. (2020) found that overexpression of StPHT1;7 in potato influenced plant growth and tolerance to drought stress. Studies on PHT1 family genes are mostly concentrated in herbaceous plants, but few are concentrated in woody plants. Although the PHT1 transporter was isolated and identified in poplar (Zhang et al. 2016a), there is no in-depth functional analysis or mechanistic research, and PHT transporter proteins have not been reported in apple.
Apple (Malus domestica) is the fourth most economically significant woody fruit plant after Citrus sp., Vitis vinifera, and Musa L. (Hummer and Janick 2009). China is the world’s largest producer of apples. In the northwestern region of China, drought and low Pi levels are the most challenging abiotic stresses for apple trees (Hayano-Kanashiro et al. 2009). Phosphate is an important nutrient for apple crops because it helps drive flowering, fruit set, quality, and yield. This element also promotes cell turgidity by maintaining high leaf water potential, which in turn increases stomatal conductance and rates of photosynthesis under drought conditions (Hassan and Sahrin 2012). Therefore, it is urgent that researchers develop plants with enhanced efficiency of soil phosphorus use under such conditions.
The MdPHT1 family contains 14 genes, MdPHT1;7, located on chromosome 7, and encodes 519 amino acid proteins. The expression of the MdPHT1;7 gene was high in roots and leaves, and MdPHT1;7 was upregulated by Pi starvation and drought stress in root (Sun et al. 2017). In this work, to study the function of MdPHT1;7 and its role in Pi uptake in plants, we transformed ‘Orin’ apple calli and ‘Micro-Tom’ tomato plants to overexpress MdPHT1;7 and investigated the responses to low Pi of transgenic calli and plants. We believe that this study may provide important insights into the behavior of MdPHT1;7 and provide a potential strategy to enhance crop Pi acquisition. Genetic engineering approaches can be applied in efforts to improve the efficiency of phosphate uptake by stressed apple plants.
Materials and Methods
Plant Materials and Treatments
Transgenic and untransformed wild-type (WT) calli of ‘Orin’ apple were placed in the dark at 25 °C on MS medium containing 1.0 mg L−1 2,4-D, 1.0 mg L−1 6-BA, and 8 g L−1 agar and were sub-cultured at 15-day intervals. To investigate the response of transgenic ‘Orin’ calli and the WT to Pi deficiencies, we shifted 0.02 g samples of 10-day-old tissue to the following treatment media: (1) MS control (subculture), with a Pi concentration of 1000 μM; and (2) Low Pi, with the Pi concentration decreasing from 100 to 20 μM. After incubating all plates in the dark at 25 °C for 20 days, the calli were photographed and weighed. The experiments were performed with three biological replicates.
Seeds of WT and transgenic ‘Micro-Tom’ tomato were suspended in 0.05% agarose, stratified in the dark at 4 °C for 3 days, and then seeded in plastic pots (7 × 7 × 7 cm). The seedlings were cultured in a growth chamber (22 °C, 100 μmol photons m−2 s−1, 70% relative humidity, and 16-h light photoperiods) with regular watering for 3 weeks. The experiments were performed with three biological replicates.
After 3 weeks, healthy and uniformly sized plants were assigned to four experimental groups. For the two groups under low Pi stress, the root system was cleaned and cultivated in perlite carefully. The control group was irrigated with standard 1/2-strength Hoagland nutrient solution supplemented with 500 μM KH2PO4 every 3 days, and the Low-Pi treatment group was irrigated with 1/2-strength Hoagland nutrient solution plus 5 μM KH2PO4 every 3 days. After 21 days of treatment, ten plants per line were used to measure the physiological indexes. Another two groups for drought stress were induced by withholding water for 15 days after full irrigation. The control group was normal watering as normal irrigation. Drought stress treatment was performed in triplicate with ten plants per line for each replicate.
Cloning and Analysis of MdPHT1;7
Full-length protein sequences from Arabidopsis, rice, and Populu strichocarpa were downloaded from the NCBI protein database (http://www.ncbi.nlm.nih.gov/guide/) to study their evolutionary relationships. Phylogenetic trees were drawn with the MEGA6.0 program (Testerink and Munnik 2011) using the neighbor-joining (NJ) method (Saitou and Nei 1987) with Poisson corrections and 1000 replications for the bootstrap analysis. The cDNA from leaves of Malus domestica cv. ‘Roya Gala’ was used as template for amplifying the MdPHT1;7 sequences. Specific primers (Table S1) for gene cloning were designed based on the revised putative MdPHT1;7 sequences. PCR was performed with PrimeSTAR® HS DNA Polymerase (TaKaRa, Dalian, China), and amplification conditions were empirically optimized. The PCR products were added to the 3-termini using TaqDNA Polymerase (Fermentas, USA) and cloned into the pMD19-T vector (TaKaRa, Dalian, China). Then, the positive clones were sequenced. The three-dimensional (3D) protein structures for MdPHT1;7 were automated by comparative modeling by SWISS-MODEL (https://swissmodel.expasy.org/). The conserved membrane-spanning domains in MdPHT1;7 were identified with an online tool, prediction of TMHMM (http://www.cbs.dtu.dk/services/TMHMM/).
Subcellular Localization Analysis
For subcellular localization, we used the pGWB405 vector and applied the homologous recombination method (Nakagawa et al. 2007). This vector was driven by the CaMV 35S promoter and included green fluorescent protein (GFP) gene fusions. The 35S:MdPHT1;7-GFP plasmid was introduced into Agrobacterium tumefaciens strain EHA105, and leaves of tobacco (Nicotiana benthaamiana) were transiently transformed as described by Yang et al. (2000). After 2 days, GFP expression was observed using confocal microscopy, and images were processed with FV10-ASW software (version 2.1a; Olympus, Berlin, Germany). The AtCBL1n:mCherry construct was used as a marker for the localization of plasma membrane proteins (Batistic et al. 2010). Primers and restriction sites are listed in Table S1.
Yeast Manipulations
The yeast mutant BY4743, which defects PHO84, a high-affinity Pi transporter gene (Qin et al. 2012; Winzeler et al. 1999), and the expression vector pYES2 were used in yeast complement experiments. The coding sequence of MdPHT1;7 was amplified from cDNAs using Prime STAR Max DNA Polymerase (TaKaRa, Dalian, China). PCR productions were subcloned into pYES2. These constructs were transformed into yeast BY4743. Transformed yeast strains grew in YNB (yeast nitrogen base) medium to the logarithmic phase and were then harvested and washed with Pi-free YNB medium. Collected yeast was first suspended in Pi-free YNB medium and cultivated until the absorbance at 600 nm was 1.0. Then, tenfold serial dilution with equal volumes was applied to solid YNB medium (the sample volume was 3 L) supplied with 20 and 100 mM KH2PO4 and incubated at 30 °C for 2 days. Primers and restriction sites are listed in Table S1.
Genetic Transformation of ‘Orin’ Apple Callus and Tomato
The callus of ‘Orin’ apple was used for gene transformation (Li et al. 2002) and growth assays. They were placed in the dark at 25 °C on MS medium containing 1.0 mg L−1 2,4-D, 1.0 mg L−1 6-BA, and 8 g L−1 agar and were sub-cultured at 15-day intervals. For transformation, 7-day-old calli grown in liquid medium were co-cultivated (10 min, gentle rotation, 25 °C) with A. tumefaciens EHA105 carrying the pGWB408-MdPHT1;7 plasmid. After 2 days of cocultivation, the callus was washed three times with sterile water containing 400 mg L−1 cefotaxime (Cef) and then transferred to subculture medium supplemented with 200 mg L−1 Cef and 30 mg L−1 Kan for transgene selection (Xie et al. 2012).
‘Micro-Tom’ tomato was transformed via the Agrobacterium-mediated method, as previously described (Guo et al. 2012). The resultant plants were PCR-confirmed to select positive transgenic lines, and they were screened with 100 mg L−1 Kan.
RNA Extraction, DNA Isolation, and qRT-PCR
Total RNA was isolated from the positive lines and the untransformed WT according to the CTAB method (Chang et al. 1993). Overexpression of MdPHT1;7 was confirmed by quantitative real-time PCR (qRT-PCR). The DNA was removed by treatment with RNase-free DNase I (Thermo Scientific, Waltham, MA, USA). Apple genomic DNA was then extracted by a modified CTAB method (Modgil et al. 2005), and first-strand cDNA was synthesized using a Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific) with the same amount of mRNA (1 μg). The qRT-PCR procedures followed the protocols for SYBR® Premix Ex Taq™ II (TliRNaseH Plus) (TaKaRa, Dalian, China), and the assay was performed using a CFX96 TM REAL-Time System C1000 Thermal Cycler (Bio-Rad Laboratories, Foster City, CA). Transcripts of the Malus elongation factor 1 alpha gene (EF-1a; DQ341381) were used to standardize the cDNA samples for different genes. Specific primer sequences for expression analysis are shown in Table S1. All experiments were repeated three times biologically based on three separate RNA extracts from three repeats.
Evaluation of Stress Tolerance
Electrolyte leakage (EL) in the leaves was measured according to the methods of Dionisio-Sese and Tobita (1998) by placing 10 uniformly sized pieces (1 × 1 cm) in a test tube containing 10 mL of distilled water. The initial electrical conductivity (EC0) was determined using another test tube that contained 10 mL of distilled water but no leaf tissue. All EL measurements were made with an electrical conductivity analyzer (DDS-307; Shanghai Precision Scientific Instrument Co., Ltd., Shanghai, China). After 3.5 h of incubation in a water bath at room temperature, the second round of electrical conductivity (EC1) of the medium was measured. Samples were then autoclaved at 100 °C for 20 min to release all electrolytes and cooled to RT before measuring the final electrical conductivity (EC2). Afterwards, the percentage of electrolyte leakage was calculated as EL = (EC1 – EC0)/(EC2 – EC0) × 100%. The hydrogen peroxide (H2O2) concentration was measured according to Patterson et al. (1984). Lipid peroxidation was measured by the determination of malondialdehyde (MDA) concentration using the 2-thiobarbituric acid method, as described in Heath and Packer (1968). Leaves were collected from 10 plants per treatment to analyze various physiological indexes. Using 80% acetone to extract chlorophyll (Chl), the Chl concentrations were determined spectrophotometrically as described by Arnon (1949).
Extraction and Measurement of Enzyme Activity
For enzyme extracts and assays, 10 seedlings were used to provide an adequate amount of root and leaf tissues in each experimental replicate (n = 3). Fresh roots (0.1 g) and leaves (0.1 g) were ground separately in liquid nitrogen and then suspended in 1 mL of solution containing 10 mM phosphate buffer (pH 7.0). The homogenate was centrifuged (4 °C, 1500 g, 10 min), and the resulting supernatant was collected. The activities of superoxide dismutase (SOD, A001-4) and catalase (CAT, A007-1) were determined with commercial assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). All data were obtained with a UV-1750 spectrometer (Shimadzu, Kyoto, Japan).
The Concentration of Phosphorus in Plant
To measure the concentrations of phosphorus (P) in plants, the whole-plant samples were washed three times with distilled water. They were fixed at 105 °C for 30 min, dried at 80 °C for 48 h, and ground into powder. Then, 0.1 g of powdered material from each tissue type was put into individual 100 mL digestion tubes, to which 5 mL of concentrated sulfuric acid was added. The tubes were placed in an electric digestion furnace (HYP-1040; Xianjian Inc., Shanghai, China) and heated at 370 °C for approximately 4 h, adding 15 drops of H2O2 per hour. After cooling, deionized water was added up to the tube’s scale mark before 5 mL of clear liquor was taken to determine the total amount of P with a segmented flow analyzer (AA3 HR Nutrient Autoanalyzer; Seal Analytical, Norderstedt, Germany).
Detection of Superoxide ion (O2–) and H2O2
Accumulations of O2– and H2O2 were examined by histochemical staining methods that used nitro blue tetrazolium (NBT) and diaminobenzidine (DAB), respectively. For O2− detection, leaves were incubated under darkness for 4 h at 25 °C in fresh NBT solution (1 mg mL−1) prepared in 10 mM HEPES (pH 7.5) with 0.1% Triton X-100. For H2O2 detection, leaves were placed in a fresh DAB (1 mg mL−1) solution prepared in 10 mM sodium phosphate buffer (pH 7.5) with 0.1% Triton X-100. The samples were then incubated in a growth chamber overnight until brown spots were visible. The chlorophyll was removed by immersing the leaves in 90% ethanol and heating in a boiling water bath for 15 min. Samples were fixed in 90% ethanol with 20% glycerol at 4 °C prior to photographing (Sun et al. 2018).
Statistical Analysis
All experiments were conducted in triplicate (n = 3), and statistical analysis of the data from plants in the control and stress treatments was performed by one-way analysis of variance using SPSS software (version 20.0; IBM Corp., Armonk, NY). To examine the effects of rootstocks and different treatments on experimental variables, all variables were analyzed by two-way ANOVAs with SPSS 20.0. A probability value of P < 0.05 indicated that the difference between treatments was statistically significant. The data are presented as the mean plus standard deviation (SD) for three replicates.
Results
Isolation and Subcellular Localization of MdPHT1;7
MdPHT1;7 (MDP0000261121) is one of 14 phosphorus transporter 1 families in Malus and has the GenBank accession number MK732926. This gene is located on chromosome 7, and the open reading frame is 1560 bp and encodes 519 amino acid residues. Phylogenetic analysis showed that MdPHT1;7, PtrPHT1;1 and PtrPHT1;2 are paralogs (Fig. 1a). SWISS-MODEL for the MdPHT1;7-amino acid sequence was used to model the 3D homology of the protein structure (Fig. 1b), indicating that the spatial structure of the transporters was composed of 12 a-helices and similar to most members of PHT1 in plant (Raghothama 1999; Smith 2001). MdPHT1;7 contains 12 conserved membrane-spanning domains, with a 62 aa-long hydrophilic region. The N-terminus of MdPHT1;7 transmembrane domains inside 12 strong transmembrane helices contains the following residues: 23–46, 73–92, 101–120, 137–154, 163–187, 214–233, 295–312, 348–365, 374–391, 404–427, 440–458 and 485–502. They are highlighted by a red background (Fig. 1c).
Bioinformatic analysis of MdPHT1;7. a Phylogenetic analysis of MdPHT1;7 and other PHT proteins. The corresponding accession numbers are as the follows: MdPHT1;7 (MK732926.1), AtPHT1;1 (Q8VYM2.2), AtPHT1;2 (Q96243.2), AtPHT1;3 (O48639.1), AtPHT1;5 (Q8GYF4.2), PtrPHT1;1 (Potri.002G038900), PtrPHT1;2 (Potri.005G223600), OsPHT1;9 (Q8H6G7.2), OsPHT1;10 (Q69T94.1), OsPHT1;11 (Q94DB8.1), and OsPHT1;13 (Q7XRH8.2). Unrooted phylogenetic tree was constructed using MEGA6.0 program with Neighbor-joining method. MdPHT1;7 is indicated in red circle. b Three-dimensional protein structures for MdPHT1;7 merged images. c The conserved membrane-spanning domains in MdPHT1;7
The molecular characteristics of MdPHT1;7 and its phylogenetic analysis suggested that it is a member of the PHT gene family in apple. Analyses of heterologous expression in tobacco leaves have indicated that the MdPHT1;7-GFP fusion proteins are located in the plasma membrane (Fig. 2).
Localization of MdPHT1;7. The fusion protein of MdPHT1;7–GFP was transiently expressed in tobacco leaves and observed with confocal microscopy. a MdPHT1;7–GFP fluorescence. b The plasma membrane protein localization marker AtCBL1n-mCherry. c Merged images. d Merged with bright field. Scale bar = 50 μm
The Inorganic Phosphate Transport Ability of MdPHT1;7
To analyze the Pi transport characteristics of MdPHT1;7, the open reading frames (ORFs) were cloned into a yeast expression vector (pYES2). The construct was transformed into the yeast Pi transport mutant (BY4743) lacking the high-affinity Pi transporter gene PHO84. Transformed yeasts were cultured in YNB with low Pi conditions (20 μM and 100 μM) and normal Pi conditions (1000 μM). The results showed that the mutant cells of the yeast BY4743 strain grew poorly under low Pi conditions, while cells of MdPHT1;7 transformants grew more or larger than that of the control (Fig. 3), suggesting that MdPHT1;7 proteins can completely complement the yeast Pi transport mutant under low Pi status.
Functional characterizations of MdPHT1;7 in a yeast inorganic phosphate (Pi) transport mutant. Under different Pi conditions, three repeated experiments were performed. Repeat experiments are shown as a1, a2, and a3 under the 1000 µM Pi condition; as b1, b2, and b3 under the 100 µM Pi condition; and as c1, c2, and c3 under the 20 µM Pi condition. WT, the yeast of wild-type; BY4743, the Pi uptake-defective yeast mutant; MdPHT1;7, MdPHT1;7 transformed into yeast BY4743
MdPHT1;7 Enhanced Tolerance to Limited Pi Nutrients in Apple Calli
We obtained two transgenic ‘Orin’ lines that overexpressed MdPHT1;7, as confirmed by PCR with gDNA (Fig. S1A) and by qRT-PCR with cDNA (Fig. S1B). The mRNA transcripts were increased by 19- and 33-fold in L1 and L2, respectively. After 14 days of growth, transgenic and WT calli of similar sizes and weights were transferred to control and low Pi media. On 20 days, the WT calli on the low Pi medium turned white and stopped growing, while those of the two transgenic lines were slightly yellow but showed better growth (Fig. 4). Under normal conditions, the Pi concentrations in WT and transgenic calli were not obviously different. However, the transgenic calli had higher Pi conditions under low Pi stress. The fresh weight of callus apple decreased under low Pi conditions, and the transgenic plants had a higher fresh weight than the WT plants under low Pi stress. These results showed that overexpressing MdPHT1;7 confers tolerance to Pi deficiency at the cellular level.
Effects on low Pi tolerance by overexpression of MdPHT1;7 in calli of ‘Orin’ apple with 1000 μM, 50 μM and 5 μM Pi for 20 days. a Low Pi tolerance in WT and MdPHT1;7-Oe plants. b–d The fresh weight of wild-type (WT) apple calli and transgenic callus. e–g The P content of WT and transgenic callus under different Pi conditions. The data are the means of 3 replicates with the SD. Different letters indicate significant differences between treatments as determined by one-way ANOVA and Tukey’s multiple range tests (P < 0.05). WT, the wild-type of apple calli; L1, L2, the different lines of MdPHT1;7-Oe transgenic callus
Overexpression of MdPHT1;7 Enhances Pi Tolerance in Tomato
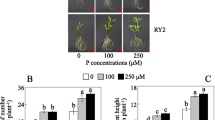
Because MdPHT1;7 expression is induced by low Pi conditions (Sun et al. 2017), we overexpressed this gene in tomato for further analysis of its biological function under low Pi stress. After its coding region was introduced into a plant-overexpressing vector under the control of the CaMV35S promoter, we obtained two transgenic tomato lines (OE-1 and OE-2). OE-1 and OE-2 were two lines that no longer segregated until the F2 generation. These two lines constitutively overexpressed MdPHT1;7 with high mRNA transcript levels (Fig. S2). Under normal conditions, performance did not differ between the WT and the transgenic lines. However, when low Pi treatment was applied, by 21 days, most of the WT leaves were short and left yellow, while those of the transgenic lines remained green and vigorous (Fig. 5a). After 21 days of Pi deprivation, the SOD activity, CAT activity, H2O2 concentration, MDA concentration and electrolyte leakage were increased in response to low Pi treatment, but the damage indicator degree of increment was lower in the transgenic plants, instead of enzyme activity. The electrolyte leakage measurements showed that values were 93.70% (OE-1) and 83.06% (OE-2) of that determined for the WT (Fig. 5f). The chlorophyll contents of OE-1 and OE-2 were 1.66- and 1.98-fold higher than that of WT under low Pi conditions (Fig. 5h).
Effects on low Pi tolerance by overexpression of MdPHT1;7 in ‘Micro-Tom’ tomato for 21 days. a Low Pi tolerance in WT and MdPHT1;7-Oe transgenic tomato. b Total P content of leaves in WT and transgenic plants under normal and low Pi conditions. c H2O2 concentration, d MDA concentrations; e SOD activity, f electrolyte leakage; g CAT activity and h total chlorophyll in WT and transgenic plants under low Pi stress. The data were recorded on 21 days of stress treatment and are presented as the means of 3 replicates with the SD. Different letters indicate significant differences between treatments as determined by one-way ANOVA and Tukey’s multiple range tests (P < 0.05). WT, the wild-type of tomato; OE-1, OE-2, the different lines of MdPHT1;7-Oe transgenic tomato
Overexpression of MdPHT1;7 in Tomato Enhances Drought Tolerance
MdPHT1;7 expression is induced by drought stress (Sun et al. 2017), and we overexpressed this gene in tomato for analysis of its resistance under drought stress. Under well-watered conditions, performance did not differ between the WT and the transgenic lines. However, when drought treatment was applied (i.e., irrigation withheld for 15 days), most of the WT leaves were necrotic and kraurotic, while those of the transgenic lines remained green and vigorous in the OE-1 and OE-2 lines after 15 days (Fig. 6a). The transgenic plants had higher Pi conditions under normal and drought conditions (Fig. 6b). In addition, the accumulation of two kinds of ROS molecules, O2− and H2O2, did not change with the overexpression of MdPHT1;7 in tomato under normal conditions. However, transgenic plants accumulated less ROS than WT plants when exposed to drought stress, as indicated by the weaker blue color upon NBT staining (Fig. 6c) and the weaker brown color upon DAB staining (Fig. 6d) in transgenic plants.
Effects on drought tolerance by overexpression of MdPHT1;7 in ‘Micro-Tom’ tomato. a Drought tolerance in WT and MdPHT1;7-Oe transgenic tomato. b Total P content in WT and transgenic plants under normal and drought conditions. c, d Histochemical staining assay with NBT (c) and DAB (d) to detect O2− and H2O2 accumulation in leaves, respectively, at 15 days. The data were recorded on 15 days of stress treatment and are presented as the means of 3 replicates with the SD. Different letters indicate significant differences between treatments as determined by one-way ANOVA and Tukey’s multiple range tests (P < 0.05). WT, the wild-type of tomato; OE-1, OE-2, the different lines of MdPHT1;7-Oe transgenic tomato
Drought damage was evaluated by the levels of H2O2, MDA, electrolyte leakage, and total chlorophyll, which are typical parameters used for assessing tolerance to abiotic stresses in crop plants (Tang et al. 2013; Wang et al. 2012; Pompelli et al. 2010). After 15 days of water deprivation, for all plant types, H2O2 and MDA concentrations and electrolyte leakage were increased in response to drought treatment, but the degree of increment was lower in the transgenic plants (Fig. 7). The activities of SOD and CAT were higher in the overexpression lines after drought stress. The SOD activity of OE-1 and OE-2 was 1.28- and 1.31-fold higher, respectively, than the level calculated for the WT (Fig. 7b). These results suggested that overexpression of MdPHT1;7 led to enhanced drought stress tolerance in tomato.
Changes in the levels of H2O2 accumulation and activities of ROS-scavenging enzymes in tomato leaves under drought stress. a H2O2 concentration, b SOD activity, and c CAT activity; d MDA concentrations; e electrolyte leakage; f total chlorophyll in WT and transgenic plants under drought stress. The data were recorded on 15 days of stress treatment and are presented as the means of 3 replicates with the SD. Different letters indicate significant differences between treatments as determined by one-way ANOVA and Tukey’s multiple range tests (P < 0.05). WT, the wild-type of tomato; OE-1, OE-2, the different lines of MdPHT1;7-Oe transgenic tomato
Discussion
Plants must acquire Pi from their environment against a step concentration gradient, as the Pi concentration in the soil seldom exceeds 10 μm, whereas intracellular Pi levels reach the millimolar range (Schachtman et al. 1998). The uptake and transport of phosphorus in soil mainly depend on phosphorus transporters. Phosphorus transporters play an important role in the response to low phosphorus stress. They can improve tolerance to low phosphorus stress by enhancing the phosphorus absorption and utilization efficiency of plants. Pi can also increase tissue osmotic pressure and enhance tolerance to drought stress (Graciano et al. 2005).
All PHT members identified in plants can be classified into five groups: PHT1, PHT2, PHT3, PHT4 and PHT5. These PHTs are responsible for phosphate transport across the membranes of cells, chloroplasts, mitochondria and Golgi bodies (Rouached et al. 2010). To date, PHT1 members are the most widely investigated because of their critical roles in Pi acquisition from growth media and in translocation within plant tissues. The expression of most PHT1 members is root-specific, with more transcripts located in root epidermis cells and root hairs (Mudge et al. 2002; Rae et al. 2003). The expression of most PHT1 members is upregulated under Pi deprivation, which is consistent with their role in nutrient acquisition (Muchhal et al. 1996). Thus, inducing the expression of high-affinity PHTs is an important adaptation strategy for plants in low-Pi soil environments (Raghothama 1999). In this study, MdPHT1;7 transcripts were highly detected in the roots and were dramatically induced once exposed to low-Pi stress.
BLAST search analysis was used to identify the homologous counterparts of MdPHT1;7 in plant species. MdPHT1;7 was highly similar to the PHT members reported in Arabidopsis, rice and poplar. MdPHT1;7 is also highly similar to AtPHT1;1, AtPHT1;2, AtPHT1;3 and AtPHT1;5 (Sun et al. 2017). AtPHT1;1 is highly expressed, and transcript levels increase substantially in response to Pi deprivation. Loss of PHT1;1 function impact Pi content and plant growth (Shin et al. 2004). AtPHT1;2 was suppressed by nitrate starvation and induced by nitrate resupply and sulfate starvation in comparison to continuous nutrient treatment (Bao et al. 2011). AtPHT1;3, an Arabidopsis Pi transporter, in shoots and roots under normal and Pi-deficient conditions (Yang et al. 2018). In the T-DNA insertion Arabidopsis mutant, the AtPHT1;5 expression level was increased under Pi-sufficient conditions and low Pi conditions compared with WT (Zhang et al. 2016a, b). MdPHT1;7 has 12 membrane-spanning domains separated by a central hydrophilic loop (62 aa) (Fig. 1c). The structural characteristics are similar to those previously reported; the PHT1 family contains 12 putative transmembrane domains, hydrophilic N- and C-termini and a hydrophilic loop between TM6 and TM7 (Karandashov and Bucher 2005).
Members of the PHT1 gene family in the plant share homologies with the yeast PHO84 Pi transporter and belong to the phosphate-H+ symporter family. The yeast PHO84 mutant lacks a functional copy of a high-affinity Pi transporter gene and thus displays deficient growth under limiting Pi conditions (Bun-Ya et al. 1991). In our present study, the yeast Pi transport mutant BY4743, which lacks the high-affinity Pi transport gene PHO84, was used to verify the complementary Pi transport abilities of MdPHT1;7. Our results indicated that MdPHT1;7 could complement the yeast Pi uptake mutant under low Pi conditions (Fig. 3), implying that MdPHT1;7 has the function of Pi uptake and transportation under low Pi conditions (Qin et al. 2012; Liu et al. 2016).
However, under low-Pi stress, the plants with MdPHT1;7 overexpression had significantly improved apple callus fresh weights and P content (Fig. 4) based on their significantly higher Pi uptake. Thus, MdPHT1;7 plays a critical role in plant adaptation against low-Pi stress as a mediator in Pi acquisition under Pi-deficient conditions. MdPHT1;7 overexpression enhanced P accumulation in transgenic apple calli under low Pi conditions. The same results were found in genetically modified tomato (Fig. 5). Low Pi damage was evaluated by measuring electrolyte leakage, levels of MDA and total chlorophyll, which are typical parameters used for assessing tolerance to abiotic stresses in plants (Tang et al. 2013; Wang et al. 2012). The resistance of transgenic tomato was obviously enhanced under low phosphorus stress, the injury index was smaller and the content of antioxidant enzymes was higher.
Drought stress is now a global phenomenon that significantly threatens crop production (Zhao and Running 2010). Drought conditions generate and accumulate ROS, leading to membrane disruption, enzyme dysfunction and protein oxidation and aggregation. In agricultural production, to alleviate the lack of available phosphorus in soil under drought stress, it is common to apply fertilizer while recovering water to regulate water and fertilizer, which can promote the improvement of plant resistance and yield. The application of phosphate fertilizer under drought conditions can increase the photosynthesis of potato and improve the plant water potential, water use efficiency and tuber yield (Motalebifard et al. 2013). Applying phosphate fertilizer can reduce the stomatal conductance of pea leaves, increase the length of the plant root system and improve the absorption and utilization efficiency of phosphorus to improve the resistance of pea to drought and low phosphorus levels (Jin et al. 2015). In dwarf bamboo, the application of phosphorus fertilizer to dwarf bamboo obviously increased the growth rate and photosynthetic rate under water-sufficient conditions. Under water stress, phosphate fertilization improved the drought stress on photosynthesis and sucrose metabolism of dwarf bamboo, slowed down nitrogen metabolism damage, improved the dwarf bamboo relative growth rate, and reduced the influence of water stress on plants (Liu et al. 2015b).
Various abiotic stresses, such as drought and nutrition stress, lead to the overproduction of ROS in plants. Plants alleviate the effects of abiotic stresses-induced oxidative stress through the upregulation of a series of antioxidant enzymes. Those enzymes include CAT and SOD to eliminate scavengers of ROS (Jithesh et al. 2006; Gill and Tuteja 2010). Overexpression of the MdPHT1;7 gene in tomato increased the resistance to drought conditions, and ROS and other damage indexes were significantly lower than those in the control (Fig. 6). The activity of antioxidant enzyme SOD and CAT was higher than that in the WT after drought stress (Fig. 7). Overexpression of the MdPHT1;7 gene not only increases the phosphorus content of plants but also increases the drought tolerance of transgenic plants. Therefore, it is of great significance to study the phosphorus absorption of plants under drought stress to improve the stress resistance, phosphorus absorption and utilization efficiency of plants and the sustainable development of agriculture.
As an essential macronutrient, Pi is important in plant growth and development. Given the low availability of Pi in arable fields, a large amount of Pi fertilizer is frequently applied to achieve high crop yields. Consequently, nonrenewable Pi resources deplete rapidly and contribute to environmental pollution (Vance et al. 2010). Improved Pi efficiency in apple and other plants through molecular breeding is crucial for sustainable agriculture and world food security (Parry and Hawkesford 2010).
Conclusion
In the current study, MdPHT1;7, a high-affinity PHT gene, was shown to be useful as an important gene resource and as a potential molecular marker for enhancing plant Pi-use efficiency based on its high affinity for external Pi under Pi-limited conditions. Under Pi-deficient and drought conditions, MdPHT1;7 overexpression significantly improved growth traits and led to the accumulation of more phosphorus, which indicates that MdPHT1;7 plays an important role in plant Pi acquisition under Pi deprivation and drought stress. The results of this study broaden our understanding of the functions of PHTs under Pi-deficient conditions in apple as well as in other plants.
Abbreviations
- Pi:
-
Inorganic phosphate
- PHT:
-
Phosphate transporter
- WT:
-
Wild-type
- OE:
-
Overexpression
- YNB:
-
Yeast nitrogen base
- Cef:
-
Cefotaxime
- qRT-PCR:
-
Quantitative real-time PCR
- EL:
-
Electrolyte leakage
- H2O2 :
-
Hydrogen peroxide
- MDA:
-
Malondialdehyde
- Chl:
-
Chlorophyll
- SOD:
-
Superoxide dismutase
- CAT:
-
Catalase
- ACP:
-
Acid phosphatase
- NBT:
-
Nitro blue tetrazolium
- DAB:
-
Diaminobenzidine
- SD:
-
Standard deviation
- ORFs:
-
Open reading frames
References
Ackerson RC (1985) Osmoregulation in cotton in response to water stress: III. Effects of phosphorus fertility. Plant Physiol 77(2):309–312
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in beta vulgaris. Plant Physiol 24(1):1–15
Bao SJ, An LJ, Su S, Zhou ZJ, Gan YB (2011) Expression patterns of nitrate, phosphate, and sulfate transporters in Arabidopsis roots exposed to different nutritional regimes. Botany 89(9):647–653
Batistic O, Waadt R, Steinhorst L, Held K, Kudla J (2010) CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J 61(2):211–222
Begum N, Ahanger MA, Zhang LX (2020) AMF inoculation and phosphorus supplementation alleviates drought induced growth and photosynthetic decline in Nicotiana tabacum by up regulating antioxidant metabolism and osmolyte accumulation. Environ Exp Bot 176:104088
Bucher M, Rausch C, Daram P (2015) Molecular and biochemical mechanisms of phosphorus uptake into plants. J Plant Nutr Soil Sci 164:209–217
Bun-Ya M, Nishimura M, Harashima S, Oshima Y (1991) The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol 11(6):3229–3238
Cao MX, Liu HZ, Zhang C, Wang DD, Liu XF, Chen Q (2020) Functional analysis of StPHT1;7, a Solanum tuberosum L. phosphate transporter gene, in growth and drought tolerance. Plants-Basel 9(10):1384
Chang S, Puryear J, Cairney J (1993) A simplified method for isolating RNA from pine trees. Plant Mol Biol Rep 11(2):113–116
Dionisio-Sese ML, Tobita S (1998) Antioxidant response of rice seedlings to salinity stress. Plant Sci 135(1):1–9
Faustino LI, Bulfe NM, Pinazo MA, Monteoliva SE, Graciano C (2013) Dry weight partitioning and hydraulic traits in young Pinus taeda trees fertilized with nitrogen and phosphorus in a subtropical area. Tree Physiol 33(3):241–251
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48(12):909–930
Glassop D, Smith SE, Smith FW (2005) Cereal phosphate transporters associated with the mycorrhizal pathway of phosphate uptake into roots. Planta 222(4):688–698
Graciano C, Guiamét JJ, Goya JF (2005) Impact of nitrogen and phosphorus fertilization on drought responses in Eucalyptus grandis seedlings. For Ecol Manag 212(1–3):40–49
Guo B, Jin Y, Wussler C, Blancaflor EB, Motes CM, Versaw WK (2008) Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol 177(4):889–898
Guo M, Zhang YL, Meng ZJ, Jiang J (2012) Optimization of factors affecting Agrobacterium-mediated transformation of Micro-Tom tomatoes. Genet Mol Res 11(1):661–671
Hassan N, Sahrin S (2012) A mathematical model of nutrient management for pineapple cultivation in Malaysia. Adv Environ Biol 6(5):1868–1872
Hayano-Kanashiro C, Calderon-Vazquez C, Ibarra-Laclette E, Herrera-Estrella L, Simpson J (2009) Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation. PLoS ONE 4(10):e7531
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts: II. Role of electron transfer. Arch Biochem Biophys 125(3):850–857
Hummer KE, Janick J (2009) Rosaceae: taxonomy, economic importance, genomics. Springer, New York
Jin J, Lauricella D, Armstrong R, Sale P, Tang CX (2015) Phosphorus application and elevated CO2 enhance drought tolerance in field pea grown in a phosphorus-deficient vertisol. Ann Bot 116(6):975–985
Jithesh MN, Prashanth SR, Sivaprakash KR, Parida AK (2006) Antioxidative response mechanisms in halophytes: their role in stress defence. J Genet 85(3):237–254
Kakar KM, Muhammad T, Taj FH, Nawab K (2001) Phosphorus use efficiency of soybean as affected by phosphorus application and inoculation. J Agron 92(1):49–50
Karandashov V, Bucher M (2005) Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci 10(1):22–29
Knappe S, Flugge UI, Fischer K (2003) Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol 131(3):1178–1190
Li D, Shi W, Deng X (2002) Agrobacterium-mediated transformation of embryogenic calluses of Ponkan mandarin and the regeneration of plants containing the chimeric ribonuclease gene. Plant Cell Rep 21(2):153–156
Li Y, Wang X, Zhang H, Wang SL, Ye XS, Shi L, Xu FS, Ding GD (2019) Molecular identification of the phosphate transporter family 1 (PHT1) genes and their expression profiles in response to phosphorus deprivation and other abiotic stresses in Brassica napus. PLoS ONE 14(7):e0220374
Liu F, Chang XJ, Ye Y, Xie WB, Wu P, Lian XM (2011) Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol Plant 4(6):1105–1122
Liu C, Rubæk GH, Liu F, Andersen MN (2015a) Effect of partial root zone drying and deficit irrigation on nitrogen and phosphorus uptake in potato. Agric Water Manag 159:66–76
Liu CG, Wang YJ, Pan KW, Jin YQ, Li W, Zhang L (2015b) Effects of phosphorus application on photosynthetic carbon and nitrogen metabolism, water use efficiency and growth of dwarf bamboo (Fargesia rufa) subjected to water deficit. Plant Physiol Biochem 96:20–28
Liu F, Xu YJ, Jiang HH, Jiang CS, Du YB, Gong C, Wang W, Zhu SW, Han GM, Cheng BJ (2016) Systematic identification, evolution and expression analysis of the zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int J Mol Sci 17(6):930
Lopez-Arredondo DL, Leyva-Gonzalez MA, Gonzalez-Morales SI, Lopez-Bucio J, Herrera-Estrella L (2014) Phosphate nutrition: improving low-phosphate tolerance in crops. Annu Rev Plant Biol 65:95–123
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290(5494):1151–1155
Mimura T, Mitsuhashi N, Sekiguchi Y, Ohnishi M (2003) Phosphate transport and intracellular distribution in plants. Tanpakushitsu Kakusan Koso 48(15 Suppl):2044–2051
Misson J, Thibaud MC, Bechtold N, Raghothama K, Nussaume L (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55(5):727–741
Modgil M, Mahajan K, Chakrabarti SK, Sharma DR, Sobti RC (2005) Molecular analysis of genetic stability in micropropagated apple rootstock MM106. Sci Hortic 104(2):151–160
Motalebifard R, Najafi N, Oustan S, Nyshabouri MR, Valizadeh M (2013) The combined effects of phosphorus and zinc on evapotranspiration, leaf water potential, water use efficiency and tuber attributes of potato under water deficit conditions. Sci Hortic 162:31–38
Muchhal US, Pardo JM, Raghothama KG (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93(19):10519–10523
Mudge SR, Rae AL, Diatloff E, Smith FW (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31(3):341–353
Nagy R, Vasconcelos MJ, Zhao S, McElver J, Bruce W, Amrhein N, Raghothama KG, Bucher M (2006) Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol 8(2):186–197
Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104(1):34–41
Pao SS, Paulsen IT, Saier MH Jr (1998) Major facilitator superfamily. Microbiol Mol Biol Rev 62(1):1–34
Parry MA, Hawkesford MJ (2010) Food security: increasing yield and improving resource use efficiency. Proc Nutr Soc 69(4):592–600
Paszkowski U, Kroken S, Roux C, Briggs SP (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99(20):13324–13329
Patterson BD, MacRae EA, Ferguson IB (1984) Estimation of hydrogen peroxide in plant extracts using titanium(IV). Anal Biochem 139(2):487–492
Pompelli MF, Barata-Luis R, Vitorino HS, Goncalves ER, Rolim EV, Santos MG, Almeida-Cortez JS, Ferreira VM, Lemos EE, Endres L (2010) Photosynthesis, photoprotection and antioxidant activity of purging nut under drought deficit and recovery. Biomass Bioenergy 34(8):1207–1215
Qin L, Guo Y, Chen L, Liang R, Gu M, Xu G, Zhao J, Walk T, Liao H (2012) Functional characterization of 14 Pht1 family genes in yeast and their expressions in response to nutrient starvation in soybean. PLoS ONE 7(10):e47726
Rae AL, Cybinski DH, Jarmey JM, Smith FW (2003) Characterization of two phosphate transporters from barley; evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Mol Biol 53(1–2):27–36
Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50:665–693
Rausch C, Bucher M (2002) Molecular mechanisms of phosphate transport in plants. Planta 216(1):23–37
Rouached H, Arpat AB, Poirier Y (2010) Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant 3(2):288–299
Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15(16):2122–2133
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Schachtman DP, Reid RJ, Ayling SM (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116(2):447–453
Schnable PS (2012) The B73 maize genome: complexity, diversity, and dynamics (November, pg 1112, 2009). Science 337(6098):1040–1040
Sekhon RS, Briskine R, Hirsch CN, Myers CL, Springer NM, Buell CR, de Leon N, Kaeppler SM (2013) Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS ONE 8(4):e61005
Shin H, Shin HS, Dewbre GR, Harrison MJ (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39(4):629–642
Smith FW (2001) Sulphur and phosphorus transport systems in plants. Plant Soil 232:109–118
Sun T, Li M, Shao Y, Yu L, Ma F (2017) Comprehensive genomic identification and expression analysis of the phosphate transporter (PHT) gene family in apple. Front Plant Sci 8:426
Sun X, Wang P, Jia X, Huo L, Che R, Ma F (2018) Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol J 16(2):545–557
Tang L, Cai H, Ji W, Luo X, Wang Z, Wu J, Wang X, Cui L, Wang Y, Zhu Y, Bai X (2013) Overexpression of GsZFP1 enhances salt and drought tolerance in transgenic alfalfa (Medicago sativa L.). Plant Physiol Biochem 71:22–30
Tariq A, Pan K, Olatunji OA, Graciano C, Zhang A (2018) Phosphorous fertilization alleviates drought effects on Alnus cremastogyne by regulating its antioxidant and osmotic potential. Sci Rep 8(1):5644
Testerink C, Munnik T (2011) Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J Exp Bot 62(7):2349–2361
Tian H, Drijber RA, Li X, Miller DN, Wienhold BJ (2013) Arbuscular mycorrhizal fungi differ in their ability to regulate the expression of phosphate transporters in maize (Zea mays L.). Mycorrhiza 23(6):507–514
Tng DY, Janos DP, Jordan GJ, Weber E, Bowman DM (2014) Phosphorus limits Eucalyptus grandis seedling growth in an unburnt rain forest soil. Front Plant Sci 5:527
Vance CP, Uhde-Stone C, Allan DL (2010) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157(3):423–447
Wang S, Liang D, Li C, Hao Y, Ma F, Shu H (2012) Influence of drought stress on the cellular ultrastructure and antioxidant system in leaves of drought-tolerant and drought-sensitive apple rootstocks. Plant Physiol Biochem 51:81–89
Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M’Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285(5429):901–906
Xie XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ 35(11):1884–1897
Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22(6):543–551
Yang WT, Baek D, Yun DJ, Lee KS, Hong SY, Bae KD, Chung YS, Kwon YS, Kim DH, Jung KH, Kim DH (2018) Rice OsMYB5P improves plant phosphate acquisition by regulation of phosphate transporter. PLoS ONE 13(3):e0194628
Ye Y, Yuan J, Chang X, Yang M, Zhang L, Lu K, Lian X (2015) The phosphate transporter gene OsPht1;4 is involved in phosphate homeostasis in rice. PLoS ONE 10(5):e0126186
Zhang C, Meng S, Li M, Zhao Z (2016a) Genomic identification and expression analysis of the phosphate transporter gene family in poplar. Front Plant Sci 7:1398
Zhang J, Zhou X, Xu Y, Yao M, Xie F, Gai J, Li Y, Yang S (2016b) Soybean SPX1 is an important component of the response to phosphate deficiency for phosphorus homeostasis. Plant Sci 248:82–91
Zhao M, Running SW (2010) Drought-induced reduction in global terrestrial net primary production from 2000 through 2009. In: Agu Fall Meeting, 2010.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31901965), and Natural Science Foundation of Inner Mongolia Autonomous Region (2019BS03037). The authors are grateful to Dr. Guomin Han at Anhui Agricultural University for providing the yeast mutant BY4743 which defects PHO84.
Author information
Authors and Affiliations
Contributions
FM and JZ designed the experiments. TS, BZ, TP and HM performed the experiments. TS, QW and FM wrote the manuscript with the help from other authors. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.

12374_2021_9309_MOESM1_ESM.jpg
Supplementary file1 (JPG 28 KB) Fig. S1 PCR and qRT-PCR analysis of the transgenic calli of ‘Orin’ apple. (A) PCR results for Oe experiments. Lanes: L1 and L2, MdNAT7-Oe transgenic lines; M, molecular marker DL2000. (B) qRT-PCR analysis of MdPHT1;7 overexpression transcripts in L1 and L2. WT, the wild-type of apple calli; L1, L2, the different lines of MdPHT1;7-Oe transgenic callus.

12374_2021_9309_MOESM2_ESM.jpg
Supplementary file2 (JPG 24 KB) Fig. S2 PCR and qRT-PCR analysis of the MdPHT1;7 overexpression transgenic in ‘Micro-Tom’ tomato. (A) PCR results for Oe experiments. Lanes: M, molecular marker DL2000; V, positive vector containing pGWB 408-MdNAT7 plasmid; OE-1 and OE-2 transgenic lines. (B) qRT-PCR analysis of MdPHT1;7 overexpression transcripts in OE-1 and OE-2. WT, the wild-type of tomato; OE-1, OE-2, the different lines of MdPHT1;7-Oe transgenic tomato.
Rights and permissions
About this article
Cite this article
Sun, T., Zhou, B., Pei, T. et al. Phosphate Transporter MdPHT1;7 Enhances Phosphorus Accumulation and Improves Low Phosphorus and Drought Tolerance. J. Plant Biol. 64, 403–416 (2021). https://doi.org/10.1007/s12374-021-09309-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-021-09309-2