Abstract
D53 is a crucial gene involved in tillering regulation in crops via the strigolactone signaling pathway. The objective of our research was to clone the homologous sugarcane (Saccharum L. spp. hybrids) gene of D53 (ScD53) by in silico cloning combined with RT-PCR and rapid amplification of cDNA ends techniques, and the physicochemical properties and structure of ScD53 protein were analyzed using bioinformatic software and tools. The relationships between the expression of ScD53 and some plant hormones, bud germination and tillering occurrence of seedlings were also detected using the real-time fluorescence quantitative PCR (FQ-PCR) technique. The homolog was named ScD53 and possessed a 3918-bp cDNA sequence. Bioinformatic analysis showed that ScD53 has a complete open reading frame of 3363 bp that encodes 1120 amino acid residues. The ScD53 protein possesses the conserved domain of the P-loop NTPase and ClpB_D2-small superfamilies, which may be unstable hydrophilic proteins and are likely to be located in the nucleus. The main secondary and tertiary structures of the ScD53 protein are composed of random coil and alpha helix structures and exhibit high similarity in structure and homology with D53 proteins from sorghum [Sorghum bicolor (L.) Moench] and maize (Zea mays L.). The FQ-PCR results showed that the expression of ScD53 is tissue specific and is differently expressed in the different tissues. The expression of ScD53 is associated with bud germination, the high expression of which inhibits bud germination. Plant hormone treatment experiments indicated that the expressions of ScD53 were induced in bud and leaf tissues by strigolactones, 3-indole acetic acid and kinetin (KT, a cytokinin analog). The high expression of ScD53 following strigolactone and auxin treatment obviously delayed occurrence of tillers and reduced the number of tillers for the seedlings. However, this was not observed during KT treatment, which implied that the negative effects of this gene on seedlings tillering phenotype were eliminated by the positive influences of other unknown pathways. Our results above help elucidate the function of ScD53 and provide important information and direction for the use of this gene in improving sugarcane yield through tiller regulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tillering is one of the most important agronomic traits for gramineous plants, such as (Oryza sativa L.), wheat (Triticum aestivum L.) and sugarcane (Saccharum L. spp. hybrids). It determines the final yield and plant plasticity by influencing the tillering and panicle number of crops (Li et al. 2012). Previous studies have shown that the tillering capability of gramineous plants is jointly regulated by multiple plant hormones, such as auxins, cytokinins and strigolactones (Leyser 2003; Tanaka et al. 2006; Kyozuka 2007; Mikihisa et al. 2008; Gallavotti et al. 2008; Brewer et al. 2013). The downward movement of auxins and the upward movement of cytokinins and strigolactones form a dynamic equilibrium that regulates crop tillering (Brewer et al. 2009; Dun et al. 2009). Interestingly, auxin is not directly involved in tillering regulation, but together with cytokinins and strigolactones executes regulatory functions (Shani et al. 2006; Kyozuka 2007).
The strigolactones are a class of novel, recently discovered plant hormones, first reported by Cook et al. (1966, 1972) in their research on the germination stimuli of seeds of parasitic plants. They were subsequently isolated from numerous plants (Fen and Chen 2011), where they are required for some plant development processes, such as branching, hypocotyl growth, lateral root formation and root hair elongation (Gao et al. 2013). Recently, some key genes from its signal transduction pathways have been successfully cloned, such as D3/MAX2/RMS4 (Stirnberg et al. 2002; Ishikawa et al. 2005), D14/D88/HTD2/AtD14/DAD2 (Arite et al. 2009; Gao et al. 2009) and D53/SMX-L6/7/8 (Jiang et al. 2013; Zhou et al. 2013). Jiang et al. (2013) and Zhou et al. (2013) showed that D53 plays a key role in the perception and response of strigolactones signaling under the influence of strigolactones. D53 proteins can interact with D14 and D3 proteins to form D53–D14–SCFD3 protein complexes. These enable the ubiquitin D53 protein to be specifically degraded by the proteasome system, thereby inducing the expression of downstream target genes and ultimately regulating plant tillering. This implies that the tillering capability of plants can be regulated by regulating the expression of D53, which can influence the signal transduction of strigolactones.
Sugarcane (Saccharum L. spp. hybrids) is an important global sugar crop, producing more than 70% of the world’s sugar, and it is also an efficient source for renewable energy production (Li and Wei 2006; Lee and Bressan 2006). Sugarcane reproduces asexually. Stalks are the primary harvested product, so the number and weight of effective (harvestable) stalks impact the ultimate tonnage yield ha−1 (Li et al. 2015a, b). Stalk number per stool is influenced by the tillering traits of the variety, and thus, tillering is regarded as one of sugarcane’s most fundamentally important agronomic traits (Vasantha et al. 2012). Improvement in sugarcane is needed through a better understanding of tiller regulation and enhancement. Key genes and metabolic pathways regulating tillering need further analysis and screening to provide the theoretical guidance for improving yields via tillering.
In view of the importance of the D53 gene in tillering regulation, our objective was to clone the homologous sugarcane gene of D53 (ScD53) by in silico cloning combined with RT-PCR and rapid amplification of cDNA ends (RACE) techniques, and the physicochemical properties and structure of ScD53 protein were analyzed using bioinformatic software and tools. The relationships between the expression of ScD53 and some plant hormones, bud germination and tillering occurrence of seedlings were also detected using the real-time fluorescence quantitative PCR (FQ-PCR) technique. Our findings should contribute toward characterizing the structure, expression and function of this gene, and also provide theoretical information for the further functional analysis and utilization of this gene in the regulation of tillering capability of sugarcane.
Materials and Methods
Materials and Treating Methods
ROC22, the primary sugarcane variety in China, was selected as the research material for this study. Apical meristem tissues were sampled for gene cloning. For all FQ-PCR experiments, three biological replicates were designed, and sampled tissues were immediately frozen in liquid N2 after sampling and stored at − 80 °C prior to analysis.
Root, stem, leaf, bud and apical meristem tissues were sampled during the tillering period for tissue-specific expression detection of ScD53. Single-bud stem cuttings were cultured in a tray containing Hoagland’s nutrient solution (Chen et al. 2013). The germinating bud tissues were sampled for expression of ScD53 at different developmental stages while dormant (day 0), sprouting (day 3), elongating (day 6) and elongating buds with a greenish-yellow sharp leaf (day 9). Other sprouting buds were continuously sprayed for 3 days with 60 mg/L kinetin (KT, a plant growth regulator with the same physiological function as cytokinin), 60 mg/L 3-indole acetic acid (IAA), 10 µmol/L of strigolactones (SLs) and water, and then, these treated bud tissues were also sampled for detecting the expression of ScD53.
Single-bud stem cuttings were planted in buckets containing sand in a greenhouse, four buds per bucket. These buds were immersed in Hoagland’s nutrient solution every 3 days to ensure adequate nutrition. Once these buds had developed into seedlings with 4–5 leaves, different concentrations of plant hormones (IAA: 100 mg/L, 200 mg/L, 400 mg/L; KT: 100 mg/L, 200 mg/L, 400 mg/L, SLs: 1 µmol/L, 10 µmol/L, 30 µmol/L) were continuously sprayed on the leaves and stem base for screening out the appropriate concentration of plant hormone treatment. Control seedlings were treated with water. The results showed that the KT 400 mg/L solution encouraged earlier tillering, with tillers after 8 days treatment, and the seedlings subsequently produced more tillers than the control. In contrast, 400 mg/L IAA solution and 10 µmol/L SLs solution effectively delayed the occurrence of tillers, which occurred after 16 days and 28 days, respectively, and the subsequent seedlings developed fewer tillers than the control. Finally, based on the above results, the seedlings were sprayed with 400 mg/L KT solution, 400 mg/L IAA solution and 10 µmol/L SLs solution for 35 days, and then, these tissues from tillering bud and leaf were sampled for detecting the expression of ScD53 after treating seedlings with different plant hormones.
Cloning of ScD53
The total RNA of these sampled tissues was extracted using the TransZol plant RNA kit (Transgen Biotech, China), and the RNA was reverse-transcribed into cDNA as the template for the RT-PCR using the TransScript one-step gDNA removal and cDNA Synthesis SuperMix reverse transcription kit (Transgen Biotech, China). Using electronic cloning technology, the D53 gene-encoding region sequences from rice and sorghum were used as probes (KF709434.1 and XM_002441614.1), and the expressed sequence tag (EST) sequences with high nucleotide identity were retrieved from the sugarcane EST library on the GenBank database. Vector NTI 11.5 software (Invitrogen, USA) and DNAMAN 5.0 (Lynnon Biosoft, USA) software were used for comparison and splicing to obtain overlapping groups. The overlapping groups were then used to continuously compare and retrieve sugarcane EST sequences until no new sequences were available for splicing and extension, thereby obtaining the final electronic clone sequences. Primer Premier 5.0 software (Premier Biosoft International, Canada) was used to design the RT-PCR primers and 5′ RACE gene-specific primers according to electron cloning sequences (Table 1). The TransTaq DNA polymerase high fidelity (HiFi) (Transgen Biotech, China) and ClonTech SMARTER™ RACE cDNA amplification kit (ClonTech, USA) were used in the RT-PCR and RACE experiments for the gene cDNA sequences, following which the coding region primer was designed to amplify the coding region sequence for verification. All of the above PCR products were recovered using the EasyPure quick gel extraction kit (Transgen Biotech, China), connected to the pEASY-T5 Zero Cloning Kit vector (Transgen Biotech, China), and then transferred into Trans1-T1 phage-resistant chemically competent cells (Transgen Biotech, China). At least three clones were selected for bidirectional sequencing by the Beijing Genomics Institute (BGI, Beijing, China).
Bioinformatic Analysis of ScD53 Protein
The online ORF Finder tool (https://www.ncbi.nlm.nih.gov/orffinder/) was used to analyze the open reading frame of the cDNA sequence of ScD53 and the presumed amino acid sequence. The protein characteristics of ScD53, including the basic physicochemical properties, hydrophilicity or hydrophobicity, conserved domain, subcellular orientation, potential signal peptide and function, were predicted using ProtScale (https://web.expasy.org/protscale/), InterPro (http://www.ebi.ac.uk/interpro/search/sequence-search), TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/), PSORT (http://psort.hgc.jp/form.html), SignalP4.1 (http://www.cbs.dtu.dk/services/SignalP/), ProtParam (https://web.expasy.org/protparam/) and Protfun (http://www.cbs.dtu.dk/services/ProtFun/).
The secondary and tertiary structures of ScD53 were analyzed using the SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) and Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) online tools. The homologs of ScD53 were retrieved by Blastp in NCBI, and their amino acid sequences were compared with ScD53 using DNAMAN5.0. The NJ (neighbor-joining) tree for all of the homologs of ScD53 was constructed with 1000 bootstrap replications using MEGA7.0 software (Kumar et al. 2016).
Expression Analysis of ScD53
The FQ-PCR primer was designed according to the coding region sequence of the ScD53 gene, using Primer Express3.0 (PE Applied Biosystems, USA). The primer sequences were 5′-AAAGCGTGCCACCGACTGTA-3′ and 5′-GGCATGAATCAGGCTCTCCT-3′. The GAPDH gene was used as the reference gene (primer sequences: 5′-CACGGCCACTGGAAGCA-3′ and 5′-TCCTCAGGGTTCCTGATGCC-3′) (Ling et al. 2014). The FQ-PCR system was configured in accordance with the instructions of SYBR premix ExTaq™ II (TaKaRa, Japan) with the following amplification procedure: pre-denaturation at 95 °C for 30 s, then 40 cycles of denaturation at 95 °C for 5 s, followed by annealing and extension at 60 °C for 34 s. All of the PCRs were run on an ABI Viia7 Real-time PCR System, and each experiment was performed with three technical replicates. The 2−ΔΔCT method (Livak and Schmittgen 2001) was used to calculate the relative expression of genes. Analysis of variance (ANOVA) was used to assess the relative expression data, and the diagrams were plotted in Excel 2007 (Microsoft, USA).
Results
Cloning the cDNA Sequence of ScD53
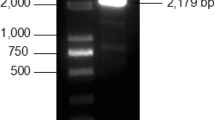
A total of 21 sugarcane EST sequences with a nucleotide identity of more than 90% were obtained using the Blast tool in the GenBank database (Table 2). A spliced sequence fragment of 3204 bp was obtained by electronic cloning splicing technology, it contained a partial 3′UTR region with a length of 422 bp and an incomplete encoding region at the 5′-end by comparing these reference sequences (KF709434.1 and XM_002441614.1) from the D53 genes of rice and sorghum, and the results showed the spliced sequence. Using the RT-PCR primers, a fragment of 2521 bp was obtained (Fig. 1a), which had 99.97% nucleotide identity with the previous spliced sequence, thereby confirming the validity of the electronic cloning sequence. The 5′ RACE experiment yielded a 1132-bp PCR product (Fig. 1b). The entire cDNA sequence (3918 bp) was obtained by splicing the above sequences using Vector NTI 11.5 and DNAMAN 5.0. Amplification of the coding region resulted in a correct fragment of 3395 bp (Fig. 1c). This cDNA sequence, named ScD53, contains a 133-bp 5′ UTR sequence, a 3363-bp coding region sequence and a 422-bp 3′ UTR sequence, and was uploaded to GenBank with the accession number: KX893542.
Bioinformatic Analysis of ScD53 Protein
Physicochemical Properties of ScD53 Protein
ORF Finder deduced that ScD53 protein encoded a total of 1120 amino acids (as shown in Fig. 2). Analysis of the physical and chemical properties of protein using the ProtParam and ProtScale tools showed that the molecular formula of ScD53 was C5284H8453N1561O1649S45, with a molecular weight of 121.68 kDa and theoretical pI of 6.94. Its instability index > 40 (54.83) indicated that ScD53 was an unstable protein with an aliphatic index of 81.60 and a total grand average of hydropathicity index (GRAVY) was − 0.324. The 141st amino acid had the highest hydrophobicity score of 2.411, thereby possessing the strongest hydrophobicity, while the 110th and 744th amino acids had the lowest hydrophobicity score of − 2.989, indicating that these two amino acids were mostly hydrophilic. A total of 61.69% of the amino acids were hydrophilic, implying that the ScD53 protein may be a hydrophilic protein.
Signal Peptide Prediction and Subcellular Localization of ScD53 Protein
The predicted results using SignaIP4.1 showed that there was a high score in the 34th amino acid position for C-score, but no high score for S-score and Y-score appeared in the same position simultaneously (Fig. 3), and the D-cutoff value of 0.222 was less than 0.450. These results implied that no signal peptide was found in ScD53 protein. In addition, because the mean S-score of 0.237 was less than 0.5, ScD53 protein was presumed to be a non-secretory protein. Localization analysis in cells using PSORT software indicated that ScD53 was probably located in the nucleus with a high probability value of 97.0%.
Structure and Function Prediction of ScD53
The conserved domain analysis using the conserved domain search tool in NCBI indicated that the structure of ScD53 included the domain of the p-loop NTPase superfamily and ClpB_D2-small superfamily, and possessed an AAA-specific binding site and three non-specific binding sites (ClpA, PRK10865, AAA_2). The secondary structure of ScD53 protein, which was predicted by using SOPMA online software, consisted mainly of random coil, alpha helix, extended strand and beta turn, respectively, accounting for 51.34%, 33.21%, 11.61% and 3.84% of the secondary structure. The tertiary structure of ScD53 was constructed according to the folds recognition model using Phyre2 software, which indicated that the spatial structure of ScD53 was dominated by random coil and alpha helix (see Fig. 4). Comparison of the spatial structure to other plant D53/D53-like proteins indicated that these D53 proteins from sorghum, maize, and rice are similar to ScD53. The functional prediction using ProtFun2.2 revealed that ScD53 might constitute an enzyme protein that participates in coenzyme biosynthesis.
ScD53 Homology Analysis
The evolutionary relationships between ScD53 and other plant D53 proteins were compared by a neighbor-joining phylogenetic tree of D53 proteins from 13 monocotyledons and five dicotyledons using MEGA7.0 (Fig. 5). The tree showed that all D53 proteins were clustered into two large groups, one of which was composed of dicotyledonous D53 proteins, while the other was composed of monocotyledonous D53 proteins, indicating that the D53 proteins from monocotyledons had high homology. This large group containing all of D53 proteins from monocotyledons was divided into two subgroups. One subgroup included the D53 proteins from Orchidaceae, Palmae and Musaceae, and the other subgroup was composed of D53 proteins from Gramineae. Thus, D53 proteins from Gramineous are closely related, and ScD53 protein was most closely related to sorghum D53 protein, followed by maize and millet D53 proteins. And homology analysis using the Blastp program in NCBI showed that ScD53 has 93%, 82%, 80% and 69% identity with these D53 proteins of sorghum, maize, foxtail [Setaria italic (L.) P. Beauv.], and rice, respectively. According to the comparison map of the amino acid sequences of D53 proteins from sugarcane, sorghum, maize, foxtail and rice (Fig. 6), the region from the 1st to 460th amino acids was relatively conserved, whereas the middle and posterior regions were less conserved.
Expression Analysis of ScD53 Gene
The ScD53 gene of ROC22 was expressed at different levels in the root, stem, leaf, bud and apical meristem (Fig. 7). The highest expression was detected in the stem, which was significantly higher than that in the other tissues (P < 0.05), followed by the apical meristem and bud, and low expression was detected in the leaves and roots, indicating that the expression of this gene exhibits certain tissue specificity.
The ScD53 gene was differentially expressed in sugarcane bud tissues according to developmental stage (Fig. 8a). The highest expression was detected in the dormant buds, which was significantly higher than sprouting buds, elongation buds and sharp leaf buds. These results indicated that the high expression of ScD53, as in dormant buds, can inhibit the germination of sugarcane buds.
Relative expression of ScD53 gene at different developmental stages of buds, and as affected by plant hormone treatment of sprouting buds and seedlings. a the relative expression at different developmental stages of buds; b the relative expression in buds after treating sprouting buds with different plant hormones; c the relative expression in tillering buds after treating seedlings with different plant hormones; d the relative expression in leaves after treating seedlings with different plant hormones; the error bars represent the standard error of each treating group, *significant difference from control at P < 0.05; **significant difference at P < 0.01
After three days of treatment with 60 mg/L KT, 60 mg/L IAA, and 10 µmol/L SLs, both IAA and SLs inhibited the continued development of the sprouting buds, whereas KT had no significant influence on the growth of sprouting buds in comparison with the control. Following hormone treatment, the expression of ScD53 appeared to increase significantly in comparison with the control (Fig. 8b). After continuously spraying the seedlings with 400 mg/L KT, 400 mg/L IAA and 10 µmol/L SLs solution for 35 days, all three plant hormones significantly induced the expression of ScD53 in the tillering buds and leaves compared to the control (Fig. 8c, d). The highest expression was observed in the KT treatment, followed by the IAA treatment. The above results indicate that these three exogenous hormones positively regulate the expression of ScD53.
Discussion
Tillering is an important component of plant phenotypic plasticity and is also an important protection mechanism for plants to adapt to the surrounding environment and avoid injury (Gao et al. 2013). In agricultural crops, tillering is largely responsible for crop biomass or yield and thus is considered to be one of the most important agronomic traits. Tillering is mainly derived from the growth and development of the axillary buds at the leaf axils of plants (Rameau et al. 2015), and thus, the development of the axillary bud plays a key role in the forming of tillering. Axillary bud development is strongly influenced by both external environmental factors and internal factors (Wang et al. 2014). In the absence of genetic factors, endogenous hormones are considered to be closely involved in the axillary bud development of plants (Mcsteen 2009). Before 2008, IAA and CTK were thought to be the main regulatory hormones of axillary bud development in plants. After that, a new class of phytohormone, named strigolactones, was discovered, and also shown to participate in the regulation of axillary bud development (Gomez-Roldan et al. 2008; Umehara et al. 2008), the regulation of axillary bud development is conserved in monocots and dicots (Stanga et al. 2013). Some mutations occurring in these genes involved in the synthetic pathway of SLs significantly affected tiller formation, including MAX3/CCD7/RMS5/D17/DAD3 (Booker et al. 2004; Drummond et al. 2009), MAX4/CCD8/RMS1/D10/DAD1 (Foo et al. 2005; Sorefan et al. 2003; Snowden et al. 2005; Arite et al. 2007) and D27 (Lin et al. 2009). In addition, some genes involved in the signal transduction pathways of SLs also played a very important role in the regulation of plant tillering or branch control, namely D3/MAX2/RMS4 (Stirnberg et al. 2002), D14/D88/HTD2/AtD14/DAD2 (Arite et al. 2009; Gao et al. 2009) and D53/SMX-L6/7/8 (Jiang et al. 2013; Zhou et al. 2013). D3/MAX2/RMS4 and D14/D88/HTD2/AtD14/DAD2 are two important SLs signaling molecules that interact with D53/SMX-L6/7/8, which is an inhibiting factor of SLs, to form protein complexes that ubiquitinate the inhibiting factor and cause it to be further specifically degraded by the proteasome, thereby inducing the expression of downstream target genes and the response of SLs signaling. The inhibiting factor thus plays a key role in the SLs signaling pathway (Wang et al. 2015; Li et al. 2015a, b).
In this study, the homolog of D53 was successfully cloned from a sugarcane variety and was found to encode 1120 amino acids, which is slightly smaller than the 1131 and 1142 amino acids of the D53 proteins from rice and sorghum, but greater than the 678 amino acids of the D53 protein from Narenga porphyrocoma (Liu et al. 2017). The conservative domain of ScD53 is similar to the D53 proteins from rice and sorghum, all of which contain a p-loop NTPase superfamily domain, a ClpB_D2-small superfamily domain, one AAA-specific binding site and two non-specific binding sites (ClpA, PRK10865) (Jiang et al. 2013; Zhou et al. 2013), except for the one non-specific AAA_2 binding site occurring only in the ScD53 and sorghum D53 protein. Homology analysis demonstrated that ScD53 has the highest homology with sorghum D53 (a 93% identity), followed by the D53 proteins of maize and millet, which corroborate the findings of Zhou et al. (2013) and Jiang et al. (2013).
ScD53 exhibited particularly high expression in the stem and low expression in the root, which is consistent with the expression of D53 in rice (Jiang et al. 2013). Previous studies have indicated that the expression of D53 in the rice d53 mutant was significantly down-regulated compared with that of wild type, and the low expression of D53 alleviated axillary bud germination inhibition and increased the number of tillers (Jiang et al. 2013; Zhou et al. 2013). In this study, ScD53 was highly expressed in dormant buds compared to germinating and developing buds, demonstrating that ScD53 may have a negative regulatory role in bud germination of sugarcane.
In addition to environmental factors, plant tillers (branches) are also regulated by plant hormones (Durbak et al. 2012). Plant IAA produced at the apical meristem can be transported to the stem base in a polar manner where they inhibit the germination of axillary buds through two purported mechanisms, namely the auxin transport channel formation hypothesis or the secondary messenger hypothesis (Kebrom 2017). Both SLs and CTK are currently thought to be important secondary messengers as they regulate axillary bud development together with IAA (Rashotte et al. 2003; Rameau et al. 2015). We demonstrated by plant hormone treatments that sugarcane bud development was regulated by KT (in a similar manner to CTK), IAA and SLs, which is consistent with results in other crops (Rameau et al. 2015). Our FQ-PCR experiments showed that the expression of ScD53 was also significantly induced by the three hormones, similar to that observed in the D53 homologs of rice and Arabidopsis (Jiang et al. 2013; Zhou et al. 2013; Soundappan et al. 2015; Wang et al. 2015). While KT induced the expression of ScD53, high expression of this gene did not inhibit the further bud development, suggesting that other pathways may eliminate the negative effect of ScD53. Our results help elucidate the function of ScD53 and provide important information and direction for the use of this gene in improving sugarcane yield through tiller regulation. Further studies are needed to illustrate the molecular mechanisms of ScD53 in tillering regulation of sugarcane, and this will help to understand the molecular basis of sugarcane tillering trait.
References
Arite, T., H. Iwata, K. Ohshima, M. Maekawa, M. Nakajima, M. Kojima, H. Sakakibara, and J. Kyozuka. 2007. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant Journal 51(6): 1019–1029.
Arite, T., M. Umehara, S. Ishikawa, A. Hanada, M. Maekawa, S. Yamaguchi, and J. Kyozuka. 2009. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant and Cell Physiology 50(8): 1416–1424.
Booker, J., M. Auldridge, S. Wills, D. McCarty, H. Klee, and O. Leyser. 2004. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Current Biology 14(14): 1232–1238.
Brewer, P.B., E.A. Dun, B.J. Ferguson, C. Rameau, and C.A. Beveridge. 2009. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiology 150(1): 482–493.
Brewer, P.B., H. Koltai, and C.A. Beveridge. 2013. Diverse roles of strigolactones in plant development. Molecular Plant 6(1): 18–28.
Chen, D.W., Y. Huang, Y.L. Lu, Y. Jiang, and Q.W. Li. 2013. Effect of different nutrient solutions on the growth of sugarcane tissue culture seedlings. Guangdong Agricultural Science 40(21): 28–31.
Cook, C.E., L.P. Whichard, W.E. Monroe, G.H. Egley, P. Coggon, P.A. Luhan, and A.T. Mcphail. 1972. Germination stimulants. II. Structure of strigol, a potent seed germination stimulant for witchweed (Striga lutea). Journal of the American Chemical Society 94(17): 6198–6199.
Cook, C.E., L.P. Whichard, B. Turner, M.E. Wall, and G.H. Egley. 1966. Germination of witchweed (Striga lutea L.): Isolation and properties of a potent stimulant. Science 154(3753): 1189–1190.
Drummond, R.S.M., N.M. Martínez-Sánchez, B.J. Janssen, K.R. Templeton, J.L. Simons, B.D. Quinn, S. Karunairetnam, and K.C. Snowden. 2009. Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiology 151(4): 1867–1877.
Dun, E.A., P.B. Brewer, and C.A. Beveridge. 2009. Strigolactones: Discovery of the elusive shoot branching hormone. Trends in Plant Science 14(7): 364–372.
Durbak, A., H. Yao, and P. Mcsteen. 2012. Hormone signaling in plant development. Current Opinion in Plant Biology 15(1): 92–96.
Fen, D., and G.L. Chen. 2011. Shoot-branching control with strigolactones: Research progress. Chinese Journal of Ecology 30(2): 349–356.
Foo, E., E. Bullier, M. Goussot, F. Foucher, C. Rameau, and C.A. Beveridge. 2005. The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17(2): 464–474.
Gallavotti, A., Y. Yang, R.J. Schmidt, and D. Jackson. 2008. The relationship between auxin transport and maize branching. Plant Physiology 147(4): 1913.
Gao, Y., Y. Li, Y.F. Xie, and Y.K. Liu. 2013. Research progress on molecular mechanism of strigolactones in regulating of plant lateral shoot development and its interaction with auxin. Journal of Plant Resources and Environment 22(4): 98–104.
Gao, Z., Q. Qian, X. Liu, M. Yan, F. Qi, G. Dong, L. Jian, and B. Han. 2009. Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Molecular Biology 71(3): 265–276.
Gomez-Roldan, V., S. Fermas, P.B. Brewer, V. Puech-Pagès, E.A. Dun, J.P. Pillot, F. Letisse, R. Matusova, S. Danoun, J.C. Portais, H. Bouwmeester, G. Bécard, C.A. Beveridge, C. Rameau, and S.F. Rochange. 2008. Strigolactone inhibition of shoot branching. Nature 455(7210): 189–194.
Ishikawa, S., M. Maekawa, T. Arite, K. Onishi, I. Takamure, and J. Kyozuka. 2005. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant and Cell Physiology 46(1): 79–86.
Jiang, L., X. Liu, G. Xiong, H. Liu, F. Chen, L. Wang, X. Meng, G. Liu, H. Yu, and Y. Yuan. 2013. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504(7480): 401.
Kebrom, T.H. 2017. A Growing stem inhibits bud outgrowth-the overlooked theory of apical dominance. Frontiers in Plant Science 8: 1874.
Kumar, S., G. Stecher, and K. Tamura. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870.
Kyozuka, J. 2007. Control of shoot and root meristem function by cytokinin. Current Opinion Plant Biology 10(5): 442–446.
Lee, T.S.G., and E.A. Bressan. 2006. The potential of ethanol production from sugarcane in Brazil. Sugar Tech 8(4): 195–198.
Leyser, O. 2003. Regulation of shoot branching by auxin. Trends in Plant Science 8(11): 541–545.
Li, S.J., J. Gao, J.Y. Li, and Y.H. Wang. 2015a. Advances in regulating rice tillers by strigolactones. Chinese Bulletin of Botany 50(5): 539–548.
Li, X.J., X.Q. Lin, H.B. Liu, C.J. Chun, C.H. Xu, and X.L. Liu. 2015b. Clone and bioinformatics analysis of the TB1 gene in sugarcane. Chinese Journal of Tropical Crops 36(11): 1978–1985.
Li, W.C., J.W. Wang, and J.J. Yu. 2012. Research overview of the tillering gene in rice. Crops 3: 19–22.
Li, Y.R., and Y.A. Wei. 2006. Sugar industry in china: R&D and policy initiatives to meet sugar and biofuel demand of future. Sugar Tech 8(4): 203.
Lin, H., R. Wang, Q. Qian, M. Yan, X. Meng, Z. Fu, C. Yan, B. Jiang, Z. Su, J. Li, and Y. Wang. 2009. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21(5): 1512–1525.
Ling, H., Q. Wu, J. Guo, L. Xu, and Y. Que. 2014. Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR. PLoS ONE 9(5): e97469.
Liu, X.H., R.H. Zhang, Y.Y. Gui, X.Q. Zhang, J.J. Wei, H. Zhou, H.P. Ou, and X.L. Liu. 2017. Cloning and bioinformatics analysis of tillering-related gene NpD53 from Narenga porphyrocoma (Hance) Bor. Journal of Southern Agriculture 48(9): 1554–1559.
Livak, K.J., and T.D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4): 402–408.
Mcsteen, P. 2009. Hormonal regulation of branching in grasses. Plant Physiology 149(1): 46–55.
Mikihisa, U., H. Atsushi, Y. Satoko, A. Kohki, A. Tomotsugu, T.K. Noriko, M. Hiroshi, K. Yuji, S. Ken, and Y. Koichi. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455 (7210): 195–200.
Rameau, C., J. Bertheloot, N. Leduc, B. Andrieu, F. Foucher, and S. Sakr. 2015. Multiple pathways regulate shoot branching. Frontiers in Plant Science 5: 741.
Rashotte, A.M., S.D. Carson, J.P. To, and J.J. Kieber. 2003. Expression profiling of cytokinin action in arabidopsis. Plant Physiology 132(4): 1998–2011.
Shani, E., O. Yanai, and N. Ori. 2006. The role of hormones in shoot apical meristem function. Current Opinion Plant Biology 9(5): 484.
Snowden, K.C., A.J. Simkin, B.J. Janssen, K.R. Templeton, H.M. Loucas, J.L. Simons, S. Karunairetum, A.P. Gleave, D.G. Clark, and H.J. Klee. 2005. The decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE 8 gene affects branch production and plays a role in leaf senescence, root growth and flower development. Plant Cell 17(3): 746–759.
Sorefan, K., J. Booker, K. Haurogne, M. Goussot, K. Bainbridge, E. Foo, S.P. Chatfield, S. Ward, C.A. Beveridge, C. Rameau, and O. Leyser. 2003. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes & Development 17(12): 1469–1474.
Soundappan, I., T. Bennett, N. Morffy, Y. Liang, J.P. Stanga, A. Abbas, O. Leyser, and D.C. Nelson. 2015. Smax1-like/d53 family members enable distinct max2-dependent responses to strigolactones and karrikins in arabidopsis. Plant Cell 27(11): 3143.
Stanga, J.P., S.M. Smith, W.R. Briggs, and D.C. Nelson. 2013. SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiology 163(1): 318–330.
Stirnberg, P., K. van de Sande, and H.M.O. Leyser. 2002. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129(5): 1131–1141.
Tanaka, M., K. Takei, M. Kojima, H. Sakakibara, and H. Mori. 2006. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant Journal for Cell & Molecular Biology 45(6): 1028–1036.
Umehara, M., A. Hanada, S. Yoshida, K. Akiyama, T. Arite, N. Takeda-Kamiya, H. Magome, Y. Kamiya, K. Shirasu, K. Yoneyama, J. Kyozuka, and S. Yamaguchi. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455(7210): 195–200.
Vasantha, S., D.E. Shekinah, C. Gupta, and P. Rakkiyappan. 2012. Tiller production, regulation and senescence in sugarcane (Saccharum species hybrid) genotypes. Sugar Tech 14(2): 156–160.
Wang, M., H.W. Chen, H.L. Chen, and K.F. Liu. 2014. Research progress in regulatory role of strigolactones in shoot branching. Acta Horticulturae Sinica 41(9): 1924–1934.
Wang, L., B. Wang, L. Jiang, X. Liu, X. Li, Z. Lu, X. Meng, Y. Wang, S.M. Smith, and J. Li. 2015. Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27(11): 3128.
Zhou, F., Q. Lin, L. Zhu, Y. Ren, K. Zhou, N. Shabek, F. Wu, H. Mao, W. Dong, and L. Gan. 2013. D14-SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504(7480): 406–410.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31760412) and Candidates of the Young and Middle Aged Academic Leaders of Yunnan Province (2014HB038), and by funding from the Yunnan Provincial Science and Technology Department for High-End Talent Program. We would like to thank LetPub (www.letpub.com) and Dr. David M. Burner for editorial assistance.
Author information
Authors and Affiliations
Contributions
ALL carried out the experiment and wrote the manuscript. XJL, CJL, HBL, QYZ, XQL participated in experimental work. XLL conceived and designed the study.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lv, AL., Li, XJ., Li, CJ. et al. Cloning and Expression Analysis of the ScD53 Gene from Sugarcane. Sugar Tech 21, 898–908 (2019). https://doi.org/10.1007/s12355-019-00730-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12355-019-00730-z