Abstract
Sugarcane mosaic resistance is established as a key criterion for the breeding program in Cuba. However, molecular characterization of Sugarcane mosaic virus (SCMV) strains occurring in Cuba has not been reported so far. Leaf samples from commercial sugarcane cultivars were collected for SCMV screening in three testing sites (the Matanzas, Camagüey and Holguin provinces). Reverse-Transcriptase polymerase chain reaction was used to identify the virus. The nucleotide sequence comparison grouped the isolates into SCMV subgroups. Sugarcane cultivars inoculated with SCMV from three testing sites displayed different symptoms. Results should be decisive to generate the highest selection pressure, increasing the efficiency of the sugarcane breeding program to the SCMV resistance in the commercial cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mosaic is one of the most important viral diseases of sugarcane. It is widely distributed around the world, producing significant losses in sugarcane yields (Padhi and Ramu 2011). This disease is commonly associated with strains of Sugarcane mosaic virus (SCMV) and Sorghum mosaic virus (SrMV). Both viruses are members of the SCMV subgroup in the genus Potyvirus of the family Potyviridae. Four other viruses, Maize dwarf mosaic virus (MDMV), Johnson grass mosaic virus (JGMV), Pennisetum mosaic virus (PenMV), and Zea mosaic virus (ZeMV), are also included in the SCMV subgroup although they have never been isolated from sugarcane (Brunt 1992; Yang and Mirkov 1997; Perera et al. 2009). Another virus, Sugarcane streak mosaic virus (SCSMV) has been identified as the major cause of mosaic symptoms in commercial sugarcane cultivars of Asian countries. SCSMV belongs to a new genus in the Potyviridae family and can simultaneously infect sugarcane with SCMV (Hall et al. 1998).
The only effective way to control mosaic disease is to introduce resistant cultivars for commercial practices. Thus, in major breeding programs, SCMV resistance is considered as a key selection criterion for releasing new cultivars. In Cuba, for 30 years, promising cultivars have been artificially tested against SCMV in the final selection stage, and mosaic is not a major problem in commercial fields (INICA 2011). Nevertheless, in the last 3 years symptomatic variations of susceptibility reactions for standard genotypes (controls) have been confirmed in the testing sites. The resistant standards get high levels of SCMV infection and the standards cannot be separated into resistance groups, providing unreliable ratings per variety. A possible explanation for these observations could be the occurrence of genomic changes in the virus population.
Reverse transcriptase polymerase chain reaction (RT-PCR) protocols are currently available to identify SCMV and SrMV using the degenerate primers 1n/2n (Marie-Jeanne et al. 2000). Direct sequencing of RT-PCR products has been used for the identification and characterization of strains virus in plants (Gaur et al. 2003; Grisham and Pan 2007). A large number of Potyvirus sequences have been reported allowing nucleotide (nt) and amino acid (aa) sequence comparisons (Marie-Jeanne et al. 2000). The whole-genome variation of Potyviridae members can be reliably reflected by the diversity in the nt sequence of their coat protein (CP) genes (Adams et al. 2005).
Variations of SCMV isolates in terms of plant response to virus and virus aggressiveness could be determined using symptom expression and infection rate of sugarcane cultivars. The principal components analysis is a method applied to detect relationships between measured variables (Annicchiarico 2002) and, an ideal model for visualizing patterns in SCMV-trial data.
The objective of this study was to conduct a genomic/symptomatic characterization of sugarcane mosaic isolates, using the reliable approaches of RT-PCR and sequencing. The results will be used to support the National Genetic Breeding Program for the selection of sugarcane mosaic resistant cultivars in the coming years.
Materials and Methods
Sample Collection
Five symptomatic and five asymptomatic fresh leaves were collected from each one of 15 sugarcane cultivars in the three testing sites for sugarcane mosaic resistance in Cuba situated in the Matanzas, Camagüey and Holguín provinces.
RNA Extraction and RT-PCR
Frozen lamina tissue (≈200 mg) was placed in liquid nitrogen, ground in a mortar and homogenized. Total RNA was extracted with RNeasy Plant Mini Kit (Quiagen) using manufacturer’s protocol. Total RNA was eluted in a final volume of 40 μl of diethylpyrocarbonate-treated (DEPC) water and stored at −20 °C. RT-PCR was performed to detect SCMV as described by Marie-Jeanne et al. (2000). Reverse transcription was performed on RNAs by using the enzyme M-MLV (Promega) reverse transcriptase primed with the reverse primer as the initial primer. Primers for detection were 1n (5′-ATGGTHTGGTGYATHGAR-3′) and 2n (5′-TGCTGCKGCYTTCATYTG-3′). PCR was carried out on cDNA with 0.5 units of Taq DNA polymerase (Promega), 30 pmol of each primer, and 0.5 mM of each dNTP in 40 μl reaction. PCR was performed by first heating at 94 °C for 5 min, followed by 30 cycles at 94 °C for 1 min, 50 °C for 2 min, and 72 °C for 1 min, and then one cycle of final extension at 72 °C for 10 min. The reaction mixture (5 μl) was analyzed on 1.5 % agarose gel stained with ethidium bromide.
DNA Sequencing, Nucleotide and Phylogenetic Analyses
The RT-PCR products were directly sequenced with both upstream and downstream primers from two directions using an automated ABI3130x1 DNA Sequencer (Hitachi). Sequences of isolates determined in this study were analyzed together with 23 reported SCMV, SrMV and MDMV isolates from GenBank (Table 1). Nucleotide sequences were aligned using the ClustalX algorithm (Thompson et al. 1997), and the phylogenetic tree was obtained using the neighbor-joining method and MEGA software.
Inoculation of Virus Isolates
To assess symptomatic variations between virus variants in sugarcane hosts, two cultivars were planted under screen-house conditions: CP31-294 [susceptible (S)], and, B42231 [resistant (R)]. The trial was arranged in a randomized complete block design with two replicates and each plot containing 20 plants spaced with 250–300 mm intervals. For inoculum preparation, each virus variant was previously spread in B34104 (susceptible to SCMV) and maintained in the pathology field. Each inoculum was then prepared by mixing juice from symptomatic leaves of the B34104 cultivar with an abrasive powder. A cotton pad was soaked in the inoculum and then rubbed onto each sugarcane seedling (2–3 leaf stage) (INICA 2011). The virus variants evaluated were (1) Matanzas inoculum, (2) Camaguey inoculum, (3) Holguín inoculum, (4) a control without inoculation. Length, diameter, symptoms and infection percentages were recorded from 15 to 60 days post-inoculation. Total RNA of the youngest leaf was extracted and RT-PCR to detect SCMV was performed as described above.
Pigment Detection
Fresh leaf samples (200 mg) were collected at 60 days post-SCMV inoculation, grounded in liquid nitrogen and homogenized with 5 ml of methanol. The tissue homogenate was incubated for 16 h at 4 °C. The mixture was then centrifuged for 6 min at 10,000g. The absorbance was measured at 470, 653 and 666 nm to obtain concentrations of chlorophyll (A), chlorophyll (B) and total carotenoids as described by Acevedo et al. (1993).
Statistical Analysis
For the statistical analysis, the infection rates were transformed using the square root. Repeated measures analysis of variance (ANOVA) was performed to assess significant differences between SCMV variants. A principal component analysis was performed to detect relationships between variables using the Statistica v6.0 software.
Results
Identification and Characterization of SCMV
A SCMV-specific fragment of 327 bp (expected size) was amplified in seven sugarcane samples (Table 2). Two cultivars were infected with SCMV in all of the three test sites (B34104 and Ja64-11). Cultivar C88-380 was infected only in asymptomatic samples collected at Holguín. The cultivar B34104 is the inoculum source for SCMV testing trials.
Three RT-PCR products chosen based on inoculum source of each testing site were directly sequenced. The partial viral CP gene sequences obtained have been submitted to GenBank (accession numbers given in Table 1). The preliminary analysis of partial sequences shows that great similarity exists between the isolates from each testing site (Fig. 1). The overall nucleotide (nt) and amino acid (aa) identity of our SCMV isolates showed a 99.0 and 97.0 % respectively.
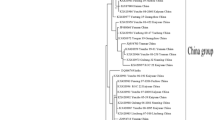
The phylogenetic tree was constructed based on the nt sequence alignment of 23 SCMV, SrMV and MDMV isolates used as references and our three SCMV isolates (Fig. 2). The analyzed isolates can be divided into five groups. The first group includes only SCMV isolates from maize. The second and fourth groups comprise SrMV isolates with greatest dissimilarity between them. The third group includes only MDMV isolates from Bulgaria. Finally, the fifth group comprises SCMV isolates from sugarcane and worldwide distribution.
Our three SCMV variants/inoculum showed great similarity between the isolates in fifth group at nt and aa sequence level ranged between 92.3 and 95.4 % (mean 93.9 %) and between 84.3 and 91.2 % (mean 88.7 %), respectively. The overall nt and aa identity between Cuban isolates and the SCMV isolates included in the first group ranged between 85.9 and 88.0 % (mean 85.9 %) and between 64.0 and 70.0 % (mean 65.6 %), respectively (Table 2).
Symptom Expression and Virus Infection Rate
The relationship between exposure to the three inoculum sources and SCMV resistance in two sugarcane genotypes was tested. Results showed that the CP31-294 cultivar had a mosaic susceptible pattern, while B42231 was SCMV resistant. The infection percentage was variable between virus inoculums (Table 3). The progression of SCMV infection in CP31-294 (susceptible pattern) is show in Fig. 3. The isolate from Matanzas was the most infective in causing SCMV infection. Significant differences were demonstrated between inoculums. The presence of virus in samples showing mosaic symptoms were confirmed using RT-PCR.
Statistical Analysis
Principal component analysis showed that the first two components (CP) accounted for 89.4 % of total data variability (Table 4). The first component (CP1) was characterized by the infection percentages (PI) from 30–60 days post-inoculation, which were negatively correlated with length, diameter, chlorophyll B (Cb) and carotenoids (Cx). Chlorophyll A (Ca) was distinguished in the second component (CP2).
Graphical representation of the CP1–CP2 components showed well differentiated groups (Fig. 4). The SCMV susceptible (S) cultivar CP31-294 was consistently separated from the resistant (R) B42231 cultivar. Inside the CP31-294 susceptible group, two separate clusters were formed corresponding to different inoculums treatments. It was showed the Matanzas inoculum as the most SCMV infective one.
Discussion
For the first time a genomic characterization of SCMV has been conducted in Cuba. Results demonstrated great similarity between the isolates from each testing site. The specific primers used to detect members of the SCMV subgroup amplified a fragment of the core region in the CP gene. The core region is the best marker to study the sequence relationship between strains of a virus in the genus Potyvirus (Viswanathan et al. 2009).
The CP is the best characterized of all gene products for SCMV and consists of the highly variable, surface-exposed amino-(N)-terminus, a highly conserved core region, and a surface-exposed carboxyl-(C)-terminus (Shukla and Ward 1988). Nucleotide sequence identities (and aa similarities) have been widely used for Potyvirus taxonomic purposes, taking into consideration that all CP gene nt identity percentages vary between 40 and 70 % for different potyviruses and are above 90 % for different strains of the same virus (Adams et al. 2005).
This study establishes that mosaic of sugarcane is induced only by SCMV. However, recent studies reported the evidence of co-infection of sugarcane by SCMV and SrMV (Xu et al. 2008; Padhi and Ramu 2011). Additionally SCSMV and SrMV cause mosaic in sugarcane, either alone or in combination with SCMV (Viswanathan et al. 2009).
In Cuba, for about 20 years, the inoculum has been replicated in the same host at each location and unexpected atypical symptoms were observed from standard controls in the resistance test. The most accepted explanation is the occurrence of genetic changes during the virus-environment-host interactions. However, a molecular characterization of the entire CP gene or the whole genome of SCMV circulating strains is necessary for corroborate such hypothesis. It has been recognized that SCMV has diverged under host selection pressure and geographical isolation for a long period of time (Xu et al. 2008).
Padhi and Ramu (2011) demonstrated that recombination is the major cause of evolution and emergence of new variants of SCMV (Chare and Holmes 2006). Otherwise variation in strain identity can be explained by changes in use of sugarcane cultivars, as illustrated by the history of SCMV strains in Louisiana, where new strains occurred when new cultivars were introduced (Koike and Gillaspie 1989; Grisham and Pan 2007).
According to the phylogenetic tree our virus isolates are nearly related to other SCMV strains with worldwide distribution. The study conducted by Alegria et al. (2003) reported that SCMV isolates were clustered into several subgroups correlated to geographical origin. Results obtained by Goodman (1999) indicated that no association exists between SCMV strain prevalence and specific cultivars or regions. Since SCMV can be transmitted through vegetative sett, the germplasm exchange could be one possible explanation for such high sequence similarities observed among the intercontinental isolates of SCMV (Padhi and Ramu 2011).
Considering the results of SCMV symptom expression under green-house conditions, genetic studies could be conducted which showed differences between the SCMV variants. Putra et al. (2003) reported that SCMV moves slower in moderately resistant cultivars than in the susceptible ones, and from the point of inoculation to younger leaves, roots and tillers, and eventually to leaves growing next to the inoculated leaves.
As expected, the infection percentage in susceptible cultivar CP31-294 indicated rapid virus multiplication, and the variation in this variable might support the severity of the virus variants. This result should be of great value, for instance, the most infective isolates should be used to generate the highest selection pressure, increasing the efficiency of the breeding program.
Differences found in variables such as length, diameter, chlorophyll A, chlorophyll B and carotenoids were in correlation to the infection percentage. In the infected susceptible cultivar the virus infection causes a decrease in plant growth showing a reduction in length and diameter. Also, pigment reduction may be caused, mainly due to a rise in chlorophylase activity or inhibition of pigment synthesis as a consequence of plastidic protein deviated for viral replication (Acevedo et al. 1993).
Summarizing, these results become crucial for the development of new strategies in the Cuban sugarcane genetic breeding program. The requirement of an enduring surveillance of disease causal agents should be enhanced with the use of powerful and reliable molecular tools. Additionally, this work could serve as a guideline for other sugarcane breeding programs worldwide.
References
Acevedo, R., B. Díaz, D. Piñón, E. Fernández, and M. Fajardo. 1993. Behavior of some physiologic indicators in sugarcane infected with Sugarcane mosaic virus (SCMV). Revista de Protección Vegetal 8: 291–299.
Adams, M.J., J.F. Antoniw, and C.M. Fauquet. 2005. Molecular criteria for genus and species discrimination within the family Potyviridae. Archives of Virology 150: 459–479.
Alegria, O.M., M. Royer, M. Bousalem, M. Chatenet, M. Peterschmitt, J.C. Girard, and P. Rott. 2003. Genetic diversity in the coat protein coding region of eighty-six Sugarcane mosaic virus isolates from eight countries, particularly from Cameroon and Congo. Archives of Virology 148: 357–372.
Annicchiarico, P. 2002. Genotype x environment interactions. Challenges and opportunities for plant breeding and cultivar recommendations. FAO Plant Production and Protection 174: 2–4.
Brunt, A.A. 1992. The general properties of Potyviruses. Archives of Virology 5: 3–16.
Chare, E.R., and E.C. Holmes. 2006. A phylogenetic survey of recombination frequency of plant RNA virus. Archives of Virology 151: 933–946.
Chen, J., J. Chen, and M.J. Adams. 2002. Molecular characterization of potyviruses from sugarcane and maize in China. Archives of Virology 147: 1237–1246.
Espejel, F., D. Jeffers, J.C. Noa-Carrazana, S. Ruiz-Castro, and L. Silva-Rosales. 2006. Coat protein gene sequence of a Mexican isolate of Sugarcane mosaic virus and its infectivity in maize and sugarcane plants. Archives of Virology 151: 409–412.
Gaur, R.K., M. Singh, and G.P. Rao. 2003. Molecular Characterization of Sugarcane Mosaic Virus isolate from north eastern region of India. Sugarcane 5: 149–153.
Goodman B S. (1999) A study of South African strains of Sugarcane Mosaic Potyvirus (SCMV) identified by sequence analysis of the 5´region of the coat protein gene. MS thesis, Department of Biotechnology, Durban University of Technology, Durban, South Africa.
Grisham, M.P., and Y.B. Pan. 2007. A genetic shift in the virus strains that cause mosaic in louisiana sugarcane. Plant Disease 91: 453–458.
Hall, J.S., B. Adams, T.J. Parsons, R. French, L.C. Lane, and S.G. Jensen. 1998. Molecular cloning, sequencing and phylogenetic relationship of a new potyvirus: Sugarcane Streak Mosaic Virus, and a reevaluation of the classification of the Potyviridae. Molecular Biology and Evolution 10: 323–332.
Handley, J.A., G.R. Smith, J.L. Dale, and R.M. Harding. 1998. Sequence diversity in the coat protein coding region of twelve Sugarcane mosaic potyvirus isolates from Australia, USA and South Africa. Archives of Virology 143: 1145–1153.
INICA. (2011) Manual de Normas y Procedimientos del Programa de Fitomejoramiento. Boletín Especial Cuba & Caña, p. 200.
Jiang, J.X., X.P. Zhou, and Y.L. Shi. 2002. Identification of the viral pathogen of sugarcane mosaic disease in Hangzhou. Journal of Agricultural Biotechnology 10: 377–380.
Koike, H., and A.G. Gillaspie Jr. 1989. Mosaic. In Diseases of sugarcane. Major diseases, ed. C. Ricaud, B.T. Egan, A.G. Gillaspie Jr, and C.G. Hughes, 400 . Amsterdam: Elsevier.
Kong, P., and H.H. Steinbiss. 1998. Complete nucleotide sequence and analysis of the putative polyprotein of Maize dwarf mosaic virus genomic RNA (Bulgarian isolate). Virology 143: 1791–1799.
Marie-Jeanne, V., R. Ioos, J. Peyre, B. Alliot, and P. Signoret. 2000. Differentiation of Poaceae Potyviruses by reverse transcription polymerase chain reaction and restriction analysis. Journal of Phytopathology 148: 141–151.
Oertel, U., J. Schubert, and E. Fuchs. 1997. Sequence comparison of the 3′-terminal parts of the RNA of four German isolates of Sugarcane mosaic potyvirus (SCMV). Archives of Virology 142: 675–687.
Padhi, A., and K. Ramu. 2011. Genomic evidence of intraspecific recombination in Sugarcane mosaic virus. Virus Genes 42: 282–285.
Perera, M.F., M.P. Filippone, C.J. Ramallo, M.I. Cuenya, M.L. García, L.D. Ploper, and A.P. Castagnaro. 2009. Genetic diversity among viruses associated with sugarcane mosaic disease in Tucumán, Argentina. Phytopathology 99: 38–49.
Putra, L.K., J.O. Helen, P.J. Anthony, and J.L. Whittle. 2003. Distribution of Sugarcane mosaic virus in sugarcane plants. Australasian Plant Pathology 32: 305–307.
Shukla, D.D., and C.W. Ward. 1988. Amino acid sequence homology of coat protein as a basis for identification and classification of the potyvirus group. Journal of General Virology 69: 2703–2710.
Thompson, J.D., T.J. Gibson, F. Plewniak, F. Jeanmouging, and D.G. Higgins. 1997. CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleid Acids Research 25: 4876–4882.
Viswanathan, R., R. Karuppaiah, and M. Balamuralikrishnan. 2009. Identification of new variants of SCMV causing sugarcane mosaic in India and assessing their genetic diversity in relation to SCMV type strains. Virus Genes 39: 375–386.
Xu, D.L., J.W. Park, T.E. Mirkov, and G.H. Zhou. 2008. Viruses causing mosaic disease in sugarcane and their genetic diversity in southern China. Archives of Virology 153: 1031–1039.
Yang, Z.N., and T.E. Mirkov. 1997. Sequence and relationships of Sugarcane mosaic and Sorghum mosaic virus strains and development of RT-PCR-based RFLPs for strain discrimination. Phytopathology 87: 932–939.
Acknowledgments
The work was supported by the Sugarcane Research Institute, Cuba. The authors would like to thank the Pathology Department for their assistance in the sample and data collection, as well the anonymous referees for their useful critical and recommendations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Puchades, Y., La O, M., Montalván, J. et al. Genetic and Symptomatic Characterization of Sugarcane mosaic virus (SCMV) in Cuba. Sugar Tech 18, 184–191 (2016). https://doi.org/10.1007/s12355-015-0375-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12355-015-0375-0