Abstract
Neurog1 is a pro-neural basic helix-loop-helix (bHLH) transcription factor expressed in progenitor cells located in the ventricular zone and subsequently the presumptive white matter tracts of the developing mouse cerebellum. We used genetic inducible fate mapping (GIFM) with a transgenic Neurog1-CreER allele to characterize the contributions of Neurog1 lineages to cerebellar circuit formation in mice. GIFM reveals Neurog1-expressing progenitors are fate-mapped to become Purkinje cells and all GABAergic interneuron cell types of the cerebellar cortex but not glia. The spatiotemporal sequence of GIFM is unique to each neuronal cell type. GIFM on embryonic days (E) 10.5 to E12.5 labels Purkinje cells with different medial-lateral settling patterns depending on the day of tamoxifen delivery. GIFM on E11.5 to P7 labels interneurons and the timing of tamoxifen administration correlates with the final inside-to-outside resting position of GABAergic interneurons in the cerebellar cortex. Proliferative status and long-term BrdU retention of GIFM lineages reveals Purkinje cells express Neurog1 around the time they become post-mitotic. In contrast, GIFM labels mitotic and post-mitotic interneurons. Neurog1-CreER GIFM reveals a correlation between the timing of Neurog1 expression and the spatial organization of GABAergic neurons in the cerebellar cortex with possible implications for cerebellar circuit assembly.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cerebellum is an ideal model system to study regulatory pathways in neurogenesis and neuronal pattern formation. Although comprised of millions of neurons, the cerebellum consists of few principal neuronal cell types arranged within a uniform cortical cyto-architecture and three pairs of deep cerebellar nuclei (DCN) embedded in the white matter [1, 2]. Cerebellar neurogenesis is protracted, spanning embryonic and postnatal development, and is characterized by spatially and temporally restricted patterns of neuronal production [3]. Cerebellar neurons are generated from two distinct germinative zones: a ventricular zone (VZ) lying dorsal to the fourth ventricle; and from the upper rhombic lip (uRL), a specialized region lying between the fourth ventricle and the roof plate. These two germinative zones express distinct basic helix-loop-helix (bHLH) transcription factor genes—Ptf1a (pancreas transcription factor 1a), Ascl1 (achaete-scute homologue 1) and neurogenins Neurog1 and Neurog2 in the VZ, and Atoh1 (atonal homologue 1) in the uRL—that have each been used in the genetic fate mapping of progenitors.
Genetic inducible fate mapping (GIFM) with a transgenic Atoh1-CreER allele reveals that uRL progenitors are fate-mapped to become glutamatergic DCN projection neurons and subsequently glutamatergic granule cell interneurons in the cerebellum [4]. Cumulative fate mapping using a Ptf1a Cre (pancreas transcription factor 1a) knock-in allele labels GABAergic Purkinje cells, DCN inhibitory interneurons and Golgi, basket and stellate cell inhibitory interneurons of the cerebellar cortex [5, 6]. More recently, GIFM of Ascl1, a bHLH gene expressed throughout the Ptf1a expression territory in the VZ [7], has been used to define the spatiotemporal production of neurons from the VZ [8, 9]. Knock-in Ascl1 CreER mice reveals that Purkinje cells are labeled during embryonic day (E) 10.5 to E12.5 and have distinct settling patterns in the adult cerebellum depending on the day they express Ascl1 [9]. GABAergic DCN neurons are the first interneurons to be labeled at E10.5–E11.5 with inhibitory interneurons of the cerebellar cortex subsequently fate-mapped to granule, Purkinje cell and then molecular layers of the cerebellar cortex [9].
Fate mapping of neurogenin-expressing progenitors reveals differing contributions from Neurog1- and Neurog2-expressing lineages to the cerebellum. GIFM using a Neurog2 CreER knock-in allele labels Purkinje cells and DCN GABAergic neurons but little contribution to GABAergic interneurons in the cortex [10]. In contrast, we have shown progenitors expressing Neurog1 contribute to Purkinje cells and multiple GABAergic interneuron cell types in the cerebellar cortex using transgenic Neurog1-EGFP and Neurog1-Cre alleles to short-term and cumulatively map cell fates, respectively [11]. A CNS-wide GIFM study using transgenic Neurog1-CreER mice extends this observation revealing Purkinje cells are fate-mapped E10.5 to E12.5 and are followed by GABAergic interneurons in the granule cell layer at E14.5 and E17.5 [12]. Thus, preliminary Neurog1 GIFM data suggests similarities between Neurog1 and Ascl1 spatiotemporal fate maps in the cerebellum and the possibility that the 2 bHLH genes act coordinately to establish cerebellar circuitry.
In this study, we used Neurog1-CreER GIFM to generate a detailed spatiotemporal fate map of Neurog1 lineages in the cerebellum. We show that Neurog1-expressing progenitors are fate-mapped to become GABAergic neuronal cell types of the cerebellum with the spatiotemporal sequence of fate mapping unique to each cell type. We confirm that Purkinje cells are the first cerebellar neurons to be labeled at E10.5 to E12.5 and are followed by inhibitory interneurons of the granule cell, Purkinje cell, and molecular layers. BrdU birth dating and markers of cell proliferation reveal that Neurog1 is expressed in Purkinje cells around the time they become post-mitotic but in mitotic and post-mitotic GABAergic interneurons suggesting distinct Neurog1 functions in Purkinje cell and interneuron cell lineages.
Material and Methods
Neurog1-CreER T2 and Cre Reporter Mouse Lines
Neurog1-CreER T2 recombinase and Cre reporter mouse lines used in this study have been described previously. See [12, 13] for details of the BAC transgenic mouse line Neurog1-CreER T2. See [14] for details of the knock-in Rosa26 loxP-STOP-td-Tomato (here referred to as R26 loxP-STOP-td-Tomato) reporter mouse line. See [15] for details of the transgenic reporter mouse line Z/EG (pCAGGS-Bgeo/EGFP). Mice were obtained from The Jackson Laboratory, Bar Harbor, ME. All procedures were approved by the Uniformed Services University of the Health Sciences Institution for Animal Care and Use Committee (IACUC). Mice were housed in the Uniformed Services University’s Center for Laboratory Animal Medicine on a standard 12 h light-dark cycle, with food and water available ad libitum. Mice were genotyped using The Jackson Laboratory Genotyping PCR Protocols or, for Z/EG only, by β-galactosidase staining in X-Gal reagent (Mirus Bio LLC). Mice were timed-mated overnight, and the presence of a vaginal plug the following morning used to denote embryonic day 0.5 (E0.5). Using this counting method, mice give birth at E19.5. The day of birth was designated as postnatal day 0 (P0).
Genetic Inducible Fate Mapping (GIFM)
Two overnight breeding strategies were employed for GIFM: (1) Neurog1-CreER T2 were mated with heterozygous (R26 loxP-STOP-td-Tomato/+) or homozygous (R26 loxP-STOP-td-Tomato/td-Tomato) reporters; (2) Neurog1-CreER T2 were mated with transgenic Z/EG reporters. Homozygous R26 loxP-STOP-td-Tomato/td-Tomato mice are viable and fertile (The Jackson Laboratory). Timed-pregnant females or postnatal pups were administered tamoxifen dissolved in sunflower oil by intra-peritoneal injection. Mice were injected with 50 mg/Kg tamoxifen at ≈9 a.m. and given a second injection of 50 mg/Kg tamoxifen 6 h later. Controls were given intra-peritoneal injections of sunflower oil only at equivalent times. Tamoxifen disrupted parturition and lactation in the mice resulting in poor progeny survival rates. To improve survival, mice were removed by terminal caesarian from E19.5 pregnant dams and fostered to lactating Swiss Webster females (Taconic).
Immunohistochemistry (IHC)
Mice were fixed for IHC as described previously [11]. Adult tissue was processed for free-floating IHC as described previously [16]. Embryonic tissue was collected on slides and processed for IHC as described previously [11]. All immunofluorescent labeling, except for Pax2, was performed using Alexa Fluor® 488, 555, or 647 dye-labeled secondary antibodies (Life Technologies) and the tissue counterstained with DAPI and mounted under Mowiol. Pax2 immunolabeling was revealed using a biotinylated anti-rabbit secondary antibody with Dy-Light 488 streptavidin (Vector Laboratories). Immunofluorescent images were captured on a Zeiss 710 laser scanning confocal microscope (LSCM), a Zeiss Pascal LSCM, and a Leica AF6000 fluorescence microscope with motorized x, y, and z planes. For bright-field microscopy, sections were immunolabeled with biotinylated secondary antibodies followed by Vectastain ABC reagent and immunolabeling revealed with diaminobenzidine tetrahydrochloride (DAB) (Vector Laboratories). Bright-field images were captured on a Zeiss A1 microscope or Hammamatsu Nanozoomer 2.0-RS digital slide scanner.
Details of the primary antibodies used for IHC are as follows: (1) 1:500 dilution of rat monoclonal anti-BrdU (Accurate Chemical & Scientific Corp.); (2) 1:2000 dilution of mouse monoclonal anti-Calbindin D28K (anti-Calb1, Swant); (3) 1:1000 dilution of chicken polyclonal anti-green fluorescent protein (anti-GFP, Abcam); (4) 1:1000 dilution of mouse monoclonal anti-glial fibrillary acidic protein (anti-GFAP, Abcam); (5) 1:2000 dilution of rabbit polyclonal anti-Hsp25 (Enzo Life Sciences); (6) 1:200 dilution of rabbit monoclonal anti-Ki67 (Thermo Scientific); (7) 1:1500 dilution of mouse monoclonal anti-metabotropic glutamate receptor 2 (anti-Grm2, AbCam); (8) 1:500 dilution of rabbit polyclonal anti-neurogranin (Abcam); (9) 1:500 dilution of mouse monoclonal anti-NeuN (clone A60, Millipore); (10) 1:500 dilution of rabbit polyclonal anti-Olig2 (Millipore); (11) 1:2500 dilution of mouse monoclonal anti-parvalbumin (anti-Pvlb, Swant); (12) 1:200 dilution of rabbit polyclonal anti-Pax2 (Zymed); (13) 1:400 dilution of rabbit polyclonal anti-phospho-histone H3 (phH3, Millipore); (14) 1:100 dilution of rabbit polyclonal anti-PLCβ4 (Santa Cruz Biotechnology); (15) 1:1000 dilution of chicken polyclonal anti-red fluorescent protein (anti-RFP, Rockland); (16) 1:500 dilution of mouse monoclonal anti-S100β (Sigma); (17) 1:250 dilution of rabbit polyclonal anti-Zebrin II/Aldolase C (gift of R. Sillitoe).
In Situ Hybridization (ISH)
Timed-pregnant Swiss Webster mice (Taconic) were fixed for ISH as described previously for IHC. The tissue was cryo-protected in sucrose and frozen sections cut on a cryostat and post-fixed in fresh fixative. ISH was performed using RNAscope® probes for DAB singleplex and fluorescent multiplex hybridizations according to the instructions from the manufacturer Advanced Cell Diagnostics (ACD). Neurog1 and Ascl1 probes were hybridized singleplex with DAB (3,3'-Diaminobenzidine) labeling on adjacent tissue sections for comparison or in duplex using ACD’s green/red fluorescent probe labeling system.
Cell Fate Quantification
Cell fate counts were recorded from tiled 20× fluorescent images of cerebellar sections processed for anti-EGFP or anti-RFP reporter IHC. We used morphological criteria validated by dual IHC using anti-EGFP/anti-RFP antisera with cell-type specific antibodies to score cell fate. Purkinje cells were identified by their large, pear-shaped soma positioned between the granule cell and molecular layers of the cerebellar cortex and by their highly branched dendritic arbors (confirmed by anti-Calb1 IHC). Golgi cells were identified by their large, irregularly shaped soma within the granule cell layer (confirmed by anti-Nrg and anti-Grm2 IHC). Candelabrum and Lugaro cells were identified by the position of their relatively small soma within the Purkinje cell layer and supporting morphology. Candelabrum cells had 1–2 sparsely branched dendrites and a characteristic beaded axonal arborization in the molecular layer [17]. Lugaro cells had a fusiform cell body and horizontal bipolar dendrites [18]. Candelabrum and Lugaro cells were counted together in this study as Purkinje cell layer (PCL) interneurons. Basket and stellate cells were identified by the position of their small, multipolar soma within the molecular layer (confirmed by anti-Pvlb IHC). The molecular layer was divided along the middle laminar axis into inner and outer sublayers, and the number of basket and stellate cell types were counted by their location within the inner and outer halves, respectively. Cell counts were recorded from 3–4 coronal sections spanning the full rostrocaudal extent of the cerebellum. When presented as the mean standard error of the mean (SEM), cell counts were obtained from at least three animals unless stated otherwise.
Bromodeoxyuridine (BrdU) Delivery
Timed-pregnant females or postnatal pups were administered with BrdU dissolved in saline by intra-peritoneal injection. Mice were injected with 50 mg/Kg BrdU at ≈9 a.m. and given a second injection of 50 mg/Kg BrdU 6 h later. RFP reporter expressing cells retaining BrdU were identified by punctate nuclear staining with anti-BrdU IHC.
Quantification of Ki67+ and BrdU+ Cells
For Ki-67 cell counts, timed-pregnant females or neonates were processed for dual anti-Ki67/anti-RFP IHC 24 h after the first tamoxifen injection. The percentage of total RFP reporter expressing cells in the cerebellum co-expressing Ki67 was quantified from 40× confocal Z stacks taken from serially spaced sections of the cerebellum.
For short-term BrdU pulse labeling, timed-pregnant females or neonates were processed for dual anti-BrdU/anti-RFP IHC 24 h after the first BrdU injection. The percentage of RFP+ cells in the cerebellum co-immunoreactive for BrdU was quantified from 40× confocal Z stacks as described for Ki67. For long-term BrdU pulse labeling, mice were processed for dual anti-BrdU/anti-RFP IHC on P42. The percentage of RFP+ cerebellar cell types co-immunoreactive for BrdU was quantified from tiled 20× fluorescent images of cerebellar sections.
Results
We used Neurog1-CreER T2 mice in this study to develop a detailed spatiotemporal fate-map of Neurog1-expressing lineages in the cerebellum. Neurog1-CreER T2 mice (throughout referred to as Neurog1-CreER) carry copies of a modified BAC in which the single Neurog1 coding exon was replaced with CreER T2. Cre mRNA in situ hybridization (ISH) patterns recapitulate Neurog1 mRNA ISH in Neurog1-CreER embryos demonstrating restricted expression of Cre to Neurog1 expression sites [12, 13]. Two reporter lines were used to identify Neurog1-CreER-expressing cell lineages: Rosa26 loxP-STOP-td-Tomato (here referred to as R26 td-Tomato) labeled cells with red fluorescent protein (RFP), and Z/EG reporters labeled cells with enhanced green fluorescent protein (EGFP).
We analyzed mice heterozygous and homozygous for the R26 td-Tomato allele in this study and Neurog1-CreER; R26 td-Tomato/+ and Neurog1-CreER; R26 td-Tomato/td-Tomato mice are collectively referred to as Neurog1-CreER; R26 td-Tomato. Although we did not observe a difference in the intensity or distribution of RFP epilfluorescence in the brain between Neurog1-CreER; R26 td-Tomato/+ and Neurog1-CreER; R26 td-Tomato/td-Tomato mice, RFP fluorescence was enhanced by immunohistochemistry (IHC) to normalize reporter signal between mice heterozygous or homozygous for R26 td-Tomato. Mice carrying both the Neurog1-CreER and Z/EG transgene are referred to as Neurog1-CreER; Z/EG, and the EGFP reporter signal was also enhanced by IHC.
Cre recombinase was activated in Neurog1-CreER; R26 td-Tomato and Neurog1-CreER; Z/EG mice by the delivery of tamoxifen (TMX) in sunflower oil. Mice were injected with tamoxifen the morning of a given embryonic day and given a second tamoxifen injection 6 h later. Recombination at loxP sites is reported to occur within 6 h of TMX delivery and continues for up to 36 h [19]. Thus E10.5 TMX delivery in Neurog1-CreER mice is predicted to label cells expressing Neurog1 ≈ E10.75–E12.25. Mice injected with sunflower oil only were used to control for leaky Cre recombinase activity not controlled by TMX administration.
Short-term Neurog1-CreER; R26 td-Tomato Labeling is Consistent with Neurog1 ISH in Sites of GABAergic Neurogenesis
We verified Neurog1-CreER marks cells of Neurog1-expressing lineages in the developing cerebellum by comparing short-term Neurog1-CreER; R26 td-Tomato labeling patterns and ISH data for Neurog1. Neurog1-CreER; R26 td-Tomato were given TMX on E11.5, E12.5, or E18.5 and fixed 24 h later. ISH was performed on sections from E12.5, E13.5, and P0 Swiss Webster mice for comparison. The data reveal corresponding patterns of ISH and RFP reporter gene expression in the cerebellum and surrounding hindbrain structures in early cerebellar development (Fig. 1). ISH signal is present at the ventricular zone (VZ) but absent from the rhombic lip (RL) and midline of the developing cerebellar anlage of E12.5 and E13.5 mice (Fig. 1a, c). In E12.5 Neurog1-CreER; R26 td-Tomato mice, RFP is localized to layers of cells adjacent and parallel to the VZ (but not to the RL or midline regions) in more dorsal aspects of the cerebellar anlage, consistent with the rapid dorsal migration of RFP marked cells from the Neurog1-expressing regions of the VZ (Fig. 1b). In E13.5 Neurog1-CreER; R26 td-Tomato mice RFP labeling is present throughout the VZ (but not in the RL or midline) suggesting a slower VZ exit of E12.5 compared to E11.5 GIFM progenitors (Fig. 1d). ISH signal at P0 is markedly reduced in the cerebellum compared to E12.5 and E13.5. At this age, ISH-labeled cells are clustered in lateral and dorsal aspects of the cerebellum parallel but deep to the external granule cell layer (EGL) (Fig. 1e). This pattern is consistent with Neurog1 expression in cells in deeper layers of the developing cerebellar cortex (Fig. 1e). RFP-expressing cells are similarly distributed in the developing cerebellar cortex in P0 Neurog1-CreER; R26 td-Tomato mice (Fig. 1f). However, dense RFP expression is also detected in the RL with E18.5 GIFM suggesting ectopic Neurog1-CreER expression in the RL during later embryogenesis (Fig. 1f arrow and inset). Combined, these data indicate that Neurog1-CreER mice can be used to fate-map Neurog1-expressing lineages from the cerebellar VZ but that later fate mapping could result in ectopic labeling of RL-derived cell lineages.
E10.5 to E12.5 Neurog1-CreER GIFM labels Purkinje Cells with Distinct Medial-lateral Settling Patterns in the Adult Cerebellum
Cerebellar Purkinje cells have been fate-mapped at E10.5, E11.5, and E12.5 using Neurog1-CreER GIFM with a Rosa26 β-glatactosidase (R26 Lacz/+ ) reporter [12]. This CNS-wide study revealed that E11.5 fate mapping was the highest contributor to Purkinje cell fate and that E12.5 Purkinje cells settled preferentially in rostral cerebellar lobes. However, no other spatiotemporal characteristics of Purkinje cell fate were reported. In contrast, E10.5–E12.5 Ascl1 CreER spatiotemporal Purkinje cell fate patterns form distinct anterior-posterior (A-P) stripes that shift in medio-lateral (M-L) axis depending on the day of TMX delivery [9]. We examined whether alternative Cre reporters would reveal Neurog1-expressing progenitors that give rise to a similarly striped distribution of Purkinje cells.
GIFM using transgenic Z/EG reporters results in sporadic Purkinje cell labeling in adult Neurog1-CreER; Z/EG mice with TMX injection at E11.5 or E12.5 (Fig. 2a, b). The frequency of EGFP appears comparable to published Neurog1-CreER; R26 Lacz/+ data [12], and differences in the distribution of labeled Purkinje cells were noticeable between Neurog1-CreER; Z/EG mice injected with TMX on E11.5 or E12.5. With TMX at E11.5, EGFP Purkinje cells were scattered across the full M-L axis of the cerebellum, whereas TMX at E12.5 marked fewer Purkinje cells restricted to the vermis and paravermis (Fig. 2b).
The frequency of Purkinje cell labeling was significantly increased when we used Neurog1-CreER GIFM with the knock-in R26 td-Tomato reporter to mark cells with RFP. Numerous dual anti-RFP/anti-Calb1 Purkinje cells were present in Neurog1-CreER; R26 td-Tomato mice given TMX at E10.5, E11.5, or E12.5 (Fig. 2c). TMX at E10.5 resulted in RFP immunolabeled Purkinje cells present across the majority of the M-L and A-P axis with only a narrow parasagittal stripe of RFP-negative cells present at the midline of the anterior lobes (Fig. 1d, arrow heads). The density of RFP Purkinje cell labeling increased with TMX at E11.5 in which marked Purkinje cells were present throughout the M-L and A-P axis of the adult cerebellum. A-P stripes of RFP-negative cells were not present with E11.5 GIFM, although the density of RFP-labeled Purkinje cells was reduced in lateral hemispheric regions compared to the vermal/paravermal regions (Fig. 2d).
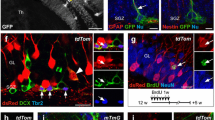
E12.5 GIFM produced a dramatic change in the distribution of fate-mapped Purkinje cells. Purkinje cells marked at this timepoint settled in a bilaterally symmetrical pattern of three parasagittal stripes along the M-L axis in the adult vermis and paravermis (Fig. 2d, arrows). A medial stripe of RFP-labeled Purkinje cells spanned the full A-P axis at the midline and was flanked by two relatively narrow bilaterally symmetrical RFP-negative A-P stripes within the vermis. These RFP-negative stripes were flanked laterally by an A-P stripe of RFP-labeled cells at the border of the vermis and paravermis, segregating the RFP labeled population in the anterior vermis into three parasagittal RFP+ stripes. The midline RFP+ Purkinje cell population expands along the M-L axis such that the vermis contains a single medial A-P RFP stripe flanked by RFP-negative Purkinje cells in the posterior cerebellum. Here, the lateral A-P stripe of RFP-labeled Purkinje cells is pushed out to the borders of the paravermis with the pontocerebellum where a parasagittal stripe of RFP-labeled Purkinje cells runs along crus 1 and medial portions of crus 2, the pyramis, and the copula. A comparison of E10.5, E11.5, and E12.5 Neurog1-CreER GIFM maps (Fig. 2d) supports the view that Neurog1 is expressed in all Purkinje cell progenitors in the developing cerebellum. Of note, the E12.5 GIFM map overlaps the pattern of Purkinje cells marked at earlier timepoints implying these late fate-mapped Purkinje cells derive from a subset of progenitors in which earlier Neurog1 expression is maintained past E13. The pattern of RFP+ axons in the deep cerebellar nuclei (DCN) of adult Neurog1-CreER; R26 td-Tomato indicates E12.5 GIFM Purkinje cells form dense projections to the medial and interposed nuclei (Fig. 2d, see Med and Int annotations).
We next asked if E12.5 Neurog1-CreER fate-mapped Purkinje cells result from VZ progenitors organized in M-L stripes of RFP+ progenitors in the VZ. We gave Neurog1-CreER; R26 td-Tomato mice TMX at E12.5 and fixed the embryos 24 hours post-TMX at E13.5, 4 days post-TMX at E16.5 or 6 days post-TMX at E18.5 (Fig. 2e). We found that the RFP-marked cells were uniformly distributed along the M-L axis of the cerebellar anlage at E13.5 arguing against any regionalization of Neurog1-expressing progenitors. Three days later, RFP-labeled cells had begun to cluster in the medial dorsal cerebellum of E16.5 mice. When mice were analyzed at E18.5, the clustering of RFP-marked cells was more distinct with 3–4 clusters of cells traceable in the dorsal cerebellum (Fig. 2e). The dispersal pattern of RFP-marked cells following TMX demonstrates that E12.5 GIFM Purkinje cells are generated uniformly from Neurog1-expressing progenitors in the VZ but aggregate into clusters along the M-L axis during late embryonic development. We also compared the adult distribution of E12.5 GIFM Purkinje cells with known Purkinje cell subtype markers Zebrin II (Aldolase C), Hsp25, and PLCβ4 using dual IHC, but this did not reveal any noticeable correlation in RFP+ Purkinje cell distribution with the three markers (Supplementary Fig. 1).
In addition to Purkinje cells, we observed RFP marked cells in the molecular layer (ML) (see Fig. 2c), and less frequently, in the granule cell layer (GL) in Neurog1-CreER; R26 td-Tomato mice given TMX at E10.5–E12.5. Such early fate mapping of basket and stellate cells was particularly unexpected, and yet dual IHC with anti-parvalbumin (anti-Pvlb) antibodies, a marker of basket and stellate cells in the cerebellum, confirms the identity of these cells (Supplementary Fig. 2). Similarly, dual IHC with Golgi cell marker anti-neurogranin (anti-Nrg) antibodies confirm Golgi cell fates in E10.5–E12.5 GIFM mice (Supplementary Fig. 2). However, RFP-expressing cells were also present in the ML and GL of adult Neurog1-CreER; R26 td-Tomato mice injected with sunflower oil on E10.5–E12.5 (Supplementary Fig. 2). In contrast, no RFP-labeled cells with Purkinje cell morphology or anti-Calb1 immunoreactivity were detected in these controls (Supplementary Fig. 2). Combined these control data indicate that GIFM is specific to Purkinje cell lineages in Neurog1-CreER; R26 td-Tomato mice but RFP reporter expression occurs independent of TMX delivery in some basket, stellate, and Golgi cells. Lastly, although E10.5 to E12.5 TMX delivery spans the period of deep cerebellar nuclei (DCN) neurogenesis [3], we did not fate-map cells in the DCN. This is in contrast to GIFM for the gene family member Neurog2 using a knock-in Neurog2 CreER/+allele in which TMX on E11.25 to E12.25 fates GABAergic projection neurons in the DCN [10]. Ascl1 GIFM at E10.5–E12.5 also labels DCN interneurons [9] in contrast to our findings.
Neurog1-CreER; R26 td-Tomato reporter gene expression patterns in the cerebellum are largely consistent with sites of Neurog1 expression revealed by in situ hybridization (ISH). a, c, e Neurog1 ISH at E12.5, E13.5, and P0, respectively. Black arrows show absence of hybridization in the rhombic lip. Boxed area of rhombic lip and lateral ventricular zone in panel e is shown at higher magnification in the inset. b, d, f Neurog1-CreER; R26 td-Tomato mice given tamoxifen E11.5, E12.5, and E18.5, respectively, and fixed for analysis 24 h later. White arrows point to the rhombic lip. Inset in panel f shows the reporter gene expression in the rhombic lip with E18.5 GIFM at higher magnification. Key: DCN deep cerebellar nuclei, EGL external granule cell layer, RL rhombic lip, VZ ventricular zone
GIFM using the Neurog1-CreER transgene labels cerebellar Purkinje cells from E10.5 to E12.5 in mice. All mice were analyzed at P42. a Dual anti-Calb1/anti-EGFP IHC confirms Purkinje cell fate (arrow) in Neurog1-CreER; Z/EG mice. b The distribution of EGFP-marked Purkinje cells differs when TMX is given at E11.5 compared to E12.5. c Dual anti-Calb1/anti-RFP IHC identifies GIFM Purkinje cells (arrows) in Neurog1-CreER; R26 td-Tomato mice given TMX at E10.5, E11.5, or E12.5. Note the increased density of marked Purkinje cells using the R26 td-Tomato compared to the Z/EG reporter. d Patterns of GIFM Purkinje cells following TMX on E10.5, E11.5, or E12.5. Arrow heads denote the narrow midline stripe of RFP-negative Purkinje cells with TMX at E10.5. E12.5 GIFM Purkinje cells settle in distinct bilaterally symmetrical parasagittal stripes in the vermis and paravermis of adult Neurog1-CreER; R26 td-Tomato mice (arrows). e E12.5 GIFM Purkinje cells are labeled across the full M-L axis of the E13.5 cerebellum and only aggregate into clusters during later embryogenesis. White dotted lines outline large RFP+ cell clusters. Key: ML molecular layer, PCL Purkinje cell layer, pia pial surface, Med medial DCN, Int interposed DCN, Cop copula of the pyramis, PM paramedian lobule, PFl paraflocculus, Sim simple lobule (lobules annotated 1–10, etc. using [43]), Cb cerebellum, Cp choroid plexus, RL rhombic lip, Tm tectum, VZ ventricular zone
Overlapping Neurog1 and Ascl1 Expression in Purkinje Cell Progenitors
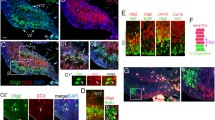
Our E10.5–E12.5 GIFM data is similar to age-matched Ascl1 CreER spatiotemporal Purkinje cell fate-map patterns produced by Sudarov and colleagues [9], suggesting co-expression of Neurog1 and Ascl1 in Purkinje cell progenitors in the VZ. Indeed an earlier ISH study has revealed overlapping expression patterns of multiple proneural bHLH transcription factors in the cerebellar VZ [7]. We examined this possibility further by performing a detailed series of ISHs for both Neurog1 and Ascl1 in E12.5 mice. Neurog1 and Ascl1 probes were hybridized singleplex on adjacent tissue sections for comparison or in duplex using fluorescent ISH. The data revealed overlapping patterns of Neurog1 and Ascl1 ISH in the E12.5 cerebellar VZ (Fig. 3). The medial and lateral boundaries of Neurog1 and Ascl1 ISH matched with both hybridization signals absent from both the rhombic lip and midline, and there was no observable difference in the rostrocaudal distribution of Neurog1 and Ascl1 hybridization signal (Fig. 3a-h, a′-h′). Dual fluorescent ISH confirms overlapping Neurog1 and Ascl1 expression in progenitor cells and suggests progenitor transition from Ascl1 to Neurog1 expression as they migrate dorsally from the VZ—the Neurog1 ISH territory overlaps but is shifted slightly dorsal to the Ascl1 ISH territory that predominates at the VZ (Fig. 3i-k, k′).
Overlapping Neurog1 and Ascl1 expression patterns in the ventricular zone of the cerebellar anlage at E12.5. a–h′ Adjacent coronal sections were analyzed by singleplex ISH. Neurog1 (a–d) is expressed in progenitor populations that overlap with regions of Ascl1 (e–h) expression. Higher magnification images (a′–h′) of the boxed regions in (a–h) show expression of Neurog1 and Ascl1 extending along the lateral (a′, e′, c′, g′) and medial (b′, f′) axis of the VZ but absent from the rhombic lip and midline. i–k′ Duplex-fluorescence ISH confirms overlapping expression of Neurog1 and Ascl1 in progenitors. Note that the Ascl1 expression territory is slightly ventral to but overlaps the Neurog1 expression territory. The boxed area in image panel k is shown at higher magnification in k′. Key: A Ascl1 ISH in a VZ progenitor, N Neurog1 ISH in a VZ progenitor, asterisk dual Ascl1/Neurog1 ISH in VZ progenitors
Neurog1-CreER GIFM Transitions from Purkinje to Golgi Cell Interneuron Fates Beginning ≈E11.75
Cell birth dating and GIFM studies report inhibitory interneuron production commences E13 in mice following the production of Purkinje cells [9, 20]. In agreement with these studies, we see a shift away from Purkinje cell to other cell fates when we give Neurog1-CreER; R26 td-Tomato mice TMX at E13.5. Only occasional, isolated RFP-labeled Purkinje cells were detected in adult mice with TMX delivery at E13.5, most of which were present at the midline and all located in the vermis and paravermis. Indeed E13.5 GIFM Purkinje cells constituted <0.5 % of all Purkinje cells fate-mapped with TMX given at E10.5 to E13.5 in Neurog1-CreER; R26 td-Tomato mice. In contrast, RFP labeling in the adult granule cell layer (GL) increased significantly with TMX at E13.5 (Fig. 4a). RFP-labeled cells in the GL contained soma larger than the surrounding granule cells and sparsely branched dendrites ascending to the ML, morphology consistent with a Golgi cell fate. This was confirmed with co-labeling with anti-Nrg (Fig. 4b, arrow). RFP-marked basket and stellate cells in the ML were also present in adult Neurog1-CreER; R26 td-Tomato mice given TMX at E12.5 (see Fig. 2c).
Neurog1-CreER; R26 td-Tomato GIFM transitions from Purkinje to Golgi cell fate with TMX injection at E13.5. All mice were analyzed at P42. a Anti-RFP IHC using ABC enhancement reveals extensive interneuron but negligible Purkinje cell fate mapping with TMX at E13.5. The left panel shows a low magnification view of the whole cerebellum with the area shown in higher magnification to the right outlined. b Dual anti-Nrg/anti-RFP IHC confirms Golgi cells (arrow) are labeled with TMX at E13.5. c Quantification of RFP marked cells with Purkinje cell morphology. Purkinje cell GIFM peaks with TMX at E11.5 in Neurog1-CreER; R26 td-Tomato and declines rapidly to less than 1 % of total labeled Purkinje cells with TMX at E13.5. d Quantification of RFP-marked cells with Golgi cell morphology. Golgi cells are first labeled with TMX at E11.5 and Golgi cell GIFM progressively increases from E11.5 to E13.5 as Purkinje cell fate declines. Histogram plots are mean plus standard error of the mean (SEM) error bar above (n ≥ 3 except E13.5 n = 2). Key: ML molecular layer, PCL Purkinje cell layer, GL Granule cell layer, WM white matter tracts
In view of the leaky RFP expression in Golgi, basket, and stellate cells, we compared the number of cells with Golgi, basket, and stellate cell morphology in Neurog1-CreER; R26 td-Tomato mice given TMX or sunflower oil to determine the contribution of leaky RFP expression to interneuron cell fate. Mice were given sunflower oil only on E10.5, E11.5, E12.5, or E13.5 and the numbers of RFP+ neurons quantified and compared to TMX-injected mice at P42. The data reveal TMX-independent RFP expression accounted for all of the Golgi cells present in Neurog1-CreER; R26 td-Tomato mice given TMX at E10.5 (7.5 ± 1.6 oil versus 7.2 ± 3.4 TMX, n = 3), 43.1 ± 8.8 % of Golgi cells present when TMX was administered at E11.5 (16.7 ± 2.2 oil versus 50.0 ± 17.7 TMX, n = 3), 9.3 ± 2.2 % of Golgi cells with TMX at E12.5 (14.7 ± 2.5 oil versus 161.8 ± 3.4 TMX, n = 3), and 6.0 ± 0.3 % of Golgi cells present with TMX at E13.5 (12 ± 4.0 oil versus 263 ± 66.2 TMX, n = 3 oil, n = 2 TMX). In contrast, basket and stellate cells were present in equal frequency in TMX or sunflower oil controls at each of the administration dates indicating TMX-independent expression accounted for all Neurog1-CreER GIFM basket and stellate cells with E10.5–E13.5 GIFM. We conclude that Golgi cells, but not basket or stellate cells, are fate-mapped during early cerebellar development in Neurog1-CreER; R26 td-Tomato mice and that Golgi cell fate commences with TMX at E11.5 and becomes the majority cell fate with TMX at E13.5.
We compared the number of Purkinje cells and Golgi cells in Neurog1-CreER; R26 td-Tomato mice given TMX at E10.5–E13.5 to measure the transition in Neurog1-CreER cell fate. Cell counts were corrected for the mean percentage leaky RFP expression in control mice injected with oil only at that age, for example, E13.5 GIFM Golgi cell numbers were reduced 6 % to generate a number representing TMX-induced RFP expression in this cell lineage. The numbers reveal that Purkinje cell GIFM increases from E10.5 to a peak with TMX at E11.5. RFP marked Purkinje cell numbers then decline by 35 % with E12.5 GIFM and Purkinje cell numbers decline further to <0.5 % total when TMX is administered at E13.5 (Fig. 4c). Thus, peak Neurog1 expression in Purkinje cell progenitors coincides with the mid-period of Purkinje cell neurogenesis at E11.75–E13.25 [3]. In contrast, Golgi cell GIFM numbers progressively increase with TMX administration from E11.5 to E13.5 (Fig. 4d). When expressed as a percentage of the two RFP cell fates in the cerebellar cortex, Golgi cells represent 2 % of the total when TMX is delivered at E11.5, 29 % with TMX at E12.5 and 99 % with TMX at E13.5. These cell counts demonstrate progenitors expressing Neurog1 transition from Purkinje to Golgi cell fates from ≈E11.75–E15.25 with the largest transition occurring from ≈E13.75–E15.25.
Temporal Neurog1-CreER GIFM Marks Exclusively Interneurons from E14.5 to P7
Short-term Neurog1 fate mapping using a Neurog1-EGFP BAC transgene labels multiple GABAergic interneuron cell types in the GL, Purkinje cell layer (PCL), and ML of the cerebellar cortex [11]. GABAergic interneurons are born in an inside-to-outside temporal pattern in the cerebellum with interneurons in the GL developing prior to those in the outer ML [20, 21]. Temporal Ascl1 CreER GIFM produces a comparable inside-to-outside pattern of GABAergic interneurons in the adult cerebellum [9]. We speculated Neurog1-CreER GIFM would similarly fate GABAergic interneurons in later embryogenesis and early postnatal development. We used Neurog1-CreER; Z/EG and Neurog1-CreER; R26 td-Tomato mice to map interneuron fate in adult mice. As was the case for Purkinje cell labeling, Neurog1-CreER; Z/EG labels interneurons with less frequency than Neurog1-CreER; R26 td-Tomato mice. However, unlike Neurog1-CreER; R26 td-Tomato mice, reporter-labeled interneurons were not detected in significant numbers in Neurog1-CreER; Z/EG mice not given TMX (≤3 EGFP cells per cerebellar section were detected in the PCL or ML). We administered TMX to Neurog1-CreER; Z/EG mice at E14.5, E16.5, E18.5, P0, or P7 and determined GIFM cell types in the cerebellum of a single adult mouse for each GIFM. Cell fate was determined by topography, key morphological characteristics and, when available, cell-type specific antibodies (see Methods).
When TMX was given at E14.5 to Neurog1-CreER; Z/EG mice, only Golgi cell fates were identified in the cerebellar cortex, confirming the transition to Golgi cell fate was complete at this stage (Fig. 5a). Golgi cells still constituted the most common labeled cell type when TMX was administered at E16.5 but EGFP-marked cells of either candelabrum or Lugaro morphology were also present in the Purkinje cell layer (PCL) (Fig. 5b). Candelabrum and Lugaro interneurons were the majority fate-mapped cell type with TMX at E18.5 (not shown). TMX at P0 primarily labeled cells in the inner ML with basket cell characteristics (Fig. 5c). In contrast, cells in the outer ML with stellate cell characteristics were the majority cell type labeled with TMX at P7 (Fig. 5d). Interestingly, GIFM from E11.5 to P7 did not label cells in the white matter tracts of the adult cerebellum and no reporter gene expressing cells exhibited astrocyte-like morphology in either Neurog1-CreER; Z/EG or Neurog1-CreER; R26 td-Tomato mice. IHC with antibodies to the oligodendrocyte marker Olig2 as well as the glial cell markers glial fibrilliary acidic protein (GFAP) and anti-S100β confirmed no oligodendrocytes or glia were fate-mapped in Neurog1-CreER; R26 td-Tomato mice with TMX delivery at E13.5 or E18.5 (not shown). Combined these data demonstrate Neurog1-CreER mice fate map interneurons but not glial cell types of the cerebellar cortex and suggest that the timing of Neurog1 expression in GABAergic interneurons correlates with their final resting position in the cerebellar cortex.
E14.5 to P7 Neurog1-CreER GIFM labels interneurons in an inside-to-outside settling pattern in the adult cerebellar cortex. All mice were analyzed at P42. a Dual IHC reveals EGFP labeled cells with TMX at E14.5 express the Golgi cell subset marker Grm2 (arrow). White hashed line indicates the Purkinje cell layer. b TMX at E16.5 labels EGFP+ cells with Lugaro cell (arrow) and candelabrum cell (crossed arrow) morphology. Note the characteristic beaded axonal arborization (asterisks) of the candelabrum cell. Both cells are not immunolabeled with antibodies to Pvlb, a marker for basket and stellate cell interneurons. c–d Dual Pvlb+/EGFP+ basket and stellate cells (arrows) with TMX at P0 or P7. Key: GL granule cell layer, ML molecular layer, PCL Purkinje cell layer
Co-expression of Neurog1 and Ascl1 in Interneuron Progenitors
The spatiotemporal sequence of Neurog1-CreER GIFM in interneurons shows similarities to the detailed spatiotemporal Ascl1 CreER fate map produced by Sudharov and colleagues and supports the finding by these authors [9] that PCL Lugaro and candelabrum interneurons follow the same spatiotemporal maturation pattern as other GABAergic interneurons. These similarities suggest co-expression of Neurog1 and Ascl1 in GABAergic interneuron progenitors. We examined this possibility by ISH in E13.5 and P0 mice. Neurog1 and Ascl1 probes were hybridized singleplex on adjacent tissue sections for comparison or in duplex using fluorescent ISH. Singleplex ISH revealed overlapping patterns of Neurog1 and Ascl1 ISH in the E13.5 cerebellar VZ (Fig. 6a-b, a′-b′) and the presumptive white matter tracts of the cerebellum of P0 mice (Fig. 6c-d, c′-d′). Co-expression of Neurog1 and Ascl1 in progenitor cells was confirmed by dual fluorescent ISH labeling of E13.5 (Fig. 6e-g, g′, g′′) and P0 (Fig. 6h-j, j′) cerebellum. Interestingly, P0 ISH patterns indicate more widespread Ascl1 expression in interneuron progenitors—Neurog1 ISH is primarily localized to the presumptive white matter tracts, whereas Ascl1 ISH extends throughout the nascent cortex to the external germinal layer, consistent with more prolonged Ascl1 expression in developing GABAergic interneuron cell lineages.
Similarities in the spatiotemporal pattern of Neurog1 with Ascl11 (see [9]) are supported by ISH data showing overlapping expression of the two bHLH transcription factor genes. a–d′ Adjacent coronal sections were analyzed by singleplex ISH. a, c Overlapping Neurog1 and b, d Ascl1 expression in the VZ and developing cerebellar cortex of E13.5 and P0 mice. a–d Higher magnification images (a′, b′, c′, d′) of the boxed regions in (a–d). e–j′ Duplex-fluorescence ISH confirms co-expression of Neurog1 and Ascl1 in E13.5 and P0 progenitors. g′ and g′′ Two boxed areas in image panel (g) at higher magnification to illustrate the transition from Ascl1 to dual Ascl1/Neurog1 to Neurog1 expression as VZ progenitors exit the VZ. X denote non-specific binding of the fluorescent red detection probe. h–j′ The boxed area in image panel (j) is shown at higher magnification in (j′). Key: A Ascl1 ISH in a VZ progenitor, EGL external granule cell layer, pWM presumptive white matter tracts, N Neurog1 ISH in a VZ progenitor, asterisk dual Ascl1/Neurog1 ISH in VZ progenitors
Neurog1 is expressed in Purkinje Cells Around the Time They Become Post-mitotic but in Mitotic and Post-Mitotic Interneurons
GIFM confirms Neurog1 expressing progenitors initially give rise to Purkinje cells and then to an overlapping sequence of multiple interneuron cell types in the cerebellum. We investigated the timing of Neurog1-CreER expression in progenitor cells, choosing the period of transition from Purkinje to Golgi cell fate (TMX E11.5–E13.5) and a later developmental period at which multiple interneuron cell types are labeled in the cerebellar cortex (TMX at E18.5). TMX was administered to pregnant dams and Neurog1-CreER; R26 td-Tomato embryos were harvested 24 h later for IHC with anti-Ki67 antibodies to determine the mitotic status of labeled cells (Ki67 is expressed during all phases of the cell cycle except the resting phase). A second set of pregnant dams were injected with sunflower oil only and Neurog1-CreER; R26 td-Tomato embryos harvested 24 h later to control for leaky RFP expression. Control mice harvested at E13.5 and E14.5 contained RFP reporter expression in peripheral ganglia, spinal cord, brain stem, and midbrain but not the cerebellum. These same CNS sites exhibited RFP expression in controls analyzed at birth (P0) plus a few (3–5 per coronal cerebellar section) scattered RFP+ cells in the intermediate zone of the cerebellum. These controls reveal leaky RFP expression in the cerebellum commences during late embryogenesis and suggest the great majority of RFP+ interneurons present in oil only adult controls derive from postnatal reporter gene expression. Accordingly, we concluded that leaky expression of the RFP reporter would not undermine our analysis.
Dual anti-Ki67/anti-RFP IHC revealed minimal Ki67 expression in RFP-marked cells in embryos given TMX at E11.5 and E12.5 with a thin zone of non-immunoreactivity separating Ki67+ cells at the VZ from RFP+ cells in the cerebellar primordium (Fig. 7a). Ki67+ and RFP+ cells abut when TMX is delivered a day later at E13.5 (Fig. 7a). In contrast, co-mingling of the two immunosignals is widespread when TMX is given at E18.5 in which dual Ki67+/RFP+ marked cells are found adjacent to the VZ and in cells streaming toward the cerebellar cortex along the presumptive white matter tracts (pWM, Fig. 7a). Quantification of the number of RFP marked cells expressing Ki67 confirms a significant increase in the percentage of mitotic cells with GIFM at E18.5 (p < 0.001) compared to earlier GIFM dates—the percentage of RFP-marked cells expressing Ki67 remains low at 2.9 to 6.8 % when TMX was administered during E11.5 to E13.5 but increases substantially to 51 % when TMX is administered at E18.5 (Fig. 7b). Dual IHC with anti-Pax2 antibodies, a marker for GABAergic interneuron progenitors in the cerebellum [22], confirms E18.5 labeled RFP+ cells express Pax2. Taken together, these results demonstrate a significant increase in Neurog1-CreER expression in mitotic GABAergic interneuron progenitor cells during late embryonic development.
Neurog1 is expressed in Purkinje cells around the time they become post-mitotic and in mitotic and post-mitotic interneurons. a, b Neurog1-CreER; R26 td-Tomato mice processed for dual anti-RFP/anti-Ki67 IHC 24 h after TMX delivery. a Dual RFP+/Ki67+ cells are indicated by arrows. b Quantification confirms a significant increase in the percentage of RFP+ cells expressing Ki67 with GIFM at E18.5 (***p < 0.001, one-way ANOVA with Tukey’s multiple comparisons test, n ≥ 3). c–e Neurog1-CreER; R26 td-Tomato mice given BrdU 24 h before TMX delivery and processed for dual anti-RFP/anti-BrdU IHC at P42. c Dual RFP+/BrdU+ cells are indicated by arrows. d A quantitative comparison of the percentage of RFP+ cells immunoreactive for BrdU reveals higher BrdU retention in RFP marked Purkinje cells (n = 3 except E13.5 n = 2). e Patterns of BrdU and RFP immunolabeled Purkinje cells in adult mice do not reveal a relationship between the two markers when BrdU is injected 24 h before a TMX delivery. Arrows indicate regions of BrdU+/RFP− Purkinje cells (slender arrows) adjacent to BrdU+/RFP+ Purkinje cells (thick arrows) within the same lobule
We hypothesized that the low percentage of dual RFP+/Ki67+ cells 24 h post E11.5–E13.5 TMX were the result of Neurog1-CreER expression in progenitors around the time they exit cell division. To test this hypothesis, we injected pregnant dams with BrdU the day before TMX and quantified the percentage of RFP-marked cells immunolabeled with anti-BrdU antisera at P42. In this analysis, long-term BrdU retention resulting in anti-BrdU immunoreactivity at P42 would identify cells undergoing terminal cell division the day before genetic marking. We found BrdU was retained in 23 ± 0.5 % of Purkinje cells marked with TMX at E11.5 and BrdU retention increased to 45 ± 10.5 % of Purkinje cells when TMX was administered at E12.5 (Fig. 7c arrows, d). Purkinje cell fating declines dramatically with TMX administration at E13.5 (<0.5 % of GIFM total) and 9.3 ± 1.2 % of these few late marked cells were BrdU+ (Fig. 7d). Thus, high percentages of BrdU retention were observed in Purkinje cells during the period of greatest genetic marking (TMX at E11.5 or E12.5) consistent with many Purkinje cells undergoing terminal cell division in the day before GIFM. BrdU retention in interneurons fate-mapped to the cerebellar cortex was at a lower rate than in Purkinje cells but more constant with 10.6 ± 1.2 % to 18 ± 7.3 % of fate-mapped Golgi cells BrdU+ for each of the genetic marking days analyzed (Fig. 7c, d). Combined Ki67 and BrdU cell counts support the view Neurog1 is expressed in Purkinje cells around the time they become post-mitotic, whereas Neurog1 is expressed in mitotic and non-mitotic interneuron progenitors.
We next mapped BrdU retention and RFP marking in Purkinje cells to determine the spatial relationship between Purkinje cell birth and Neurog1 expression. BrdU is retained in Purkinje cells located in the vermis and hemispheres of the adult cerebellum of mice injected at E10.5 or E11.5 using our injection protocol. A comparison of the adult patterns of BrdU and RFP immunolabeled Purkinje cells reveals BrdU+ and RFP+ Purkinje cell distributions do not match when BrdU is injected the day before TMX delivery on either E11.5 (not shown) or E12.5 (Fig. 7e). Dual RFP+/BrdU+ Purkinje cells (Fig. 7e, thick arrows) are located adjacent to RFP−/BrdU+ Purkinje cells (Fig. 7e, slender arrows) within the same lobules. These data reveal that not all Purkinje cells born around the same time express Neurog1 the following day.
GABAergic Interneuron Progenitors in the Presumptive White Matter Tracts Enter the Cell Cycle Around the Time They Express Neurog1
Our analysis of Ki67 expression in GIFM cells suggests large numbers of E18.5 fate-mapped GABAergic interneuron progenitors proliferate around the time they express Neurog1. To test this hypothesis, mice were pulsed with BrdU the day following TMX administration and fixed 24 h later for dual anti-RFP/anti-BrdU IHC. We chose two GIFM timepoints for BrdU delivery 24 h post-TMX, E18.5, and E11.5 GIFM that fate interneurons and Purkinje cells, respectively. As expected from the Ki67 data, cell counts reveal low numbers of 7.3 ± 1.5 % (n = 2) of RFP+ cells at E13.5 immunopositive for BrdU following TMX delivery on E11.5 and BrdU on E12.5 (Fig. 8a). In contrast, 41.5 ± 5.6 % (n = 3) of RFP+ cells in P1 neonates were immunolabeled with anti-BrdU antisera following TMX on E18.5 and BrdU on P0 (Fig. 8b). Triple IHC with Pax2 antisera confirms the majority of the RFP+/BrdU+ cells in the cerebellum of P1 neonates were GABAergic interneuron progenitors migrating to final destinations in the cerebellar cortex (Fig. 8c). These data support the view that Neurog1 is expressed in cycling interneuron progenitor cells during later phases of neurogenesis in the pWM of the cerebellum. In contrast, long-term BrdU retention in adult Purkinje cells and low rates of either anti-BrdU or anti-Ki67 immunolabeling in RFP+ Purkinje cell progenitors are all consistent with the idea that Neurog1 is expressed in VZ Purkinje cell progenitors around the time they become post-mitotic.
Mitotic GABAergic interneuron progenitors express Neurog1. Neurog1-CreER; R26 td-Tomato mice were administered TMX at E11.5 or E18.5 and BrdU the following day. Mice were fixed the day after BrdU injection. a BrdU labels multiple progenitors in the E13.5 ventricular zone (VZ) but few RFP-marked cells are immunoreactive for BrdU (arrows). b Multiple RFP+/BrdU+ interneuron progenitors (arrows) in the presumptive white matter tracts (pWM) of a P1 Neurog1-CreER; R26 td-Tomato mouse. c Co-immunolabeling with Pax2 antibodies confirms the majority of the RFP+/BrdU+ cells in the cerebellum of P1 neonates are GABAergic interneuron progenitors. Panels b and c are taken from the same section processed for triple anti-RFP/anti-BrdU/anti-Pax2 IHC
Neurog1-CreER GIFM Labels DCN Interneurons During Late Embryogenesis
Neurog1-CreER; Z/EG GIFM resulted in low-level fate mapping of cells with neuronal morphology in the DCN with TMX injected at E14.5 or older with the majority of cells fate marked with TMX E14.5 to E18.5. This finding was unexpected as DCN fates were not observed in short-term and long-term fate mapping studies using Neurog1-EGFP and Neurog1-CreER; R26 Lacz/+ mice, respectively [11, 12]. DCN fates with Neurog1-CreER GIFM were confirmed in Neurog1-CreER; R26 td-Tomato mice given TMX at E18.5 in which RFP-marked cells were present in the medial, intermediate, and lateral DCN (Fig. 9a). As was the case for other cerebellar cell fates, the frequency of reporter gene marking in the DCN was greater with the R26 td-Tomato allele compared to the Z/EG transgene with TMX delivery at E18.5. DCN labeling was minimal in control Neurog1-CreER; R26 td-Tomato mice injected with sunflower oil only and accounted for 5.7 ± 4.4 % of RFP-marked cells in the medial DCN, 3.1 ± 3.1 % of RFP-marked cells in the intermediate, and 2.2 ± 1.1 % in the lateral DCN (n = 3 for TMX and oil administered mice). The temporal sequence of RFP marking in the DCN (E14.5 GIFM onwards) is consistent with an interneuron cell fate. To identify DCN fates, we conducted dual IHC with antibodies to the DCN neuron marker NeuN, the projection neuron marker SMI32, and the interneuron subtype marker Calb2 [20]. Dual IHC revealed RFP-labeled cells express NeuN, a subset express Calb2 but none were immunolabeled with anti-SMI32 antibodies (Fig. 9b). When combined with the relatively small size of RFP+ cells compared to SMI32+ cells in the DCN, these data indicate that Neurog1 is expressed during late embryogenesis in VZ progenitors destined to become DCN interneurons. Interestingly, BrdU labeling 24 h prior to TMX injection at E18.5 resulted in co-labeling of many BrdU+/RFP+ cells in the medial, intermediate, and lateral DCN (Fig. 9c), consistent with cell birth around E17.5 and Neurog1 expression at E18.75-P0 in these DCN interneurons.
Late embryogenesis Neurog1-CreER GIFM labels interneurons in the deep cerebellar nuclei (DCN). Mice were analyzed at P42. a Low magnification view of RFP fate-mapped cells in the DCN of adult Neurog1-CreER; R26 td-Tomato mice with TMX administration at E18.5. b RFP+ DCN cells express NeuN (arrows) and a subset Calb2 (arrows) but do not express SMI32. c Co-immunolabeling of anti-BrdU/anti-RFP-marked cells (arrows) in all three DCN following BrdU pulse labeling at E17.5 and GIFM at E18.5
Discussion
Neurog1-CreER GIFM Labels Exclusively Neurons in the Cerebellum
A principal finding of this study is that Neurog1-CreER GIFM labels all GABAergic neuronal cell types of the cerebellar cortex. Temporal Neurog1-CreER GIFM reveals that GABAergic neurons are fate-mapped over a prolonged time-period spanning embryonic and postnatal stages of development. Neurog1-CreER labeling shifts from initially Purkinje cell to interneuron fate early in cerebellar development and GABAergic interneurons destined for the cerebellar cortex are fate-mapped in a temporal sequence that correlates with their final inside-to-outside resting position. Similar spatiotemporal features of GABAergic cell fate were observed with GIFM using knock-in Ascl1 CreER mice [9]. However, Ascl1 CreER GIFM additionally marked Bergmann glia, astrocyte, or oligodendrocyte cell lineages with TMX delivery during embryonic development. Similarly, Bergmann glia are labeled with GIFM using knock-in Neurog2 CreER mice [10] and cumulative fating using a Ptf1a Cre knock-in allele fates GFAP-positive astrocytes in the cerebellum [5]. Combined these studies provide compelling evidence radial glia cell types derive from VZ progenitors (for a review on VZ origins for glia see [23]). The absence of cerebellar glial cell fates in Neurog1-CreER mice therefore signifies Neurog1 is distinct among the many pro-neural bHLH genes expressed at the VZ [7] in that progenitors expressing this gene exclusively give rise to neurons. As such Neurog1-CreER mice provide a useful tool to study neuronal cell types originating from the cerebellar VZ.
Lineage tracing reveals that GABAergic interneurons and glia are generated from progenitors residing in the VZ that translocate to the presumptive white matter (pWM) during later embryonic development to generate the majority of interneurons [5, 8, 9, 24, 25]. Clonal analysis, GIFM, null, and conditional mutants all support an ontogenetic relationship between astrocytes and neurons in the pWM [5, 8, 9, 26–28], and there is good evidence that bHLH programs govern the lineage choice between interneuron and glial fate in pWM progenitors [23]. For example, conditional Ascl1 deletion in mice depletes the number of GABAergic interneurons in the cortex but increases the number of astrocytes [9]. Our data implicate Neurog1 as part of the transcription factor code specifying interneurons from pWM progenitors. Interestingly, cell grafting studies have shown that the laminar placement and cell fate of GABAergic interneurons are related to intrinsic cell cycle dynamics and cell birth date but fate is only determined when cells become post-mitotic via local environmental cues within the pWM [21, 29]. Previous expression studies in our lab reveal that Neurog1 is expressed in mitotic and non-mitotic progenitors as they migrate along the pWM. Neurog1 expression transiently increases as cells exit the pWM and enter the granule cell layer before being rapidly downregulated as cells disperse to their final location [11]. It is therefore tempting to speculate cues within the local pWM environment signal through the regulation of Neurog1 expression levels in progenitors to instruct interneuron cell fate.
E12.5 GIFM-Labeled Purkinje Cells Settle in Distinct Parasagittal Stripes in the Vermis and Paravermis
E10.5, E11.5, or E12.5 GIFM each robustly labeled Purkinje cells in the adult cerebellum of Neurog1-CreER; R26 td-Tomato mice with the highest number of Purkinje cells fate-mapped with TMX at E11.5. Purkinje cells were similarly labeled with E10.5 to E12.5 GIFM in Ascl1 CreER mice with maximal Purkinje cell fating with TMX delivery at E11.5 [9]. A comparison of Ascl1 CreER and Neurog1-CreER Purkinje cell fate patterns indicates these two spatiotemporal maps converge with increasing age. E10.5 GIFM patterns differ substantially with Purkinje cells settled across the full M-L axis in Neurog1-CreER; R26 td-Tomato mice but restricted to the lateral cerebellum in Ascl1 CreER ; R26 Lacz/+ mice. In contrast, GIFM patterns are remarkably alike with TMX at E12.5 producing similar parasagittal stripes in the adult vermis and paravermis of Neurog1-CreER; R26 td-Tomato and Ascl1 CreER ; R26 Lacz/+ mice. Similarities in the E12.5 fate maps suggest Neurog1- and Ascl1-expressing Purkinje cell lineages converge to a distinct cohort destined for the vermis and paravermis. This observation is supported by ISH that reveal overlapping Neurog1 and Ascl1 mRNA territories in the E12.5 cerebellum (patterns that suggest progenitor transition from Ascl1 to Neurog1 expression). E12.5 GIFM patterns are also reminiscent of the parasagittal distribution of marked Purkinje cells in the vermis and paravermis following the labeling of E12.5 VZ progenitors with replication-defective adenoviral vectors [30]. Thus, CreER and viral vector-labeling studies provide 3 independent lines of evidence that late-generated Purkinje cells are programmed for distinct parasagittal fates in the vermis and paravermis in the adult cerebellum.
Our GIFM data resulted in considerable overlap in Neurog1-CreER; R26 td-Tomato Purkinje cell fate maps with TMX at E10.5, E11.5, or E12.5. This suggests that individual Purkinje cells were labeled with TMX delivery on more than one date, consistent with sustained Neurog1 expression during Purkinje cell specification. In particular, the relative distribution of RFP+ Purkinje cells with E11.5 and E12.5 GIFM indicates that E12.5 marked cells are derived from a subset of Purkinje cells in which earlier Neurog1 expression is maintained past E13. The molecular consequences of extended Neurog1 expression in this Purkinje cell cohort are unknown, but it seems plausible that these cells require a distinct molecular identity linked to function.
The timing of Neurog1 Expression in Cerebellar Progenitors Differs Between Early and Later Phases of GABAergic Neurogenesis
Our data suggest that the timing of Neurog1 expression in cerebellar progenitors differs between early and later phases of GABAergic neurogenesis. E11.5 to E13.5 GIFM primarily marked non-mitotic cells in the VZ and long-term BrdU retention indicates that Neurog1 is expressed in Purkinje cells around the time they become post-mitotic. In contrast, E18.5 GIFM fates interneuron progenitors in roughly equal ratios of mitotic and non-mitotic states in the presumptive white matter tracts (pWM). Differences in the timing of expression relative to mitotic status implies distinct Neurog1 functions in VZ and pWM GABAergic progenitor cells, with probable neuronal differentiation functions in VZ Purkinje cell progenitors and cell cycle regulatory functions in pWM progenitors. One possibility is that the progressive lengthening of the progenitor cell cycle that occurs during development contributes to the increase in Neurog1 expression in mitotic progenitors. Cell cycle lengthening regulates the transition from progenitor maintenance to differentiation in the CNS [31] and cell cycle regulators have been shown to modulate Neurog2 activity [32]. Thus, changes in cell cycle protein dynamics could trigger changes in Neurog1 expression timing in later generated progenitors.
Differing Purkinje and interneuron cell functions for Neurog1 are consistent with the multiple roles ascribed to Neurog1/2 activity in the literature. Neurogenins control neuronal commitment [33], progenitor cell cycle exit [34–36], neuroblast migration [37, 38], and neuronal differentiation [39–41]. Furthermore, neuronal subtype specific functions for neurogenins have recently been described in the developing piriform cortex [39]. Determining the precise differences in Neurog1 function in VZ and pWM progenitor cells will require the development of a conditional Neurog1 mutant model to avoid the problem of neonatal lethality associated with the conventional knockout [42].
Conclusion
In this GIFM study, we have shown Neurog1 is expressed in progenitor cells fate-mapped to become Purkinje cells and GABAergic interneuron cell types of the cerebellum. We find that the timing of Neurog1 expression is unique to each neuronal cell type and determines the final resting positions of neurons in the adult cerebellar cortex. Differences in the timing of Neurog1 expression relative to cell cycle progression suggest distinct Neurog1 functions in GABAergic progenitor cells in the VZ and pWM.
Abbreviations
- M-L:
-
Medio-lateral
- A-P:
-
Anterior-posterior
- VZ:
-
Ventricular zone
- pWM:
-
Presumptive white matter tracts
- TMX:
-
Tamoxifen
References
Palay SL, Chan-Palay V. Cerebellar cortex—cytology and organization. New York, Heidelberg, Berlin: Springer; 1974.
Altman J, Bayer SA. Development of the cerebellar system. Boca Raton, FL: CRC Press; 1997.
Carletti B, Rossi F. Neurogenesis in the cerebellum. Neuroscientist. 2008;14:91–100.
Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24.
Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, et al. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–13.
Pascual M, Abasolo I, Mingorance-Le Meur A, Martinez A, Del Rio JA, Wright CV, et al. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc Natl Acad Sci U S A. 2007;104:5193–8.
Zordan P, Croci L, Hawkes R, Consalez GG. Comparative analysis of proneural gene expression in the embryonic cerebellum. Dev Dyn. 2008;237:1726–35.
Kim EJ, Battiste J, Nakagawa Y, Johnson JE. Ascl1 (Mash1) lineage cells contribute to discrete cell populations in CNS architecture. Mol Cell Neurosci. 2008;38:595–606.
Sudarov A, Turnbull RK, Kim EJ, Lebel-Potter M, Guillemot F, Joyner AL. Ascl1 genetics reveals insights into cerebellum local circuit assembly. J Neurosci. 2011;31:11055–69.
Florio M, Leto K, Muzio L, Tinterri A, Badaloni A, Croci L, et al. Neurogenin 2 regulates progenitor cell-cycle progression and Purkinje cell dendritogenesis in cerebellar development. Development. 2012;139:2308–20.
Lundell TG, Zhou Q, Doughty ML. Neurogenin1 expression in cell lineages of the cerebellar cortex in embryonic and postnatal mice. Dev Dyn. 2009;238:3310–25.
Kim EJ, Hori K, Wyckoff A, Dickel LK, Koundakjian EJ, Goodrich LV, et al. Spatiotemporal fate map of neurogenin1 (Neurog1) lineages in the mouse central nervous system. J Comp Neurol. 2011;519:1355–70.
Koundakjian EJ, Appler JL, Goodrich LV. Auditory neurons make stereotyped wiring decisions before maturation of their targets. J Neurosci. 2007;27:14078–88.
Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40.
Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–55.
Radomski KL, Zhou Q, Yi KJ, Doughty ML. Cortical contusion injury disrupts olfactory bulb neurogenesis in adult mice. BMC Neurosci. 2013;14:142.
Laine J, Axelrad H. The candelabrum cell: a new interneuron in the cerebellar cortex. J Comp Neurol. 1994;339:159–73.
Laine J, Axelrad H. Morphology of the Golgi-impregnated Lugaro cell in the rat cerebellar cortex: a reappraisal with a description of its axon. J Comp Neurol. 1996;375:618–40.
Sgaier SK, Millet S, Villanueva MP, Berenshteyn F, Song C, Joyner AL. Morphogenetic and cellular movements that shape the mouse cerebellum; insights from genetic fate mapping. Neuron. 2005;45:27–40.
Leto K, Carletti B, Williams IM, Magrassi L, Rossi F. Different types of cerebellar GABAergic interneurons originate from a common pool of multipotent progenitor cells. J Neurosci. 2006;26:11682–94.
Leto K, Bartolini A, Yanagawa Y, Obata K, Magrassi L, Schilling K, et al. Laminar fate and phenotype specification of cerebellar GABAergic interneurons. J Neurosci. 2009;29:7079–91.
Maricich SM, Herrup K. Pax-2 expression defines a subset of GABAergic interneurons and their precursors in the developing murine cerebellum. J Neurobiol. 1999;41:281–94.
Buffo A, Rossi F. Origin, lineage and function of cerebellar glia. Prog Neurobiol. 2013;109:42–63.
Zhang L, Goldman JE. Developmental fates and migratory pathways of dividing progenitors in the postnatal rat cerebellum. J Comp Neurol. 1996;370:536–50.
Zhang L, Goldman JE. Generation of cerebellar interneurons from dividing progenitors in white matter. Neuron. 1996;16:47–54.
Grimaldi P, Parras C, Guillemot F, Rossi F, Wassef M. Origins and control of the differentiation of inhibitory interneurons and glia in the cerebellum. Dev Biol. 2009;328:422–33.
Mathis L, Bonnerot C, Puelles L, Nicolas JF. Retrospective clonal analysis of the cerebellum using genetic laacZ/lacZ mouse mosaics. Development. 1997;124:4089–104.
Mathis L, Nicolas JF. Progressive restriction of cell fates in relation to neuroepithelial cell mingling in the mouse cerebellum. Dev Biol. 2003;258:20–31.
Leto K, Bartolini A, Di Gregorio A, Imperiale D, De Luca A, Parmigiani E, et al. Modulation of cell-cycle dynamics is required to regulate the number of cerebellar GABAergic interneurons and their rhythm of maturation. Development. 2011;138:3463–72.
Hashimoto M, Mikoshiba K. Mediolateral compartmentalization of the cerebellum is determined on the "birth date" of Purkinje cells. J Neurosci. 2003;23:11342–51.
Miyata T, Kawaguchi D, Kawaguchi A, Gotoh Y. Mechanisms that regulate the number of neurons during mouse neocortical development. Curr Opin Neurobiol. 2010;20:22–8.
Ali F, Hindley C, McDowell G, Deibler R, Jones A, Kirschner M, et al. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development. 2011;138:4267–77.
Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, et al. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–76.
Britz O, Mattar P, Nguyen L, Langevin LM, Zimmer C, Alam S, et al. A role for proneural genes in the maturation of cortical progenitor cells. Cereb Cortex. 2006;16 Suppl 1:i138–51.
Lacomme M, Liaubet L, Pituello F, Bel-Vialar S. NEUROG2 drives cell cycle exit of neuronal precursors by specifically repressing a subset of cyclins acting at the G1 and S phases of the cell cycle. Mol Cell Biol. 2012;32:2596–607.
Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, Morris K, et al. Smaller inner ear sensory epithelia in Neurog1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–50.
Heng JI, Nguyen L, Castro DS, Zimmer C, Wildner H, Armant O, Skowronska-Krawczyk D, Bedogni F, Matter JM, Hevner R and Guillemot F. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 2008.
Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62.
Dixit R, Wilkinson G, Cancino GI, Shaker T, Adnani L, Li S, et al. Neurog1 and Neurog2 control two waves of neuronal differentiation in the piriform cortex. J Neurosci. 2014;34:539–53.
Ma Q, Fode C, Guillemot F, Anderson DJ. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999;13:1717–28.
Shaker T, Dennis D, Kurrasch DM, Schuurmans C. Neurog1 and Neurog2 coordinately regulate development of the olfactory system. Neural Dev. 2012;7:28.
Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL and Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 1998: 20:469–82.
Franklin KBJ and Paxinos G. The mouse brain in stereotaxic coordinates, third edition. Academic Press; 2008.
Acknowledgments
We are grateful to the staff of USU’s Biomedical Instrumentation Center for technical assistance.
Funding
This work is supported by the National Science Foundation (NSF) Award #1121839.
Coflicts of Interest
The authors disclose that no competing financial interests or personal relationships exist that might bias this work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Patterns of GIFM Purkinje cells following TMX on E12.5 do not coincide with expression patterns of Zebrin II or Hsp25. Coronal sections through the rostro-caudal axis of the cerebellum were immunostained using ABC enhancement. All mice were analyzed at P42. Note there are some detachments from whole cerebellar sections during the IHC process such as the paraflocculus (e’, f), simple lobule and Crus 1 (e’, l, l’), or middle cerebellar peduncle region (e). Missing fragments do not interfere with our expression pattern analysis, which is focused on the vermis. E12.5 GIFM Purkinje cells settle in three parasagittal stripes along the medio-lateral axis of the adult vermis and paravermis (a-l). Consecutive slices were analyzed for Zebrin II and Hsp25 expression patterns. Multiple Zebrin II negative stripes (arrow heads) appear in lobules VIII and IX (a’-b’) and three distinct Zebrin II positive stripes (arrows) appear in lobules I-V (d’-f’). Three Hsp25 positive stripes (arrows) appear in lobules VI, VII, IX and X (g’-j’) and Hsp25 is not expressed in rostral sections (k’-l’). Dual IHC reveals RFP labeled Purkinje cells expressing Zebrin II (m) or Hsp25 (n) indicated with arrows. (o) Although PLCβ4 expression patterns do not compliment GIFM Purkinje cells, dual anti-RFP/anti-PLCβ4 Purkinje cells are present in lobule VIII. (GIF 381 kb)
Supplementary Figure 2
GIFM is specific to Purkinje cell lineages in Neurog1-CreER;R26td-Tomato/+ mice but RFP reporter expression occurs independent of TMX delivery in some basket, stellate and Golgi cells. Neurog1-CreER;R26td-Tomato/+ mice administered with TMX or sunflower oil only on E12.5 were analyzed at P42. Dual IHC for RFP with the basket and stellate cell marker Pvlb or the Golgi cell marker Nrg co-labels RFP interneurons in mice injected with TMX or sunflower oil (arrows). Leaky RFP labeling in sunflower oil controls is more numerous in basket and stellate cell interneurons compared to Golgi cells. In contrast, RFP is not detected in Calb1+ Purkinje cells in the absence of TMX administration. (GIF 110 kb)
Rights and permissions
About this article
Cite this article
Obana, E.A., Lundell, T.G., Yi, K.J. et al. Neurog1 Genetic Inducible Fate Mapping (GIFM) Reveals the Existence of Complex Spatiotemporal Cyto-Architectures in the Developing Cerebellum. Cerebellum 14, 247–263 (2015). https://doi.org/10.1007/s12311-014-0641-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12311-014-0641-9