Abstract
Senescence is the ultimate phase in the life cycle of leaves which is crucial for recycling of nutrients to maintain plant fitness and reproductive success. The earliest visible manifestation of leaf senescence is their yellowing, which usually commences with the breakdown of chlorophyll. The degradation process involves a gradual and highly coordinated disassembly of macromolecules resulting in the accumulation of nutrients, which are subsequently mobilized from the senescing leaves to the developing organs. Leaf senescence progresses under overly tight genetic and molecular control involving a well-orchestrated and intricate network of regulators that coordinate spatio-temporally with the influence of both internal and external cues. Owing to the advancements in omics technologies, the availability of mutant resources, scalability of molecular analyses methodologies and the advanced capacity to integrate multidimensional data, our understanding of the genetic and molecular basis of leaf ageing has greatly expanded. The review provides a compilation of the multitier regulation of senescence process and the interrelation between the environment and the terminal phase of leaf development. The knowledge gained would benefit in devising the strategies for manipulation of leaf senescence process to improve crop quality and productivity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
An organism completes its life cycle by following a specific pattern of development; it grows, matures, and ultimately dies. The plants and their organs die likewise, and the decline of plant physiological events is called senescence which represents the ultimate stage of plant growth and development (Lim et al. 2007b). This physiological decline may be initiated and impactful at various specific cells, tissues, organs, or whole organism level. Individual organs, like leaves and flowers, possess a noticeably short lifespan. Briefly, plants grow for a limited period and start to senesce after the completion of reproduction. The degradation of chlorophyll and macromolecules such as proteins and nucleic acids often accompanies the senescence process in plants. Nevertheless, this deterioration during senescence is not necessarily terminal; instead, it is a reversible process that allows senescing leaves to re-green and restore their photosynthetic capacity under certain circumstances (Rapp et al. 2015). It is believed that the alterations that occur during senescence could constitute a trans-differentiation process which distinguishes it from programmed cell death (PCD). Cell death during senescence happens slowly, at the organelle or organism level, for efficient recycling of nutrients to the growing organs and offsprings which ensures fitness and reproductive success (Kim et al. 2016). In contrast, PCD involve localized, acute and rapid cell death. A frequently used alternative term for ‘senescence’ in plant biology that often creates confusion is ‘ageing’. However, ageing refers to the changes that occur with time and throughout the life cycle of the plant (Thomas 2013). Plant senescence is controlled by intrinsic developmental factors, such as age and phytohormones, and also by environmental (extrinsic) signals, including biotic or abiotic agents, namely pathogens attack, extreme temperatures, drought, nutrient deprivation, and exposure to ozone (Lim et al. 2003; Fig. 1A). The leaf senescence process can be divided into three phases: initiation, degenerative and terminal (Yoshida 2003; Fig. 1B). During the early leaf developmental stages, the younger leaves serve as sink, and the older ones act as the source. On maturation, the demand of younger leaves declines, which results in sugar accumulation in the older leaves and it initiates senescence. This transition from sink to source and the drop in anabolic activity is designated as the initiation phase of senescence. The subsequent catabolism of macromolecules, actuation of salvage and/or remobilization pathways and the dismantling of cellular components (organellar redifferentiation) are associated with the degenerative phase of senescence. The final stage of leaf senescence, the terminal phase, is characterized by the dissipation of ROS and loss of cell integrity and cell death (Yoshida 2003; Lim et al. 2007b).
Leaf senescence paradigm. Leaf senescence is primarily governed by developmental age; however, leaf senescence onset and progression are also regulated by various internal and external stimuli (A). These stimuli engage different signaling pathways, activating transcription factors that control leaf senescence. The senescence process is associated with the down-regulation of SDGs, including photosynthesis-related and metabolic process-related genes. In addition, enhanced expression levels of SAGs (hydrolytic enzymes, TFs, transporters, etc.) are also reported during leaf senescence. The leaf senescence can be divided into three phases: initiation, degenerative and terminal (B). The initiation phase includes the sink/source transition and the decline in photosynthesis. The second stage, the degenerative phase is the degradation phase, where macromolecules and cell organelles undergo degradation. And the ultimate terminal phase involves cell death
Broadly, the plant senescence process can be categorized as natural or induced senescence (Miryeganeh 2022). Although both types of senescence are identical at the morphological level, they exhibit notable differences in the signalling and regulatory mechanisms (Wollaston et al. 2005). Natural senescence in plants has been widely studied with respect to the ageing of leaves, flowers, and coleoptiles. Leaf senescence is usually an organ level senescence but is also strongly associated with cellular or organismal death. Leaves serve as nutrient sources during the early stages of development as this source status is taken over by flowers during the very late stage of plant development (Guo et al. 2021). This review, emphasizes on how the external and internal factors influence the ageing of leaves, and the molecular basis of the evolutionarily acquired leaf developmental process. Efforts have been made to provide critical and up-to-date information on the recent advances in the study of leaf senescence, which would be beneficial in devising strategies for its manipulation to improve crop quality and productivity.
Leaf Senescence: A terminal stage of leaf development
Like plants, leaves follow a specific pattern of development wherein they grow, expand rapidly, import carbon and nitrogen and undergo rapid protein synthesis until they achieve full photosynthetic competence. After this photosynthetically active period, their contribution to photosynthate production declines, and leaves enter the terminal developmental stage of programmed cell death (Lim et al. 2003). In monocarpic plants, leaf senescence is developmentally associated with other organs or whole plant senescence and is triggered by reproductive development. However, in many tree species, leaf senescence can also occur regardless of other plant organs. Overall, leaf senescence is a degradation process that eventuates in a well-orchestrated manner and involves recycling of both macro- and micronutrients.
Induced Senescence
Plants encounter harsh growing conditions owing to exposure to several biotic and abiotic stress factors, which adversely affect their growth, development and productivity. Unlike animals, plants cannot move and escape such unfavourable environmental conditions and thus have evolved mechanisms to rapidly respond to and complete their life cycle even under deteriorating stressful situations (Wollaston et al. 2003). One such response is the onset of senescence that involves the mobilization of nutrients from the dying, no longer essential parts of the plant (e.g., a diseased leaf or an older leaf) to other growing parts (young leaves) or reproductive organs. The onset of senescence also ceases water consumption by older or diseased leaves and allows the plant to complete its life cycle. Several external factors that influence leaf senescence and their mode of action are discussed below. Table 1 presents information on important SAGs associated with natural and induced senescence in two model plants, Arabidopsis and rice.
Dark-induced senescence
The effect of light on triggering senescence is somewhat complicated and essentially relies on the intensity and wavelength of the incident light. High or sub-optimal intensities or dark conditions can induce premature leaf senescence in plants (Wollaston et al. 2003). Recently, the physiological, cytological and transcriptomic changes during dark-induced leaf senescence (DILS) in barley have been extensively reviewed by Nowicka et al. (2018). As reported, the DILS is characterized by prominent upregulation of macromolecular and metabolite degradation with a concomitant decline in photosynthesis. Weaver and Amasino (2001) have shown that senescence was delayed when whole Arabidopsis plants were placed in darkness. Nevertheless, when individual leaves were covered, senescence was induced. These results indicate that senescence is a highly localized phenomenon depending on the light status of the entire plant.
To pinpoint the conserved molecular forces driving the senescence process, microarray expression analysis of natural, dark-induced and sucrose starvation-induced suspension cultures was performed in Arabidopsis (Wollaston et al. 2005). The salicylic acid (SA), jasmonic acid (JA), and ethylene response pathways were involved in regulating natural, dark-induced and cell-suspension senescence, respectively. Comparative transcriptome studies of developmental, dark-induced (detached leaves) and dark-induced senescence of leaves attached to the plant revealed noticeable differences in the expression pattern of few senescence associated genes (SAGs) such as transcription factors, transporters, receptor like kinases, and hormone pathway genes among the different senescence processes. Developmental leaf senescence showed a higher accumulation of amino acid and oligopeptide transporters (Graaff et al. 2006). Several differentially expressed genes linked to plant hormone signal transduction pathways, TFs (WRKYs, NACs, HSFs, PIFs and bHLHs), and protein processing machinery were discovered by transcriptome analysis of dark-induced senescence in bermudagrass (Fan et al. 2019).
A delay in dark-induced senescence was reported in the transgenic rice plants overexpressing SUBMERGENCE 1A (SUB1A), an ethylene response factor (Schippers et al. 2015). The submergence tolerance gene SUB1A suppresses the effect of dark-induced senescence (DIS) by repressing the phytohormone signalling pathways, especially ethylene, JA and SA pathways in rice. Additionally, Sakuraba et al. (2014a) found that the transcript and protein levels of PHYTOCHROME INTERACTING FACTOR3, 4 and 5 (PIF3, 4 and 5) were substantially increased during age-triggered and dark-induced senescence of Arabidopsis leaves. A recent study by Hao et al. (2022) showed the implication of plastocyanin or PCY-SAG14 module in copper homeostasis under DIS in Arabidopsis. PCY-SAG14 is an endomembrane localized module that promotes DIS and is post-transcriptionally regulated by miR408. Under prolonged dark conditions, PIF3/4/5 are activated, release PCY-SAG14 module from miR408 regulation and subsequently promote DIS (Hao et al. 2022). Zareen et al. (2022) demonstrated the involvement of a multifunctional WD-40 repeat protein, HOS15 (high expression of osmotically responsive genes 15) as a positive regulator of dark-induced and natural leaf senescence in Arabidopsis. It was proposed that since HOS15 is the component of PWR-HDA9 repressor complex, it functions as a transcriptional corepressor and inhibits the acetylation of negative regulators of senescence, NPX1, PDG9 and WRKY57.
Salt stress-induced senescence
Salinity stress adversely impacts plant growth and productivity as it causes reduced growth, sink-source imbalance, leaf senescence and eventually the death of plants (Albacete et al. 2014). Salt-induced premature leaf senescence in sweet potato was shown to be associated with chlorophyll degradation, decreased photosynthesis and accumulation of ROS (Chen et al. 2012). Salt stress mainly causes osmotic stress by accumulating toxic sodium ions (Na+) in leaves (Schippers et al. 2015). Elaborately, this imbalance in the sink-source relationship leads to plant growth decline during salt stress conditions. Studies on salt-tolerant versus sensitive wheat varieties revealed that salt-tolerant variety exhibits a comparatively delayed senescence phenotype due to an increased sink strength (Schippers et al. 2015). RNA-seq analysis of salt stress-induced leaf senescence in Medicago truncatula revealed regulation of more than 4000 SAGs. Of these, 1546 were also reported to express commonly in dark and salt-induced senescence (Dong et al. 2021). Studies on the rice early leaf senescence mutant, bilateral blade senescence 1 (bbs1), demonstrated its hypersensitivity to salt stress indicating a close relation between leaf senescence and salt stress pathways (Zeng et al. 2018). Detailed analysis of the bbs1 mutant identified an insertion in the coding region of a receptor-like cytoplasmic kinase, OsRLCK109. Furthermore, it was found that the gene coding for OsRLCK109 was induced during salt stress. A recent study by Park et al. (2022) revealed the intersection of salt stress response with leaf senescence through the characterization of Arabidopsis ETHYLENE RESPONSIVE FACTOR34 (ERF34). ERF34 exhibited a differential expression pattern during developmental senescence, and its negative regulatory role in salt stress-induced and developmental senescence is also reported. The crosstalk between salt stress and senescence is supported by ER34-mediated transcriptional activation of salt-stress responsive genes, EARLY RESPONSIVE TO DEHYDRATION10 (ERD10) and RESPONSIVE TO DESICCATION29A (RD29A) (Park et al. 2022).
Drought stress and senescence
Drought stress triggers responses ranging from altered gene expression to changes in plant metabolism and growth, including leaf senescence which causes a substantial decrease in canopy size and reduced yield. Some commonality has been observed in the symptoms of senescence induced by different stressors such as drought, salinity as well as developmental age-related leaf senescence. For example, drought-induced senescence involves stomatal closure, the most notable phenomenon in older senescing leaves for allowing reduced water loss through transpiration at the whole plant level (Bosch and Alegre 2004). Carbon/nitrogen balance is another critical factor for regulating drought-induced leaf senescence that was studied in Sorghum bicolor leaves (Chen et al. 2015). A direct role of cytokinins and ABA in gene reprogramming during drought-induced senescence has also been proposed given that the high ABA enhanced carbon mobilization from senescing leaves to grains in the drought-stressed rice and wheat (Reguera et al. 2013). A study on wheat landraces subjected to drought stress revealed that the biosynthesis of stem-specific proteins halts and to compensate for the lower assimilate synthesis rate, stem senescence and remobilization processes are triggered (Bazargani et al. 2011). In tobacco, the expression of Isopentenyltransferase (IPT) driven by stress and maturation-inducible promoter (pSARK) enhanced drought tolerance by delaying leaf senescence (Rivero et al. 2007).
Nutrient limitation and senescence
Plants require both macro- and micro-nutrients for proper growth and development. The unavailability of these nutrients often causes starvation, which eventually leads to premature leaf senescence in plants. The nitrogen-limiting condition triggers chloroplast degradation to initiate nitrogen recycling in plants. A field experiment with sixteen tropical maize varieties exhibiting wide variation for grain yield at a low nitrogen supply (N) showed a negative correlation between leaf senescence at the grain filling stage and N-use efficiency (Schulte et al. 2007). The authors also pinpointed the possibility of screening these genotypes for N deficiency-induced senescence survival as a parameter for identifying N-use efficient maize cultivars. Another indispensable macroelement necessary for optimal growth and development of all crops is sulfur (S), which is available as sulphate (SO42−) in soil and as sulphur dioxide (SO2) in the environment. Interestingly, the effects of S deprivation are influenced by the availability of N as observed in oilseed rape wherein delay in leaf senescence was reported under the low S-high N condition (Dubousset et al. 2009). Delay in senescence under S-deficient conditions has been observed in barley, where the process is accompanied by down-regulation of two cysteine- and one serine-protease gene (Veliz et al. 2020). These results highlight the requirement of S for the proper onset of leaf senescence, which ultimately affects the grain quality.
Iron (Fe) is an essential micronutrient indispensable for the functioning of key biological processes, including photosynthesis and respiration (Morrissey and Guerinot 2009). Studies on wheat seedlings revealed that Fe and N deficiency induces leaf senescence. A significant reduction in Fe concentration was observed under − N / + Fe compared to + N/ + Fe condition indicating a possible interplay between varied levels of N supply and Fe accumulation. Furthermore, inhibition of Fe export from senescing leaves to younger leaves under high N supply and an enhanced Fe export under N-deprived conditions was also reported (Parveen et al. 2018). Zakari et al. (2020) investigated the relationship between N deficiency-induced leaf senescence and ABA concentration in psf (premature senescence of flag leaf) mutant versus wildtype (WT) rice plants. They further demonstrated a significant level of ABA accumulation and up-regulation of ABA biosynthesis genes (9-cis-epoxycarotenoid dioxygenases or NCEDs), ROS burst and enhanced expression of SAGs in psf mutants compared to the WT plants under N deficiency condition. In addition, ABA accumulation showed a reversible pattern on N supplementation with an enhanced expression of ABA catabolic genes. Based on these results, it was concluded that adequate N supply has an inhibitory effect on ABA levels resulting in low ROS levels and delayed leaf senescence. Optimal levels of another macronutrient, Magnesium (Mg2+) are also crucial for plant growth and development given that it is required for several essential cellular processes, including photosynthesis, protein and nucleic acid synthesis and energy metabolism (Guo et al. 2016). As per the study by Kocourkova et al. (2021), a phospholipase Dα1 (PLDα1) acts as a negative regulator of leaf senescence induced by high Mg2+ levels in Arabidopsis. Higher accumulation of ABA and JA were detected in pldα1 mutant under high Mg2+ conditions. These studies carried out on pldα1 and psf mutants highlight the overlap between the pathways regulating nutrient stress-induced senescence and natural ageing in plants.
Biotic stress and senescence
Various biotic stress factors, including pests and pathogens, challenge the plants during their growth and development. The pathogenic infection mechanisms interact with the developmental pathways to mutually influence each other owing to the cross-talk between their signalling pathways and convergence at several regulatory nodes (Guo et al. 2021). The complexity increases with the highly variable lifestyle of different pathogens influencing the plant developmental program in an unusual manner leading to an adverse effect on plant productivity. Necrotrophs induce premature senescence, while biotrophs delay host plants' ageing progression. WRKY TFs (WRKY6, 53, 70 and 30) are associated with the senescence program and are also well-known regulators of defense responses (Guo et al. 2021). Expression analysis of genes encoding for WRKY TFs indicated that five senescence-inducible OsWRKY genes (OsWRKY 2, 6, 14, 26, and 93) were also upregulated in rice infected with Magnaporthe oryzae (Wei et al. 2013). Dark-induced leaf senescence was delayed in Magnaporthe-resistant transgenic rice lines overexpressing OsWRKY93, whereas an opposite phenotype was observed in oswrky93 mutant. It is thus believed that OsWRKY93 is a potential candidate for the breeding of rice cultivars with enhanced yield and resistance to Magnaporthe infection (Li et al. 2021b). Remarkably, the mutants of positive regulators of senescence, ein2, ore1 and nac055 (stay-green mutants), showed an altered age-related resistance against Pseudomonas syringae pv tomato (Schippers et al. 2015). In Arabidopsis, infection with Botrytis cinerea induced several senescence-associated genes (SAGs) and suppressed the expression of photosynthesis and starch metabolism genes (Windram et al. 2012). Interestingly, several phytohormones play a crucial role in regulating senescence and host defense responses. Several factors and signalling pathways regulate pathogen-induced senescence as the host needs to maintain developmental homeostasis and simultaneously activate defense responses for achieving resistance to infections. For instance, ethylene, ABA and SA signalling were activated upon Botrytis cinerea infection in Arabidopsis, indicating a crosstalk between developmental and biotic stress-induced senescence processes (Windram et al. 2012).
Oxidative stress and senescence
Most of the abiotic and biotic stresses trigger the accumulation of reactive oxygen species (ROS) and oxidative stress (Jajic et al. 2015). Resultantly, a high level of ROS mainly drives the oxidation of lipid membranes and damage to cellular biomolecules, which ultimately culminates into cellular, structural and functional damage. During oxidative stress plants activate the gene expression pathways of antioxidative enzymes, such as catalases, superoxide dismutases (SODs) and ascorbate–glutathione to maintain the redox homeostasis. Studies on plant chloroplasts by Munné-Bosch and Alegre (2002) revealed that a higher accumulation of ROS occurs in chloroplasts upon ageing which act as signalling molecules to activate the expression of several TFs and SAGs that are key for the progression of senescence (Garapati et al. 2015). Similarly, the singlet oxygen has been shown to induce the expression of WRKY6, which is vital for the senescing process (Jajic et al. 2015).
Moreover, a senescence-associated metallothionine protein, LSC54, has been shown to accumulate and correlate with the rising ROS levels during oxidative stress in Arabidopsis. Application of catalase inhibitors on leaves increased the expression of LSC54, while the treatment of leaves with quenchers of ROS downregulated the expression of LSC54 (Navabpour et al. 2003). Additionally, the analysis of delayed leaf senescence in Arabidopsis mutants, ore1, ore2 and ore9, revealed that they were highly tolerant to oxidative stresses (Woo et al. 2004). REV or REVOLUTA is a redox-sensitive HD-ZIPIII TF that has been shown to positively regulate age-triggered leaf senescence in Arabidopsis (Xie et al. 2014). A recent study on oxidative stress-sensitive rice T-DNA mutant, RLS1 (reactive oxygen species-sensitive leaf senescence1) further highlighted the relationship between oxidative stress and senescence given that several SAGs and autophagy-related genes were upregulated during oxidative stress (Chen et al. 2018).
Heat stress and senescence
Generally, the elevation in temperature above the threshold level adversely impacts plant growth and crop yield (He et al. 2021). Severe heat stress induces cellular senescence by chloroplast disruption, photosynthesis impairment, initiation of DNA damage, ROS accumulation, and cell death (Fedyaeva et al. 2014). Heat stress-induced premature senescence has been attributed to accumulation of soluble sugars, decline in starch levels, degradation of soluble proteins, and accumulation of H2O2 with a concomitant decrease in antioxidant activity in sunflower primary leaves (Haba et al. 2014). Recently, PSL50 (Premature senescence leaf 50) was shown to play an important role during heat-induced premature leaf senescence by modulating the H2O2 signaling pathway in rice (He et al. 2021). Nevertheless, the involvement of phytohormones in regulating stress responses in plants has also been well-documented (Guo et al. 2021). While there is an elevated production of both ethylene and ABA, a decline in cytokinin levels during senescence triggered by high temperature has been recorded in leaves of bentgrass (Xu and Huang 2007). Remarkably, a transcriptome-based study also revealed differential regulation of several heat shock transcription factors (HSFs) during leaf senescence in Arabidopsis (Raxwal et al. 2012). More details on HSFs and senescence are provided in the following section.
Cold stress and senescence
Cold stress affects plant growth and development by indirectly imposing osmotic and oxidative stresses as well as by changing the metabolic reactions and membrane properties, which eventually triggers plant senescence. Cold stress-induced leaf senescence indeed is an effective strategy for plants growing in a low temperature area to overcome extreme low temperatures (Caselles et al. 2021). Initiation of senescence in the aboveground tissues of Iris pseudacorus was documented during the winter seasons. In contrast, the underground rhizome remains dormant and re-establishes growth when favourable conditions return. Interestingly, the cold-acclimated Arabidopsis plants showed delayed senescence in rosette leaves with a marked recovery of Fv/Fm ratio after an initial decline (Daubresse et al. 2007). Cold stress induces the accumulation of SAGs in different plant species (Yang et al. 2017). Chilling stress treatment, alone or in combination with Alternaria sps. infection, resulted in pronounced leaf senescence in cotton (Zhao et al. 2012). This was evident by an increase in malondialdehyde activity, electrolyte leakage, and a decline in chlorophyll and soluble protein content. In Iris pseudacorus, induction of leaf senescence and an appreciable increase in ABA/cytokinin ratio has been observed during winters (Caselles et al. 2021). Transcriptome analysis of Arabidopsis plants exposed to cold and dark conditions showed that the ABA pathway positively regulates the leaf senescence process, while it is negatively regulated by the brassinosteroid pathway (Panigrahy et al. 2021).
Regulation of Leaf Senescence
Owing to the importance of leaf senescence in crop productivity, extensive physiological, biochemical, molecular and genetic studies have been conducted to unravel the complex, multi-tiered regulatory processes in action under the influence of several internal and external factors during senescence to improve crop yield, an ultimate research goal (Schippers et al. 2015). Understanding these regulatory pathways during senescence could open up new avenues in senescence research, which can further facilitate agronomic applications for breeding new crop cultivars with stable and improved yield. This review compiles the information available on the transcriptional, post-transcriptional, epigenetic, and hormonal level regulation of leaf senescence which may further aid in devising strategies for genetic enhancement of multiple crop species.
Gene Programming Changes During Leaf Senescence
Several experimental approaches, including microarray, mRNA sequencing, Northern blot analyses, and differential screening studies using subtractive hybridisation techniques have been employed to study the gene expression changes during senescence in various plant species. All these studies demonstrated that extensive changes in the expression of specific genes, commonly referred to as SAGs, accompany the senescence programme (Lim et al. 2003). The initial differential screening studies in Arabidopsis thaliana have identified approximately 800 cDNA clones representing SAGs; of which, 130 were found to be non-redundant genes and 70 as new SAGs, which exhibit either leaf senescence-specific expression pattern or upregulation during the process (Gepstein et al. 2003). Efforts have been made to provide up-to-date information on the significant classes of SAGs (Table 1) that have been demonstrated to act as regulators of leaf senescence in model plant systems Arabidopsis and rice.
The transcriptional control mechanism is crucial in coordinating the senescence process through massive reprogramming of gene expression, including transcription factors (TFs) that regulate gene expression by binding to the specific cis-regulatory elements and triggering their activation or suppression. Microarray analysis of senescing leaves revealed that 100 putative TFs belonging to approximately 20 different families, especially NAC, WRKY, C2H2 type zinc finger, AP2/EREBP, bZIP, CCAAT binding, Leu zipper, MADS-box, HSFs, kinases and MYB proteins family were upregulated at least three-fold in Arabidopsis (Wollaston et al. 2005; Balazadeh et al. 2008). A similar expression pattern of several of these TFs was also observed in the transcriptomic studies of leaf senescence conducted in other plant species such as maize, cotton and rice (Woo et al. 2019). A survey of available literature revealed a huge amount of information on the TFs associated with leaf senescence, which is beyond the scope of this review. A brief enumeration of the selected class of TFs that are key regulators of leaf senescence is given below.
NAC (NAM, ATAF and CUC) proteins constitute one of the most prominent families of plant TFs that play important regulatory roles in the development, senescence and stress responses of various plant species (Broda et al. 2021; Nie et al. 2021). More than 30 NAC genes are upregulated during leaf senescence in Arabidopsis (Breeze et al. 2011). Manipulation in the expression of NAC genes resulted in the alteration of leaf senescence. AtNAP or ANAC029 in particular regulates leaf senescence by binding to the promoter of SAG113, a negative regulator of the ABA pathway, resulting into inhibition of stomatal closure and the eventual initiation of leaf senescence (Zhang and Gan 2012). ORE1 or ANAC092 is another positive regulator of leaf senescence in Arabidopsis, which regulates at least 170 genes, including the 78 known SAGs and is induced by EIN2 (ETHYLENE INSENSITIVE 2 (Balazadeh et al. 2010). It is believed that with progression in ageing, ORE1 expression elevates and in turn, physically sequesters GLKs (chloroplast activity maintainer) resulting in their reduced transcriptional activity with an ultimate effect on chloroplasts (Rauf et al. 2013). The ORE1, therefore, plays a dual role in controlling leaf senescence: firstly, by triggering the transcription of SAGs and secondly, by physically interacting with other senescence-associated TFs, thereby modulating their activity (Fig. 2). A novel regulator mode constituted by NAC075, CATALASE (CAT) and ROS has been shown to actively govern the initiation and progression of leaf senescence (Kan et al. 2021). The studies on transgenic Arabidopsis plants with altered expression of NAC075 reported its negative regulatory role in senescence as NAC075 promotes the expression of CAT2 by directly binding to the promoter region, thereby reducing the reactive oxygen species levels in the system and delaying the senescence process. Similarly, other NAC transcription factors which are positive regulators of senescence have also been reported in Arabidopsis (ATAF2, SNAC-As, ANAC017, ORE1/ANAC092), maize (ZmNAC126) and rice (OsNAC2 and ONAC011) (Mao et al. 2017; Guo et al. 2021; Zhang et al. 2021). A positive regulator of leaf senescence, ANAC046 directly binds to the promoter regions of the genes involved in the breakdown pathway of chlorophyll [NON-YELLOW COLORING1, Stay-Green 1, Stay-Green 2, and PHEOPHORBIDE α OXYGENASE] (Yamamizo et al. 2016). In Arabidopsis, the ANAC019/055/072 and NAC016 activate the chlorophyll degradation during leaf senescence by directly binding to the promoter region of the chlorophyll catabolic gene SGR1 (Zhang et al. 2021). Apart from the positive regulatory role, few of the NAC TFs are also reported to function as negative regulators in the senescence process. For example, DRL1 was shown to negatively regulate plant senescence in grapevine by fine-tuning the ABA biosynthesis pathway (Zhu et al. 2019). JUNGBRUNNEN 1 (JUB1 or ANAC042), which is induced by H2O2, activates the expression of DREB2A and several ROS-responsive genes. Indeed, it is also believed to lower the cellular H2O2 levels and reduce the effects of positive regulators of senescence (Wu et al. 2012). It is conceivable that JUB1 is a possible negative regulator of leaf senescence. On the other hand, overexpression of a NAC TF, TaNAC-S, resulted in the delay in senescence with a concomitant increase in grain yield and grain protein concentration in wheat indicating its role as a negative regulator of senescence (Zhao et al. 2015).
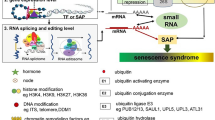
An overview of multi-tiered natural leaf senescence regulatory network. The signalling pathways of phytohormones JA, ethylene, ABA, SA, and GA initiate and promote leaf senescence in leaves. By binding to the promoters of important chlorophyll (Chl) catabolic genes (NYE1, NYC1 and PAO), MYC proteins regulate JA-induced Chl degradation downstream of JAZs in the JA signalling pathway. Moreover, MYCs indirectly regulate Chl degradation through the transcription factors ANAC019/055/072, which can trigger the activation production of the same Chl catabolic genes (CCGs). AtNAP positively regulates leaf senescence by promoting ABA production and SAG113 expression. ATAF1 contributes to ABA-induced senescence by activating the expression of genes involved in ABA biosynthesis and transport (NCED3 and ABCG40). ATAF1 enhances and suppresses the expression of ORE1 and GLK1, respectively, by directly binding to their promoter region. As a result, the expression of GLK target genes is hindered, resulting in an age-dependent drop in the expression of GLKs, whilst the expression of ORE1 target genes is increased, triggering senescence. ABA-induced senescence is mediated by the action of ABFs downstream of ABA signaling modules. EIN3, which is activated by EIN2, represses miR164 transcription by binding directly to the its promoter region which results in elevation of ORE1 transcript levels, thereby promoting leaf senescence as ORE1 activates the transcription of SAG29, SINA1 and SWEET15 and represses the expression of GLKs. PRR9 activates ORE1 and suppresses miR164 indirectly during leaf senescence. PIFs, whose expression is inhibited by ELF3, promote chloroplast deterioration by suppressing GLKs. Age and GA-induced WRKY45 promotes the expression of a number of SAGs. WRKY75 is involved in a tripartite amplification loop where it promotes SA synthesizing gene SID2. JA-induced TF, TCP4, positively regulates leaf senescence by enhancing the expression of LOX2. Whereas bHLHs, negatively regulate leaf senescence by repressing SAG29 expression. Leaf senescence is also regulated at epigenetic level. Expression of WRKY53 is partially mediated by methylation of histones via SUVH2. WRKY53, in turn along with PWR and HDA9, removes the acetylation marks from the histones of WRKY57 thereby leading to its suppression which influences the antagonistic regulation of leaf senescence by auxin and JA. Expression of WRKY57 protein level is positively mediated by auxin, whereas JA represses its expression at transcript level. The age-induced Ca2+ levels promote leaf senescence by activating the Ca-dependent protein kinase (CPK1), which in turn phosphorylates and activates the master regulator of senescence, ORE. The blue boxes represent the cytoplasmic/nuclear regulatory component of senescence; green boxes represent chloroplastic regulatory components; red boxes represent phytohormones associated with senescence. LOX: Lipoxygenase; IAA: INDOLE-3-ACETIC ACID INDUCIBLE; JAZ: JASMONATE ZIM-DOMAIN protein; ANAC: Arabidopsis NAC transcription factor; PIF: Phytochrome interacting factor; ACS: 1-aminocyclopropane-1-carboxylate synthase; EIN: Ethylene insensitive; ORE: Oresara; CPK: calcium dependent protein kinase; GLK: Golden2-like transcription factor; ATAF: Arabidopsis thaliana ACTIVATING FACTOR; ABCG40: ARABIDOPSIS THALIANA ATP-BINDING CASSETTE G40; NCED: 9-cis-epoxycarotenoid dioxygenase; ABA: Abscisic acid; AAO: Arabidopsis aldehyde oxidase; ABI: ABSCISIC ACID INSENSITIVE; PRR: PSEUDO-RESPONSE REGULATOR; ELF: early-flowering; PYL: pyrabactin resistance-like; ABF: ABRE-binding factors; NYC: NON-YELLOW COLORING; SGR: STAY GREEN; PPH: PHEOPHYTINASE; PAO: pheide a oxygenase; SOC: SUPPRESSOR OF OVEREXPRESSION OF CO; SAG: Senescence-associated gene: PCY: Plastocyanin; SUVH: SU(VAR)3–9 homolog; HDA: histone deacetylase; GID: GA INSENSITIVE DWARF; SID: SALICYLIC ACID INDUCTION DEFICIENT; ESP: Epithiospecifier protein; ERF: Ethylene-responsive element binding factors; JA: Jasmonic acid; GA: Gibberellic acid; SA: Salicylic acid
The WRKY family of TFs is comprised of many members implicated in multiple plant processes, including developmental and stress responses (Zhang et al. 2021). WRKY proteins usually interact with other proteins, including regulatory factors, to constitute an essential component of the kinases signalling cascade. Notably, a complex regulatory network involving combinatorial interactions of WRKY TF family members fine-tune the leaf senescence process. For instance, the EPITHIOSPECIFYING SENESCENCE REGULATOR (ESR) inhibits the DNA-binding activity of WRKY53 and acts as a negative regulator of senescence (Woo et al. 2019). Similarly, WRKY13-A, a partial functional homolog of AtWRKY53, positively regulates both dark-induced and natural leaf senescence by promoting JA biosynthesis in wheat (Qiao et al. 2021). WRKY54 and WRKY70 both regulate leaf senescence and the positive regulator WRKY53 possibly interacts with WRKY30 and targets WRKY22. Together they constitute a major part of the regulatory network that integrates with internal and external signals to regulate the initiation and progression of leaf senescence (Jiang et al. 2014). A model that depicts the tripartite amplification loop involving WRKY75, SA, and ROS has been proposed by Guo et al. (2017) in Arabidopsis. The positive regulator of senescence, WRKY75, induces the transcription of SA INDUCTION-DEFICIENT2 (SID2) and promotes SA production during senescence. At the same time, WRKY75 suppresses the H2O2 scavenging by repressing CAT2 expression. Auxin and JA are shown to act antagonistically on WRKY57, a negative regulator of JA-induced leaf senescence (Jiang et al. 2014). The JAZ4/8 and IAA29 function as negative and positive regulators of JA-induced leaf senescence, respectively, and interact competitively with the zinc-finger domain of WRKY57. The WRKY57 binds directly to the promoters of SEN4 and SAG12 and suppress their transcription. WRKY45 positively regulates leaf senescence by activating the transcription of SAG12, 13, 113 and SEN4. RGL1 (a repressor of the GA signalling pathway) interacts with WRKY45, resulting in the loss of its transcriptional activation potential (Woo et al. 2019). WRKY93 has been shown to play a dual role in regulating flag leaf senescence and response to fungal pathogen infection in rice (Li et al. 2021b).
Besides NAC and WRKY TFs, another TF family associated with the regulation of leaf senescence is MYB. Overexpression of R-R type MYB-like transcription factor (MYBL) in Arabidopsis displayed an enhanced senescence phenotype (Zhang et al. 2021). The OsMYB102 demonstrated a negative role in plant senescence via regulating the ABA accumulation and the signalling cascades by activating and repressing the ABA catabolic enzyme ABSCISIC ACID 8′-HYDROXYLASE and ABA-responsive genes (OsABF4 and OsNAP), respectively (Guo et al. 2021). In Arabidopsis, RAV1, a related to ABI3/VP1 (RAV) TF family member was found to act as a positive regulator of leaf senescence (Woo et al. 2010). Several research groups have also reported the involvement of various basic helix-loop-helix (bHLH) TFs, such as MYC2, MYC3, MYC4, to antagonistically interact with bHLH03, bHLH13, bHLH14 and bHLH17 and activate SAG29 expression for initiating JA-triggered leaf senescence (Woo et al. 2019). The phytochrome-interacting bHLH TFs, such as PIF4 and PIF5, positively regulate the dark-induced and natural leaf senescence in Arabidopsis (Li et al. 2021a). PIF4 binds to the promoters of NYE1 (a chlorophyll degradation regulatory gene) and GLK2, which eventually initiates chlorophyll degradation machinery (Woo et al. 2019).
Several genes encoding HSFs have also shown significant expression changes during leaf senescence in Arabidopsis and rice (Raxwal et al. 2012). Particularly, the AtHSFB1 and AtHSFA6a were upregulated during leaf senescence in Arabidopsis (Balazadeh et al. 2008). As a proof, the hsfB1 mutants exhibited early leaf senescence and significant upregulation of several SAGs, including SAG12, WRKY and peroxidase gene. The transcriptomic studies in maize, cotton, rice, petunia and bermudagrass also showed differential expression of several HSFs during age-dependent leaf senescence (Lin et al. 2015; Wang et al. 2018; Fan et al. 2019). All these studies demonstrate the complexity of signaling networks associated with leaf senescence and demand the identification of more TFs to gain a better understanding of transcriptional regulation during the last stage of leaf development process.
Epigenetic Control of Senescence
Epigenetic regulation involves modifications in gene expression without any change in the genomic sequence. This involves chemical alterations of DNA such as methylation, changes in chromatin modelling, post-translational modifications of histones, and involvement of non-coding RNAs (Guo et al. 2021). The developmental switches associated with the transition from cell survival to cell death in leaves are also tightly controlled by epigenetic and genetic mechanisms, which ultimately regulate the changes in SAGs expression. Here, we summarize the epigenetic mechanisms that regulate leaf senescence in plants.
DNA methylation
DNA methylation ensures the silencing of transposons, DNA repeats, and gene body, and thereby controls the gene expression and protects the genome from superfluous mutations. The methylation of cytosine nucleotides resulting in the formation of 5-methylcytosine is the most frequent phenomenon in plants (Law and Jacobsen 2011). In Arabidopsis, the CG and CHG type methylations are controlled by MET1 (METHYLTRANSFERASE 1) and CHROMOMETHYLASE 3 (CMT3), respectively, while the DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) catalyzes the symmetric and asymmetric DNA methylation (Feng et al. 2010). Mutations in these methyltransferases (MET1, DRM2 and CMT3) trigger pleiotropic developmental abnormalities due to genome-wide hypomethylation (Moritoh et al. 2012). Currently, the clarity on senescence-specific DNA methylation changes in plants, however, is lacking, but few reports indicate that the status of DNA methylation alters as the plant ages (Zhang et al. 2021). During senescence, the retrotransposon controlling epigenetic mechanisms ceases to function which results in the demethylation of transposable elements (TEs) and thereby facilitating their transcription and transposition (Guo et al. 2021). A study by He et al. (2018) identified a retrotransposon, NMR19 (naturally occurring DNA methylation variation region 19), whose methylation level and genomic location varied among different accessions of Arabidopsis thaliana. It was thus concluded that NMR19-4 is a novel naturally occurring epiallele that regulates leaf senescence by controlling the expression of PHEOPHYTIN PHEOPHORBIDE HYDROLASE (PPH) and eventually tuning the levels of chlorophyll. Henceforth, it would be interesting to investigate the role of transposable elements (TEs) in plant senescence. Ogneva et al. (2016) demonstrated a decline in the transcription of DNA methyltransferase genes such as CMT3 and MET1 during ageing, while the expression of demethylase genes like ROS1, DME, DML2 and DML3 were elevated at certain stages of development in Arabidopsis. Yuan et al. (2020) also revealed an epigenetic regulatory mechanism controlling leaf senescence in Arabidopsis. The DML3 expression level increases exponentially and activates the SAGs by demethylating their promoters, gene body and 3’ UTRs (upstream regulatory regions) during senescence. Arellano et al. (2020) showed that genes mediating chromatin silencing were down-regulated during dark-induced senescence, which disrupted the silencing of TEs and finally led to the reactivation of young TEs. All these studies demonstrate the relevance of DNA methylation in epigenetic reprogramming during the later phases of plant development, including leaf senescence.
Histone modifications
Histones are post-translationally modified at their N-terminal tail through covalent modifications, including methylation, ubiquitination, acetylation, SUMOylation and phosphorylation, which influences the transcriptional activity of chromatin (Guo et al. 2021; Zhang et al. 2021). Of these, histone acetylation and methylation are the two key modifications associated with leaf senescence. Acetylation of lysine residue has been correlated with transcriptional activation of chromatin. The Arabidopsis mutants for HISTONE DEACETYLASE19 (HDA19) and HDA6 showed pleiotropic developmental defects, including increased leaf longevity and delayed flowering (Wu et al. 2008). While the expression of jasmonate-responsive genes (PDF1.2, VSP2, JIN1 and ERF1) and SAGs (SAG12 and SEN4) was downregulated, the transcript level of FLOWERING LOCUS C (FLC) was elevated in the HDA6-RNAi plants. HDA6 turns off the expression of FLC, a floral repressor by deacetylating the histones associated with FLC (Wu et al. 2008). AtHDA9 (an RPD3-like histone deacetylase) promotes the initiation of leaf senescence in Arabidopsis by interacting with the transcription factor WRKY53 and the SANT domain-containing chromatin-binding protein POWERDRESS (PWR). The genome wide HDA9 occupancy profiling has shown that it directly binds to the promoters of critical negative regulators of senescence with a requirement of PWR (Woo et al. 2019). In addition to histone acetylation, histone methylation also plays a significant role in epigenetic control of gene expression. During the onset of senescence, a higher expression level of WRKY53 has been observed because of the establishment of active marks of H3K4me3 at WRKY53-associated histones (Zhang et al. 2021). A ChIP-seq study of non-senesced and senesced leaves revealed that H3K4me3 is linked to active chromatin, whereas the H3K27me3 mark is associated with repressed chromatin. Moreover, overexpression of histone methylases demonstrated ectopic heterochromatinisation and delayed senescence phenotypes in Arabidopsis. JMJ16, a JmjC domain-containing protein, is particularly an H3K4 demethylase that inhibits leaf senescence through its enzymatic activity in Arabidopsis. Genetic studies have shown that JMJ16 is a negative regulator of leaf senescence which inhibits the expression of positive regulators, WRKY53 and SAG201. JMJ16 binds to WRKY53 and SAG201 and reduces H3K4me3 levels at these loci leading to suppression of their early expression in mature leaves (Liu et al. 2019). As Arabidopsis plants mature, the double-strand breaks (DSBs) increase due to the decline in efficacy of DNA repair mechanisms. A premature senescence phenotype results from the generation of DSBs through the inducible expression of an intron-encoded endonuclease (I-PpoI). The histone lysine methylation regulated by ATM (ATAXIA TELANGIECTASIA MUTATED) represses the DSB-induced expression of senescence-associated genes, including those encoding WRKY and NAC TFs, the essential components of the leaf senescence process (Li et al. 2020).
Chromatin alterations
ATP-dependent chromatin remodeling factors can recognise the histone modifications through their histone-binding motifs, bromo or chromodomains, and can non-covalently restructure nucleosomes by disrupting or destabilising their structure (Brusslan et al. 2012). A mutation in DRD1 (a SWI2/SNF2 chromatin remodeling protein encoding gene) affects the progression of dark-induced leaf senescence given an observed decline in SAGs induction (Zhang et al. 2021). The precise underlying mechanisms by which DRD1 is implicated in leaf senescence, however, remains unknown. AT-hook proteins are among the many ATP-dependent chromatin remodeling factors that are involved in leaf senescence and regulation of chromatin structure. The overexpression and activation-tagged mutants of ORESARA 7 (ORE7), an AT-hook protein exhibited a delayed leaf senescence phenotype in Arabidopsis (Lim et al. 2007a). Additionally, a genome-wide transcriptome of senescing Arabidopsis leaves showed upregulation of chromatin remodeling factors, CHR10 and CHR19 indicating their possible positive role during the progression of senescence (Breeze et al. 2011).
Small non-coding RNAs
Small RNAs are known to regulate the senescence process through fine-tuning the expression of genes involved in signalling pathways, including TFs, phytohormone metabolism and kinases (Xu et al. 2014). For instance, a decline in the level of miR164 has been confirmed, while there is up-regulation in the ORE1 expression during leaf ageing. The ORE1/AtNAC2 positively controls ageing-induced cell death by enhancing the expression of SAGs in the Arabidopsis leaves (Kim et al. 2009). Elaborately, ORE1 is negatively regulated by miR164 during the initial stages of senescence, but this inhibition is released by EIN2 (ETHYLENE INSENSITIVE 2), which governs miR164 levels during later stages of senescence. A trifurcate feed-forward pathway was thus proposed with ORE1, miR164 and EIN2 for fine regulation of leaf senescence in an ethylene-dependent manner (Kim et al. 2009). Later studies by Kim et al. (2018) showed that PSEUDO-RESPONSE REGULATOR (PRR9), a vital component in the circadian clock of plants, directly activates the transcription of ORE1 and indirectly suppresses miR164, thereby establishing the link between ageing and circadian clocks in plants (Fig. 2). In Arabidopsis, miR319 regulates the expression of a TF, TCP (TEOSINTE BRANCHED 1, CYCLOIDEA, PCF1), which in turn, controls LOX2, a lipoxygenase that controls JA biosynthesis (Schommer et al. 2008). miR319 is also known to target another positive regulator of senescence, WRKY53 (Guo et al. 2021). ARF2, a positive regulator of senescence, targets tasiRNAs (trans-acting siRNAs) generated from TAS3 precursor transcript, whose cleavage is mediated by miR390 (Lin and Wu 2004). The miR390-TAS3-ARF2 node is, therefore, hypothesized to play a significant role in regulating leaf longevity. In Arabidopsis, the overlapping 3’-UTR of PPR (Pentatricopeptide repeat-containing protein) and WHY3 genes possess a miRNA gene locus MIR840, and both of these genes happen to be the predicted targets of miR840 and miR840*. Ren et al. (2022) reported that an increased accumulation of pre-MIR840 transcripts correlated with a decrease in PPR transcripts, but not with that of WHY3 transcript levels as its protein levels were significantly reduced at the onset of senescence. Overall, the miR840*–PPR and miR840–WHY3 pair modulate the expression of some SAGs with an effect on senescence. Moreover, the overexpression of SlymiR208, which targets two cytokinin biosynthesis genes (Slipt2 and Slipt4) results in reduced cytokinin levels and accelerated leaf senescence (Zhang et al. 2021). The overexpression and short tandem target mimic (STTM) lines of miR408 also exhibited contrasting dark-induced senescence phenotypes via modulating PCY-SAG14 module and copper reallocation in the chloroplast (Hao et al. 2022). The advent of high-throughput next-generation sequencing (NGS) has further enabled the identification of several small RNAs associated with senescence. For example, six miRNA families, osa-miR159, osa-miR160, osa-miR164, osa-miR167, osa-miR172 and osa-miR1848 were found to be involved in leaf senescence possibly by regulating the phytohormone signalling in rice (Xu et al. 2014). Exploration of miRNAs in stay-green and early leaf senescence lines of maize revealed differential expression of 16 senescence-associated miRNAs (SA-miRNAs) which mainly target TFs and chlorophyll degradation pathway genes (Wu et al. 2016). Sasi et al. (2019) provided the repository of differentially expressed miRNAs during rice flag leaf senescence, wherein 116 novel and 21 known miRNAs were differentially expressed, and their predicted targets encoded for TFs and phytohormone homeostasis pathway components.
Hormonal Regulation of Senescence
Phytohormones, in combination with both developmental and environmental signals, play a significant role in fine-tuning the leaf senescence process. A few of these act as positive regulators while some are negative regulators, and together they comprise a complex network of signalling pathways. Some of these found to control the senescence process are mentioned below:
Oxylipins, including jasmonic acid and its derivatives, are known as jasmonates (JAs) and are vital signalling molecules critical for the development and stress responses in plants (Guo et al. 2021). COI1 (CORONATINE INSENSITIVE 1; a JA receptor) triggers JA signalling pathway by activating ubiquitin-mediated proteasome degradation of JAZ proteins, a repressor of JA response (Woo et al. 2019). Several studies demonstrated a higher level of JA accumulation due to the induction of JA biosynthesis genes during leaf senescence. Research on Arabidopsis revealed that TCP TFs control JA biosynthesis under the tight control of miR319 (Guo et al. 2021). Studies on knock-out mutant of rice COI1 (oscoi1b‐1) demonstrated a stay-green phenotype and down-regulation of SAGs suggestive of a crosstalk between ethylene and JA signalling pathways during senescence (Lee et al. 2015). Antagonistic regulation of JA-induced senescence pathway by bHLH subgroup IIIe (MYC2, MYC3, and MYC4) and IIId (bHLH03, bHLH13, bHLH14, and bHLH17) factors have also been reported (Qi et al. 2015). The MYC2/MYC3/MYC4 proteins reportedly activate the NAC transcription factors (ANAC019/055/072), which in turn trigger chlorophyll catabolic genes (NYC1 and NYE1/SGR1) expression. Very recently, studies on the knockout mutants of JAZ7 revealed that it represses the expression of MYC2 leading to the suppression of JA-induced leaf senescence pathway (Guo et al. 2021). JA also regulates the level of H2O2 by repressing CAT2 expression and thus, triggering leaf senescence.
Ethylene is a gaseous plant growth regulator. Its accumulation has been reported during leaf senescence (Woo et al. 2019). Transcriptomic studies identified many ethylene response factors (ERFs) controlling leaf senescence in many plant species (Zhang et al. 2021). Plants deficient in ACC synthase showed a delayed senescence phenotype, which suggests a positive role of ethylene during leaf senescence (Wang et al. 2002). In Arabidopsis, EIN2 positively regulate leaf senescence by controlling the expression of ORE1/NAC2 (Kim et al. 2009). Further, the over-expression of EIN3 caused early senescence, whereas its silencing resulted in a delayed senescence phenotype (Li et al. 2013). Henceforth, EIN3 was said to act downstream of EIN2 and control the ORE1/NAC2 transcript levels in Arabidopsis. The ORE1/NAC2 is an established positive regulator of leaf senescence given it activates a large set of SAGs.
Cytokinin is a negative regulator of senescence. It is also involved in various plant developmental processes such as apical dominance, shoot and root branching, leaf expansion, the growth of lateral buds, photosynthesis, seed germination, floral transition, and leaf senescence (Lim et al. 2007b). In Nicotiana, the expression of IPT under the control of a senescence-inducible promoter significantly enhanced the leaf longevity, while under a stress-inducible promoter, it delayed drought-induced senescence (Rivero et al. 2007). Higher accumulation of extracellular invertase (CIN1) and hexose transporters were moreover reported upon cytokinin treatments. The cytokinin treatment-based delayed senescence is usually attributable to the consequence of enhanced sink strength of the tissue (Schippers et al. 2015).
Auxin is considered as a senescence-delaying hormone. Its accumulation transiently increases during the onset of senescence. Jiang et al. (2014) demonstrated the repressive role of auxin on SAG12 expression. In auxin treated Arabidopsis, WRKY57 was shown to accumulate and negatively regulate the expression of SAG12 by directly binding to its promoter. Further, WRKY57 was found to competitively bind to JA and auxin pathway repressors JAZ4/8 and IAA29, respectively. These results indicate that the JA-induced senescence process is antagonised by auxin via WRKY57 (Jiang et al. 2014). An elevated level of auxin response pathway repressor ARF2 has also been observed during senescence. Mutants deficient in ARF2 exhibited delayed senescence phenotype, indicating its possible positive role in senescence (Lim et al. 2010). Asides from this, ARF7 and ARF19 have also been reported to regulate the onset of leaf senescence in Arabidopsis (Ellis et al. 2005).
Abscisic acid is most crucial for plant development and responses to various environmental stresses (Guo et al. 2021). ABA has been reported to be an enhancer of senescence. It functions by promoting the expression of SAGs that are mainly related to chloroplast degradation. These include NON-YELLOW COLORING1 (NYC1), STAY-GREEN (SGR), PHEOPHYTINASE (PPH) and PHEIDE A OXYGENASE (PAO) (Pruzinska et al. 2005). Additionally, the expression of many NAC TF family members, including VND-INTERACTING2 (VNI2), A SUBFAMILY OF STRESS-RESPONSIVE NAC (SNAC-A), ORE1, Oryza sativa NAC-LIKE, ACTIVATED BY APETALA3/PISTILLATA (OsNAP) and OsNAC2 were also found to be enhanced upon ABA treatment in Arabidopsis and rice (Woo et al. 2019). A study by Mao et al. (2017) in rice demonstrated that OsNAC2 was actively involved in leaf senescence by altering the expression of chlorophyll degradation genes (OsSGR and OsNYC3). Also, the overexpression of OsNAC2 resulted in the upregulation of ABA biosynthesis genes and down-regulation of ABA catabolic genes. A positive ABA-mediated regulatory role of ATAF1 has also been reported during Arabidopsis leaf senescence. The binding of ATAF1 to the promoters of NAC092 and GLK1 triggers chlorophyll degradation and leaf senescence. Additionally, ATAF1 regulates the ABA-mediated leaf senescence by maintaining the expression of ABA biosynthesis (NCED3) and transport (ABCG40) genes, but the operational upstream transcriptional network remains unclear (Garapati et al. 2015).
Salicylic acid is a phenolic compound that functions as a signalling compound with a primary role in plant immune responses. Salicylic acid (SA) has been known to accumulate in leaves during leaf senescence (Breeze et al. 2011). SA signalling mutants, npr1 and pad4, exhibited a delayed senescence phenotype indicating the participation of SA in leaf senescence (Schippers et al. 2015). Transcriptome studies have revealed that the SA hormone response pathway was exclusive to developmental senescence (Wollaston et al. 2005). A study on senescence-associated ubiquitin ligase1 (saul1) mutants with an early senescence phenotype have also shown a higher-level expression of SA response pathway genes under low-light conditions, which was related to PHYTOALEXIN-DEFICIENT4 (PAD4)-dependent SA biosynthetic pathway (Vogelmann et al. 2012).
Strigolactone (SL) is a recently identified phytohormone involved in many developmental and stress response processes (Guo et al. 2021). The SL-deficient mutants exhibited a ‘stay-green phenotype upon dark treatment. The application of GR24 (a synthetic SL analogue) and ethylene drastically induced SAGs expression and enhanced senescence indicating a positive regulatory role of SL during leaf senescence. The double mutants of max1 and ein2 exhibited an enhanced stay-green phenotype which suggests that SL moderately augments leaf senescence in an ethylene-independent manner (Ueda and Kusaba 2015).
Other Regulators of Senescence
Nitric oxide (NO) is a gaseous free radical and an essential signalling molecule in plants. Transcriptome studies of plants over-expressing NO degrading dioxygenase (NOD) showed a massive gene-expression change, including the down-regulation of photosynthetic genes and up-regulation of several SAGs and ethylene biosynthesis genes. Literature suggests that ethylene and NO control leaf senescence antagonistically. The early senescence phenotype is moreover attenuated when NOD over-expression plants are subjected to NO treatment. Additionally, an induced expression of NOD results in the accumulation of SA in Arabidopsis (Mishina et al. 2007). The gene expression studies with NO-deficient Arabidopsis mutants also revealed the profound upregulation of chlorophyll catabolic pathway genes resulting in an early senescence phenotype. Also, NO was shown to be essential for the stability of thylakoid membranes (Liu and Guo 2013). Reportedly, an altered expression of the amino acid transporter gene, OsAAP3, results in a high-level accumulation of amino acids (arginine and lysine), which facilitates change in NO signalling pathways leading to the formation of lesions and senescence in rice (Wei et al. 2021).
Polyamines (PAs) are ubiquitous polycationic compounds with multiple developmental and physiological functions. The major PAs include putrescine (Put), spermidine (Spd), spermine (Spm), and thermo-Spm (t-Spm). PAs were shown to inhibit senescence in oat, barley and petunia (Nowicka 2017). This inhibition has been demonstrated to be mediated by the reduced activity of RNase, chlorophyll degradation and LHCII protein degradation. Dark-induced senescence results in a higher accumulation of Put, Spd and Spm at the initial stages, but their level declines during the later stage of senescence (Nowicka 2017). The association of PA accumulation with ROS scavenging capacity at the onset of the senescence process is yet unknown.
Melatonin is one of the primitive biomolecules, ubiquitously found from photosynthetically autotrophic bacteria to mammals and higher plants and has been well characterized as an anti-ageing agent in animals since its discovery. In recent years, melatonin has been linked to delayed leaf senescence (Zhao et al. 2021). The emerging research in melatonin biosynthesis indicates its inhibitory role in leaf senescence. The delayed leaf senescence phenotype obtained upon melatonin application is attributed to the cuticular structure maintenance, balancing of redox homeostasis and prevention of chlorophyll catabolism (Zhao et al. 2021). However, the genetic and molecular basis of melatonin treatment-based delayed leaf senescence is yet to be explored.
Conclusions and future perspectives
The terminal phase of plant development, including leaf senescence or aging, ensures nutrient recycling, plant fitness and reproductive success. This is an enormously intricate, yet highly orchestrated and regulated process. Decades of research encompassing several physiological and biochemical studies, application of genomics and other multi-omics approaches, etc., have answered some of the fundamental questions on the components associated with leaf senescence. These include the contribution of internal and external factors and multi-layered regulation occurring along the progression of senescence. The identified commonalities and interactions among the molecular components and hormones regulating various stress responses and leaf senescence highlight the crosstalk between development and several stress-responsive pathways. However, limited information is available on how plants or their organs coordinate these complex diverse processes together to decide when and how to die. Most of the studies have been carried out at the organ level, which is a culmination of coordinated cell development. Nevertheless, it is pertinent to investigate senescence at the cellular level. Further, the development of new assay systems and monitoring protocols for validating the molecular basis of senescence across different plant systems is also imperative. Few new regulators of senescence have been identified such as melatonin, polyamines and nitric oxide, however, the genetic and molecular basis of their action needs to be explored. In this regard, systems biology-based approaches would be instrumental in obtaining an accurate and detailed knowledge of the molecular networks associated with the regulation of leaf senescence. All this information would benefit us in designing strategies for fine-tuning senescence in crops species to improve their quality and productivity.
References
Albacete AA, Martínez-Andújar C, Pérez-Alfocea F (2014) Hormonal and metabolic regulation of source-sink relations under salinity and drought: from plant survival to crop yield stability. Biotechnol Adv 32:12–30
Ay N, Irmler K, Fischer A et al (2009) Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J 58(2):333–346
Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol 10:63–75
Balazadeh S, Siddiqui H, Allu AD et al (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62(2):250–264
Barros JAS, Cavalcanti JHF, Pimentel KG et al (2022) The significance of WRKY45 transcription factor in metabolic adjustments during dark-induced leaf senescence. Plant Cell Environ 45(9):2682–2695
Bazargani MM, Sarhadi E, Bushehri AAS et al (2011) A proteomics view on the role of drought-induced senescence and oxidative stress defense in enhanced stem reserves remobilization in wheat. J Proteom 74(10):1959–1973
Breeze E, Harrison E, McHattie S et al (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23(3):873–894
Broda M, Khan K, O’Leary B et al (2021) Increased expression of ANAC017 primes for accelerated senescence. Plant Physiol 186:2205–2221
Brusslan JA, Alvarez-Canterbury AM, Nair NU et al (2012) Genome-wide evaluation of histone methylation changes associated with leaf senescence in Arabidopsis. PLoS One 7(3):e33151
Brychkova G, Alikulov Z, Fluhr R et al (2008) A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J 54(3):496–509
Buchanan-Wollaston V, Earl S, Harrison E et al (2003) The molecular analysis of leaf senescence–a genomics approach. Plant Biotechnol J 1:3–22
Buchanan-wollaston V, Page T, Harrison E et al (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42:567–585
Caselles V, Casadesús A, Munné-Bosch S (2021) A dual role for abscisic acid integrating the cold stress response at the whole-plant level in Iris pseudacorus L. growing in a natural Wetland. Front Plant Sci 12:2738
Chen MK, Lee PF, Yang CH (2011) Delay of flower senescence and abscission in arabidopsis transformed with an FOREVER YOUNG FLOWER homolog from oncidium orchid. Plant Signal Behav 6(11):1841–1843
Chen GH, Chan YL, Liu CP et al (2012) Ethylene response pathway is essential for Arabidopsis A-FIFTEEN function in floral induction and leaf senescence. Plant Signal Behav 7(4):457–460
Chen LJ, Wuriyanghan H, Zhang YQ et al (2013) An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol 163(4):1752–1765
Chen M, Maodzeka A, Zhou L et al (2014) Removal of DELLA repression promotes leaf senescence in Arabidopsis. Plant Sci 219–220:26–34
Chen D, Wang S, Xiong B et al (2015) Carbon/nitrogen imbalance associated with drought-induced leaf senescence in sorghum bicolor. PLoS One 10:e0137026
Chen X, Lu L, Mayer KS et al (2016) POWERDRESS interacts with HISTONE DEACETYLASE 9 to positive aging in Arabidopsis. eLife 5:e17214
Chen L, Xiang S, Chen Y et al (2017) Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol Plant 10(9):1174–1189
Chen G, Wu C, He L et al (2018) Knocking out the gene RLS1 induces hypersensitivity to oxidative stress and premature leaf senescence in rice. Int J Mol Sci 19:1–16
Danisman S, van der Wal F, Dhondt S et al (2012) Arabidopsis class i and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159(4):1511–1523
Dong S, Sang L, Xie H et al (2021) Comparative transcriptome analysis of salt stress-induced leaf senescence in Medicago truncatula. Front Plant Sci 12:1378
Dubousset L et al (2009) Remobilization of leaf S compounds and senescence in response to restricted sulphate supply during the vegetative stage of oilseed rape are affected by mineral N availability. J Exp Bot 60:3239–3253
Ellis CM, Nagpal P, Young JC et al (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132(20):4563–4574
Fan J, Lou Y, Shi H et al (2019) Transcriptomic analysis of dark-induced senescence in bermudagrass (Cynodon dactylon). Plants 8:614
Fanata WI, Lee KH, Son BH et al (2013) N-glycan maturation is crucial for cytokinin-mediated development and cellulose synthesis in Oryza sativa. Plant J Cell Mol Biol 73(6):966–979
Fedyaeva AV, Stepanov AV, Lyubushkina IV et al (2014) Heat shock induces production of reactive oxygen species and increases inner mitochondrial membrane potential in winter wheat cells. Biochem 79:1202–1210
Feng S, Jacobsen SE, Reik W (2010) Epigenetic reprogramming in plant and animal development. Science (New York, N.Y.) 330(6004):622–627
Gao S, Gao J, Zhu X et al (2016) ABF2, ABF3, and ABF4 positive ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol Plant 9(9):1272–1285
Garapati P, Xue GP, Munné-Bosch S et al (2015) Transcription factor ATAF1 in arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol 168:1122–1139
Gepstein S, Sabehi G, Carp M-J et al (2003) Large-scale identification of leaf senescence-associated genes. Plant J Cell Mol Biol 36(5):629–642
Guo FQ, Crawford NM (2005) Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-. Plant Cell 17(12):3436–3450
Guo Y, Gan S (2011) AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol 156(3):1612–1619
Guo W, Nazim H, Liang Z et al (2016) Magnesium deficiency in plants: an urgent problem. Crop J 4:83–91
Guo P, Li Z, Huang P et al (2017) A tripartite ampli fi cation loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29(November):2854–2870
Guo Y, Ren G, Zhang K et al (2021) Leaf senescence: progression, regulation, and application. Mol Horticult 1(1):1–25
Hao C, Yang Y, Du J et al (2022) The PCY-SAG14 phytocyanin module regulated by PIFs and miR408 promotes dark-induced leaf senescence in Arabidopsis. Proc Natl Acad Sci U S A 119:1–10
He Y, Fukushige H, Hildebrand DF et al (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128(3):876–884
He L, Wu W, Zinta G et al (2018) A naturally occurring epiallele associates with leaf senescence and local climate adaptation in Arabidopsis accessions. Nat Commun 9(1):1–11
He Y, Zhang X, Shi Y, Xu X, Li L, Wu JL (2021) PREMATURE SENESCENCE LEAF 50 promotes heat stress tolerance in rice (Oryza sativa L.). Rice 14(1):1–7
Hensel LL, Grbić V, Baumgarten DA et al (1993) Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5(5):553–564
Hinckley WE, Keymanesh K, Cordova JA et al (2019) The HAC1 histone acetyltransferase promotes leaf senescence and regulates the expression of ERF022. Plant Direct 3(8):e00159
Hsu CY, Chou ML, Wei WC et al (2022) Chloroplast protein Tic55 involved in dark-induced senescence through AtbHLH/AtWRKY-ANAC003 controlling pathway of Arabidopsis thaliana. Genes 13(2):1–23
Huang J, Cai M, Long Q et al (2014) OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity. Transgen Res 23(4):643–655
Huang D, Lan W, Li D et al (2018) WHIRLY1 occupancy affects histone lysine modification and WRKY53 transcription in Arabidopsis developmental manner. Front Plant Sci 871:1503
Huang D, Lan W, Ma W et al (2022) WHIRLY1 recruits the histone deacetylase HDA15 repressing leaf senescence and flowering in Arabidopsis. J Integr Plant Biol 64:1411–1429
Ichikawa M, Nakai Y, Arima K et al (2015) A VAMP-associated protein, PVA31 is involved in leaf senescence in Arabidopsis. Plant Signal Behav 10(3):e990847
Jajic I, Sarna T, Strzalka K (2015) Senescence, stress, and reactive oxygen species. Plants 4:393
Jaradat MR, Feurtado JA, Huang D et al (2013) Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol 13(1):1–19
Jiang Y, Liang G, Yang S et al (2014) Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26:230–245
Jing S, Zhou X, Song Y et al (2009) Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul 58(2):181–190
Kamranfar I, Xue GP, Tohge T et al (2018) Transcription factor RD26 is a key regulator of metabolic reprogramming during dark-induced senescence. New Phytol 218(4):1543–1557
Kan C, Zhang Y, Wang HL et al (2021) Transcription factor NAC075 delays leaf senescence by deterring reactive oxygen species accumulation in Arabidopsis. Front Plant Sci 12(February):1–11
Kim HJ, Ryu H, Hong SH et al (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103(3):814–819
Kim M, Ohr H, Lee JW et al (2008) Temporal and spatial downregulation of Arabidopsis MET1 activity results in global DNA hypomethylation and developmental defects. Mol Cells 26(6):611–615
Kim JH, Woo HR, Kim J et al (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323(5917):1053–1057
Kim YS, Sakuraba Y, Han SH et al (2013) Mutation of the Arabidopsis NAC016 transcription factor Negatives leaf senescence. Plant Cell Physiol 54(10):1660–1672
Kim J, Woo HRR, Nam HGG (2016) Toward systems understanding of leaf senescence: an integrated multi-omics perspective on leaf senescence research. Mol Plant 9(6):813–825
Kim H, Kim HJ, Vu QT et al (2018) Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc Natl Acad Sci USA 115(33):8448–8453
Kim T, Kang K, Kim SH et al (2019) OsWRKY5 promotes rice leaf senescence via senescence-associated NAC and abscisic acid biosynthesis pathway. Int J Mol Sci 20(18):4437
Kim I, Kim E-H, Choi Y et al (2022) Fibrillin2 in chloroplast plastoglobules participates in photoprotection and jasmonate-induced senescence. Plant Physiol 189:1363–1379
Kocourková D, Kroumanová K, Podmanická T et al (2021) Phospholipase Dα1 acts as a negative regulator of high Mg2+-induced leaf senescence in Arabidopsis. Front Plant Sci 12:2645
Koyama T, Nii H, Mitsuda N et al (2013) A regulatory cascade involving class II ETHYLENE RESPONSE FACTOR transcriptional repressors operates in the progression of leaf senescence. Plant Physiol 162(2):991–1005
la Haba PD, la Mata LD, Molina E et al (2014) High temperature promotes early senescence in primary leaves of sunflower (Helianthus annuus L.) plants. Can J Plant Sci 94(4):659–669
Law JA, Jacobsen SE (2011) Establising, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11(3):204–220
Lee S, Seo PJ, Lee HJ et al (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 70(5):831–844
Lee SH, Sakuraba Y, Lee T et al (2015) Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J Integr Plant Biol 57(6):563–576
Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90(5):929–938
Li Z, Peng J, Wen X et al (2013) ETHYLENE-INSENSITIVE3 Is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25(9):3311
Li L, Kubiszewski-Jakubiak S, Radomiljac J et al (2016) Characterization of a novel β-barrel protein (AtOM47) from the mitochondrial outer membrane of Arabidopsis thaliana. J Exp Bot 67(21):6061–6075
Li Z, Kim JH, Kim J et al (2020) ATM suppresses leaf senescence triggered by DNA double-strand break through epigenetic control of senescence-associated genes in Arabidopsis. New Phytol 227(2):473–484
Li N, Bo C, Zhang Y et al (2021a) PHYTOCHROME INTERACTING FACTORS PIF4 and PIF5 promote heat stress induced leaf senescence in Arabidopsis. J Exp Bot 72:4577–4589
Li Y, Liao S, Mei P et al (2021b) OsWRKY93 dually functions between leaf senescence and in response to biotic stress in rice. Front Plant Sci 12:327
Liang C, Wang Y, Zhu Y et al (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci 111(27):10013–10018
Lim PO, Woo HR, Nam HG (2003) Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci 8:272–278
Lim PO, Kim Y, Breeze E et al (2007a) Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J 52(6):1140–1153
Lim PO, Kim HJ, Nam HG (2007b) Leaf senescence. Annu Rev Plant Biol 58:115–136
Lim PO, Lee IC, Kim J et al (2010) Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J Exp Bot 61(5):1419–1430
Lin JF, Wu SH (2004) Molecular events in senescing Arabidopsis leaves. Plant J Cell Mol Biol 39(4):612–628
Lin M, Pang C, Fan S et al (2015) Global analysis of the Gossypium hirsutum L. Transcriptome during leaf senescence by RNA-Seq. BMC Plant Biol 15:43
Liu F, Guo F-Q (2013) Nitric oxide deficiency accelerates chlorophyll breakdown and stability loss of thylakoid membranes during dark-induced leaf senescence in Arabidopsis. PLoS One 8:e56345
Liu J, Shen J, Xu Y et al (2016) Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J Exp Bot 67(19):5785–5798
Liu P, Zhang S, Zhou B et al (2019) The histone H3K4 demethylase JMJ16 represses leaf. Plant Cell 31:430–443
Lohman KN, Gan S, John MC et al (1994) Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol Plant 92(2):322–328
Luan WJ, Shen A, Jin ZP et al (2013) Knockdown of OsHox33, a member of the class III homeodomain-leucine zipper gene family, accelerates leaf senescence in rice. Sci China Life Sci 56(12):1113–1123
Mao C, Lu S, Lv B et al (2017) A rice nac transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol 174:1747–1763
Masclaux-Daubresse C, Purdy S, Lemaitre T et al (2007) Genetic variation suggests interaction between cold acclimation and metabolic regulation of leaf senescence. Plant Physiol 143(1):434–446
Matsuoka D, Yasufuku T, Furuya T et al (2015) An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol Biol 87(6):565–575
Miryeganeh M (2022) Epigenetic mechanisms of senescence in plants. Cells 11(2):251
Mishina TE, Lamb C, Zeier J (2007) Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ 30(1):39–52
Moritoh S, Eun CH, Ono A et al (2012) Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J 71(1):85–98
Morris K, MacKerness SA, Page T et al (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J Cell Mol Biol 23(5):677–685
Morrissey J, Guerinot ML (2009) Iron uptake and transport in plants: the good, the bad, and the ionome. Chem Rev 109(10):4553–4567
Munné-Bosch S, Alegre L (2002) Plant aging increases oxidative stress in chloroplasts. Planta 214:608–615
Munné-Bosch S, Alegre L (2004) Die and let live: leaf senescence contributes to plant survival under drought stress Sergi. Funct Plant Biol 31:203–216
Nagahage ISP, Sakamoto S, Nagano M et al (2020) An Arabidopsis NAC domain transcription factor, ATAF2, promotes age-dependent and dark-induced leaf senescence. Physiol Plant 170(2):299–308
Nakabayashi K, Ito M, Kiyosue T et al (1999) Identification of clp genes expressed in senescing Arabidopsis leaves. Plant Cell Physiol 40(5):504–514
Navabpour S, Morris K, Allen R et al (2003) Expression of senescence-enhanced genes in response to oxidative stress. J Exp Bot 54:2285–2292
Nie G, Yang Z, He J et al (2021) Genome-wide investigation of the NAC transcription factor family in miscanthus sinensis and expression analysis under various abiotic stress. Front Plant Sci 12:2464
Oda-Yamamizo C, Mitsuda N, Sakamoto S et al (2016) The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci Rep 6(January):1–13
Ogneva ZV, Dubrovina AS, Kiselev KV (2016) Age-associated alterations in DNA methylation and expression of methyltransferase and demethylase genes in Arabidopsis thaliana. Biol Plant 60(4):628–634
Panchuk II, Zentgraf U, Volkov RA (2005) Expression of the Apx gene family during leaf senescence of Arabidopsis thaliana. Planta 222(5):926–932
Panigrahy M, Singh A, Das S et al (2021) Co-action of ABA, brassinosteriod hormone pathways and differential regulation of different transcript isoforms during cold-and-dark induced senescence in Arabidopsis. J Plant Biochem Biotechnol 31(3):489–510
Park SJ, Park S, Kim Y et al (2022) Ethylene responsive factor34 mediates stress-induced leaf senescence by regulating salt stress-responsive genes. Plant Cell Environ 45(6):1719–1733
Parveen S, Ranjan RK, Anand A et al (2018) Combined deficiency of nitrogen and iron increases senescence induced remobilization of plant immobile iron in wheat. Acta Physiol Plant 40:1–12
Patel S, Kumar DSP (2008) Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4(1):20–27
Pham G, Shin DM, Kim Y et al (2022) Ran-GTP/-GDP-dependent nuclear accumulation of nonexpressor of pathogenesis-related genes1 and tgacg-binding factor2 controls salicylic acid-induced leaf senescence. Plant Physiol 189(3):1774–1793
Piao W, Kim SH, Lee BD et al (2019) Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J Exp Bot 70(10):2699–2715
Pruzinska A, Tanner G, Aubry S et al (2005) Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction 1. Plant Physiol 139:52–63
Qi T, Wang J, Huang H et al (2015) Regulation of Jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in arabidopsis. Plant Cell 27(June):1634–1649
Qiao H, Liu Y, Cheng L et al (2021) TaWRKY13-A serves as a mediator of jasmonic acid-related leaf senescence by modulating Jasmonic acid biosynthesis. Front Plant Sci 12:717233
Raines T, Shanks C, Cheng CY et al (2016) The cytokinin response factors modulate root and shoot growth and Positive leaf senescence in Arabidopsis. Plant J Cell Mol Biol 85(1):134–147
Rapp YG, Ransbotyn V, Grafi G (2015) Senescence meets dedifferentiation. Plants (Basel, Switzerland) 4(3):356–368
Rauf M, Arif M, Dortay H et al (2013) ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Rep 14(4):382–388
Raxwal V, Katiyar-Agarwal S, Agarwal M (2012) Structural and functional diversity of plant heat shock factors. In Plant Stress ©2012 Global Science Books, pp 89–96.
Reguera M, Peleg Z, Abdel-Tawab YM et al (2013) Stress-induced cytokinin synthesis increases drought tolerance through the coordinated regulation of carbon and nitrogen assimilation in rice. Plant Physiol 163(4):1609–1622
Ren Y, Li M, Wang W et al (2022) MicroRNA840 (MIR840) accelerates leaf senescence by targeting the overlapping 3′UTRs of PPR and WHIRLY3 in Arabidopsis thaliana. Plant J 109(1):126–143
Rivero RM, Kojima M, Gepstein A et al (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104(49):19631–19636
Robatzek S, Somssich IE (2001) A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J Cell Mol Biol 28(2):123–133
Sakuraba Y, Jeong J, Kang M-Y et al (2014a) Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun 5:4636
Sakuraba Y, Kim D, Kim YS et al (2014b) Arabidopsis STAYGREEN-LIKE (SGRL) positives abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett 588(21):3830–3837
Sakuraba Y, Kim YS, Han SH et al (2015a) The arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27(6):1771–1787
Sakuraba Y, Piao W, Lim JH et al (2015b) Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol 56(12):2325–2339
Sakuraba Y, Kim D, Han SH et al (2020) Multilayered regulation of membrane-bound ONAC054 is essential for abscisic acid-induced leaf senescence in rice. Plant Cell 32(3):630–649
Sasi JM, Kumar CV, Mani B et al (2019) Identification and characterization of miRNAs during flag leaf senescence in rice by high-throughput sequencing. Plant Physiol Reports 24(1):1–14
Sato Y, Morita R, Katsuma S et al (2009) Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J 57(1):120–131
Schippers JHM, Schmidt R, Wagstaff C et al (2015) Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiol 169:914–930
Schommer C, Palatnik JF, Aggarwal P et al (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6(9):e230
Schulte G, Begum N, Worku M et al (2007) Leaf senescence induced by nitrogen deficiency as indicator of genotypic differences in nitrogen efficiency in tropical maize. J Plant Nutr Soil Sci 170(1):106–114
Sharabi-Schwager M, Lers A, Samach A et al (2010) Overexpression of the CBF2 transcriptional activator in Arabidopsis delays leaf senescence and extends plant longevity. J Exp Bot 61(1):261–273
Smykowski A, Zimmermann P, Zentgraf U (2010) G-Box Binding Factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis. Plant Physiol 153(3):1321–1331
Sobieszczuk-Nowicka E (2017) Polyamine catabolism adds fuel to leaf senescence. Amino Acids 49(1):49–56
Sobieszczuk-Nowicka E, Wrzesiński T, Bagniewska-Zadworna A et al (2018) Physio-genetic dissection of dark-induced leaf senescence and timing its reversal in Barley1. Plant Physiol 178:654–671
Springer A, Kang C, Rustgi S et al (2016) Programmed chloroplast destruction during leaf senescence involves 13-lipoxygenase (13-LOX). Proc Natl Acad Sci USA 113(12):3383–3388
Thomas H (2013) Senescence, ageing and death of the whole plant. New Phytol 197(3):696–711
Thompson AR, Doelling JH, Suttangkakul A et al (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138(4):2097–2110
Trejo-Arellano MS, Mehdi S, de Jonge J, Dvorák Tomastíková E, Köhler C, Hennig L (2020) Dark-Induced Senescence Causes Localized Changes in DNA Methylation. Plant Physiol 182(2):949–961
Ueda H, Kusaba M (2015) Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol 169(1):138–147
Van der Graaff E, Schwacke R, Schneider A et al (2006) Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141(2):776–792
Veliz CG, Criado MV, Galotta MF et al (2020) Regulation of senescence-associated protease genes by sulphur availability according to barley (Hordeum vulgare L.) phenological stage. Ann Bot 126:435
Vogelmann K, Drechsel G, Bergler J et al (2012) Early senescence and cell death in arabidopsis saul1 mutants involves the PAD4-dependent salicylic acid pathway. Plant Physiol 159(4):1477–1487
Wang KL-C, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14(suppl 1):S131–S151
Wang SH, Lim JH, Kim SS et al (2015) Mutation of SPOTTED LEAF3 (SPL3) impairs abscisic acid-responsive signalling and delays leaf senescence in rice. J Exp Bot 66(22):7045–7059
Wang Z, Qian C, Guo X et al (2016) ELS1, a novel MATE transporter related to leaf senescence and iron homeostasis in Arabidopsis thaliana. Biochem Biophys Res Commun 476(4):319–325
Wang H, Chang X, Lin J et al (2018) Transcriptome profiling reveals regulatory mechanisms underlying corolla senescence in petunia. Hortic Res 5:16
Weaver LM, Amasino RM (2001) Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol 127(3):876–886
Wei T, Ou B, Li J et al (2013) Transcriptional profiling of rice early response to Magnaporthe oryzae identified OsWRKYs as important regulators in rice blast resistance. PLoS One 8(3):e59720
Wei Q, Yan Z, Xiong Y, Fang Z (2021) Altered expression of OsAAP3 influences rice lesion mimic and leaf senescence by regulating arginine transport and nitric oxide pathway. Int J Mol Sci 22(4):2181
Wei YQ, Yuan JJ, Xiao CC et al (2022) RING-box proteins regulate leaf senescence and stomatal closure via repression of ABA transporter gene ABCG40. J Integrat Plant Biol 64:979–994
Windram O, Madhou P, Mchattie S et al (2012) Arabidopsis defense against botrytis Cinerea: chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 24(9):3530–3557
Woo HR, Kim JH, Nam HG et al (2004) The delayed leaf senescence mutants of Arabidopsis, ore1, ore3, and ore9 are tolerant to oxidative stress. Plant Cell Physiol 45(7):923–932
Woo HR, Kim JH, Kim J et al (2010) The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J Exp Bot 61(14):3947–3957
Woo HR, Kim HJ, Lim PO et al (2019) Leaf senescence: systems and dynamics aspects. Annu Rev Plant Biol 70:347–376
Wu K, Zhang L, Zhou C et al (2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59(2):225–234
Wu A, Allu AD, Garapati P et al (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. The Plant Cell Online 24(2):482–506
Wu X, Ding D, Shi C et al (2016) microRNA-dependent gene regulatory networks in maize leaf senescence. BMC Plant Biol 73:16
Xie Y, Huhn K, Brandt R et al (2014) Revoluta and wrky53 connect early and late leaf development in Arabidopsis. Dev 141:4772–4783
Xu Y, Huang B (2007) Heat-induced leaf senescence and hormonal changes for thermal bentgrass and turf-type bentgrass species differing in heat tolerance. J Am Soc Hortic Sci 132:185–192
Xu F, Meng T, Li P et al (2011) A soybean dual-specificity kinase, GmSARK, and its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol 157(4):2131–2153
Xu X, Bai H, Liu C et al (2014) Genome-wide analysis of microRNAs and their target genes related to leaf senescence of rice. PLoS One 9(12):e114313
Xu P, Chen H, Cai W (2020) Transcription factor CDF4 promotes leaf senescence and floral organ abscission by regulating abscisic acid and reactive oxygen species pathways in Arabidopsis. EMBO Rep 21(7):1–20
Yamatani H, Sato Y, Masuda Y et al (2013) NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll - protein complexes during leaf senescence. Plant J Cell Mol Biol 74(4):652–662
Yamatani H, Kohzuma K, Nakano M et al (2018) Impairment of Lhca4, a subunit of LHCI, causes high accumulation of chlorophyll and the stay-green phenotype in rice. J Exp Bot 69(5):1027–1035
Yan Z, Jia J, Yan X et al (2017) Arabidopsis KHZ1 and KHZ2, two novel non-tandem CCCH zinc-finger and K-homolog domain proteins, have redundant roles in the regulation of flowering and senescence. Plant Mol Biol 95(6):549–565
Yang J, Worley E, Udvardi M (2014) A NAP-AAO3 regulatory module promotes chlorophyll degradation via aba biosynthesis in arabidopsis leavesw open. Plant Cell 26(12):4862–4874
Yang HF, Lu XY, Chen HB et al (2017) Low temperature-induced leaf senescence and the expression of senescence-related genes in the panicles of Litchi chinensis. Biol Plant 61:315–322
Yoshida S (2003) Molecular regulation of leaf senescence. Curr Opin Plant Biol 6:79–84
Yu J, Zhang Y, Di C et al (2016) JAZ7 negatively regulates dark-induced leaf senescence in Arabidopsis. J Exp Bot 67(3):751–762
Yuan L, Wang D, Cao L et al (2020) Regulation of leaf longevity by DML3-Mediated DNA demethylation. Mol Plant 13(8):1149–1161
Zakari SA, Asad MAU, Han Z et al (2020) Relationship of nitrogen deficiency-induced leaf senescence with ROS generation and ABA concentration in rice flag leaves. J Plant Growth Regul 39:1503–1517
Zareen S, Ali A, Lim CJ et al (2022) The transcriptional corepressor HOS15 mediates dark-induced leaf senescence in Arabidopsis. Front Plant Sci 13:828264
Zeng DD, Yang CC, Qin R et al (2018) A guanine insert in OsBBS1 leads to early leaf senescence and salt stress sensitivity in rice (Oryza sativa L.). Plant Cell Reports 37(6):933–946
Zhang K, Gan SS (2012) An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol 158(2):961–969
Zhang K, Xia X, Zhang Y et al (2012) An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J 69(4):667–678
Zhang S, Li C, Wang R et al (2017) The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol 173(4):2294–2307
Zhang YM, Guo P, Xia X et al (2021) Multiple layers of regulation on leaf senescence: new advances and perspectives. Front Plant Sci 12:2741
Zhang Y, Tan S, Gao Y et al (2022a) CLE42 delays leaf senescence by antagonizing ethylene pathway in Arabidopsis. New Phytol 235(2):550–562
Zhang Z, Liu C, Li K et al (2022b) CLE14 functions as a “brake signal” to suppress age-dependent and stress-induced leaf senescence by promoting JUB1-mediated ROS scavenging in Arabidopsis. Mol Plant 15(1):179–188
Zhao L, Zhang H, Zhang B et al (2012) Physiological and molecular changes of detached wheat leaves in responding to various treatments. J Integr Plant Biol 54:567–576
Zhao D, Derkx AP, Liu DC et al (2015) Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol 17(4):904–913
Zhao Y, Chan Z, Gao J et al (2016) ABA receptor PYL9 Positives drought resistance and leaf senescence. Proc Natl Acad Sci USA 113(7):1949–1954
Zhao YQ, Zhang ZW, Chen YE et al (2021) Melatonin: a potential agent in delaying leaf senescence. Crit Rev Plant Sci 40(1):1–22
Zhou X, Jiang Y, Yu D (2011) WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol Cells 31(4):303
Zhou Y, Zhang X, Chen J et al (2022) Overexpression of AHL9 accelerates leaf senescence in Arabidopsis thaliana. BMC Plant Biol 22(1):1–12
Zhu X, Chen J, Xie Z et al (2015) Jasmonic acid promotes degreening via MYC2 3 4- and ANAC019 055 072-mediated regulation of major chlorophyll catabolic genes. Plant J 84:597–610
Zhu Z, Li G, Yan C et al (2019) DRL1, encoding a NAC transcription factor, is involved in leaf senescence in grapevine. Int J Mol Sci 20:1–16
Acknowledgements
Research fundings from PURSE (Promotion of University Research and Scientific Excellence) and CRG (Core Research Grant) grants of Department of Science and Technology (DST), Government of India; DU-IoE (Delhi University-Institute of Eminence, IoE/FRP/LS/2020/27), Delhi, India; UGC (University Grants Commission, 41-512/2021(SR)) and DBT (Department of Biotechnology), Government of India are acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sasi, J.M., Gupta, S., Singh, A. et al. Know when and how to die: gaining insights into the molecular regulation of leaf senescence. Physiol Mol Biol Plants 28, 1515–1534 (2022). https://doi.org/10.1007/s12298-022-01224-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-022-01224-1