Abstract
GSTP1 involved in the metabolism of carcinogens and toxins, reduces damage of DNA and act as a suppressor of carcinogenesis. Many studies have reported that 313 A > G polymorphism is associated with different cancer in Indian population, but the results remain conflicting rather than conclusive. Therefore, we have performed meta-analysis to clarify the more precise association of GSPT1 313 A > G polymorphism with cancer risk in Indian population. We retrieved all relevant published literature from PubMed (Medline) and Google scholar web database and included those study only based on the established inclusion criteria. Pooled ORs and 95% CIs were used to appraise the strength of association. Publication bias and sensitivity analysis was also evaluated. A total of 6581 confirmed cancer cases and 8218 controls were included from eligible thirty nine case–controls studies. Pooled analysis suggested that the variant genotypes significantly increased the risk of cancer in allele (G vs. A: OR 1.266, 95% CI 1.129–1.418, p = 0.001), heterozygous (AG vs. AA: OR 1.191, 95% CI 1.047–1.355, p = 0.008), homozygous (GG vs. AA: OR 1.811, 95% CI 1.428–2.297, p = 0.001), dominant (GG + AG vs. AA: OR 1.276, 95% CI 1.110–1.466, p = 0.001) and recessive (GG vs. AG + AA: OR 1.638, 95% CI 1.340–2.002, p = 0.001) genetic models. The stability of these observations was confirmed by a sensitivity analysis. Begger’s funnel plot and Egger’s test did not reveal any publication bias. This meta-analysis suggests that the GSTP1 313 A > G polymorphism may contribute to genetic susceptibility to cancer in Indian population. However, larger studies and randomized clinical trial will be required to elucidate the biological and molecular mechanism of GSTP1 gene in cancer.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Cancer is the most dreadful disease and is the leading cause of high morbidity and mortality in worldwide [1]. Approximately 70% of deaths from cancer occur in developing countries. In India, cancer incidence is predicted to reach 1,148,757 cases in the year 2020 that may lead to a huge socio-economic burden [2]. Cancer is considered as a polygenic disease, whose pathogenesis and molecular mechanism are still intricated and difficult to resolve [3]. Epidemiological studies indicate that interaction between genetic susceptibility genes with an environmental factors and metabolism dysfunction play a key role in development of cancer [4]. Host genetic factors make it even more complex as all individuals who are exposed to these risk factors will not develop the disease since inter-individual differences in genetic susceptibility exist. Identification of host genes and genetic variation in an individual patient may contribute to new approaches to treatment and prevent cancer adeptly [5].
The genes responsible for metabolizing the tobacco carcinogens appear to be prime candidates for the investigative search of cancer susceptibility genes. Glutathione S-Transferases (GSTs) superfamily consists of the broadly expressed phase II xenobiotic metabolizing enzymes located in cytosol. GSTs mediate the conjugation of reduced glutathione with a variety of endogenous and exogenous electrophilic compounds, including several potentially toxic carcinogens and chemotherapeutic drugs, thereby reducing the reactivity of the compounds by making them water soluble and facilitating their elimination from the body for critical defense against carcinogens [6].
Pi-class glutathione-S-transferase (GSTP1) gene spanning approximately 2.84 kb is located on chromosome 11q13 encodes a phase II metabolic enzyme [7], play a key role in the inactivation of toxic and carcinogenic electrophiles [8]. Several single nucleotide polymorphisms (SNPs) has been reported in GSTP1 that lead to changes in amino acids. Among them one is characterized by an A → G transition at nucleotide 313, which replaces ATC (isoleucine) at codon 105 with GTC (valine) (I105V) within the active site of the enzyme [9]. This substitution results in a lower enzymatic activity and is associated with higher hydrophobic adduct levels and higher levels of polycyclic aromatic hydrocarbon-DNA adducts in human lymphocytes [10]. Recent genome-wide association studies (GWAS) have clearly unveiled that SNPs is the most common forms of human genetic variation have an important role in defining individual susceptibility to cancer [11].
Considering the importance of GSTP1 in the detoxification process and protect cell from various carcinogens, the possible influence of 313 (A > G) (rs1695) polymorphism in GSTP1 gene on different cancer risk in Indian population has been investigated extensively [12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50]. However, the results from these studies are inconsistent. Individual published studies contained small number of subjects and may have been underpowered to detect the modest effects of the GSTP1 313 (A > G) polymorphism on cancer susceptibility. To overcome this situation, nowadays meta-analysis statistical tool is used to explore the host risk factors associated with the complex diseases, because it employs a quantitative method to combine the data drawn from individual studies where sample sizes are small to provide reliable conclusions [51]. Given these inconclusive results and the limits of a single study with a small sample size, we performed the present meta-analysis on all eligible published studies in Indian populations to estimate the cumulative association of GSTP1 313 (A > G) gene polymorphism and overall cancer susceptibility.
Materials and Methods
Identification and Eligibility of Studies
The relevant research studies were searched in PubMed, Medline and Google Scholar electronic databases updated in February 2018. The search key words were ““cancer”, “carcinoma,” “malignancy”, and “tumor”, “Glutathione-S-transferase”, “GSTP1”, “Glutathione-S-transferase P1”, and “genetic polymorphism”, “single nucleotide polymorphism”, “genetic variants”, and ‘‘Indian”, “India’’. Furthermore, manual retrieval was undertaken additionally by browsing the references from retrieved articles for other eligible studies. If the same study was researched by more than one study, only the one with the largest sample size was included in our study. If one study investigated multiple cancers, each cancer type was counted as a separate comparison in the group stratified by cancer type. All retrieved articles were downloaded and further screened to identify potentially eligible studies.
Inclusion and Exclusion Criteria
Studies included in the meta-analysis had to meet the following inclusion criteria: (1) original case–control or cohort studies; (2) Cancers cases should have been confirmed by histology or pathology; (3) must have investigated the association between GSTP1 313 A > G polymorphism and cancer susceptibility in Indian population; (4) provided detailed frequency of genotype distribution in the cases and controls. The criteria for exclusion were (1) case reports, editorial, reviews, overlapped data, animal or mechanism studies; (2) no genotype frequency or genotype information provided; (3) no usable data reported.
Data Extraction and Quality Assessment
To minimize the bias and improve the reliability, the methodological quality assessment and data extraction were independently extracted from all eligible studies by two researchers according to the inclusion–exclusion criteria mentioned above. The data collected from each study were as follows: first author’s name, publication year, cancer type, genotyping method, and genotype distribution in cases and controls. Based on the main cancer type of the included studies, cancer types were classified. Disagreement was solved by full discussion until a consensus was reached.
Statistical Analysis
The strength of relationships between GSTP1 313 (A > G) polymorphism and cancer risk was estimated by calculating pooled ORs and their corresponding 95% CIs. Heterogeneity assumption between studies was evaluated by the Chi square-based Q-statistic and I2 statistic [52]. The random effects model (DerSimonia and Laird method) was used to assess pooled OR when there was a significant difference in terms of heterogeneity (if p < 0.05) [53]. Otherwise fixed effects model (the Mantel–Haenszel method) was used [54]. Potential publication bias was estimated by funnel plots and Egger’s test [55]. Moreover, the stability of the results was assessed using sensitivity analysis by deleting each single study involved in the meta-analysis one at a time to reflect the influence of the individual study to the pooled ORs. All p values were two sided and statistical significance level was considered for any test was p value < 0.05.The statistical analysis involved in this meta-analysis was performed by Comprehensive meta-analysis (CMA) version 2 software program (Biostat Inc., USA). To ensure the reliability and accuracy of the statistical analysis, two researchers entered the data into the software program independently and reached a consensus.
Results
Literature Search and Meta-analysis Databases
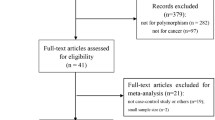
We have identified one hundred thirty three studies through literature search from the PubMed (Medline) and Google scholar for detailed evaluation. As per the pre-set selection (inclusion–exclusion) criteria, a total thirty nine published studies on association with the GSTP1 313 A > G gene polymorphism and susceptibility to multiple cancers were included in the meta-analysis (Fig. 1). Studies either showing GSTP1 polymorphism to predict survival OR as an indicator for response to therapy of patients were excluded straightaway. Similarly, research articles investigating the levels of GSTP1 mRNA or protein expression and relevant review articles were also excluded. We included only case–control or cohort design studies having frequency of all three genotypes. Eligible studies, publication year, cancer types, total numbers of controls and cases, genotyping methods, distribution of genotypes and minor allele frequency (MAF) in the controls have been shown in Tables 1 and 2.
Publication Bias
Begg’s funnel plot and Egger’s linear regression test were conducted to estimate the possible publication bias among the included studies for this meta-analysis. In the funnel plots, standard error of the log (OR) of each study was plotted against it log (OR). The appearances of funnel plot were symmetrical in all of the comparison models. Furthermore, Egger’s regression test, a linear regression approach for measuring funnel plot on the natural logarithm scale of the OR was used to provide statistical evidence to the funnel plot symmetry and showed no publication bias for all the genetic models (Table 3).
Evaluation of Heterogeneity
In order to test heterogeneity among the selected studies, Q-test and I2 statistics were employed. Heterogeneity was observed in all genetic models, i.e., allele (G vs. A), homozygous (GG vs. AA), heterozygous (AG vs. AA), recessive (GG vs. AG + AA) and dominant (GG + AG vs. AA). Thus, random effects model was applied to synthesize the data for above models (Table 3).
Association of GSTP1 313 A > G Polymorphism and Overall Cancer Susceptibility
We pooled all thirty nine studies together and it resulted into 6581 confirmed cancer cases and 8218 healthy controls to examining the overall association between GSTP1 313 A > G gene polymorphism and cancer risk. The pooled OR from overall analysis indicated that it was significantly associated with increased risk of cancer in allele (G vs. A: OR 1.266, 95% CI 1.129–1.418, p = 0.001), heterozygous (AG vs. AA: OR 1.191, 95% CI 1.047–1.355, p = 0.008), homozygous (GG vs. AA: OR 1.811, 95% CI 1.428–2.297, p = 0.001), dominant (GG + AG vs. AA: OR 1.276, 95% CI 1.110–1.466, p = 0.001) and recessive (GG vs. AG + AA: OR 1.638, 95% CI 1.340–2.002, p = 0.001) comparison models (Figs. 2, 3, 4, 5, 6).
Sensitivity Analysis
To evaluate the stability of the pooled results, sensitivity analysis was conducted. The influence of each study on the pooled OR was checked by repeating the meta-analysis while omitting each study, one at a time. The result of sensitivity analysis showed the corresponding pooled OR value did not significantly change when omitting any single study (figure not shown). This revealed that our results were statistically robust.
Discussion
Diagnosis and prevention of cancer have become one of the most important challenges of this era. Potent markers for screening high-risk populations are urgently needed for early detection and preventive actions. It is therefore, important to identify molecular markers that may help in the diagnosis of this dreadful disease in Indian populations. Several studies have supported an important role for genetics in determining the risk for cancer, and association studies are pertinent for searching susceptibility genes involved in cancer [56].
Metabolism is a cellular process required for the survival and proliferation of all cells, and increased proliferation and sustained survival are hallmarks of cancer [57]. As detoxifying enzyme, GSTs plays an important role in protecting cells from cytotoxic and carcinogenic agents in the defense system. Evidence suggests that the level of expression of GST is a crucial factor in determining the sensitivity of cells to a broad spectrum of toxic chemicals. The altered GST activity associated with the polymorphisms is expected to affect cancer risk through decreased protection against DNA damage from reactive electrophiles.
GSTP1 is widely expressed in normal epithelial cells and metabolize large hydrophobic electrophiles, such as polycyclic aromatic hydrocarbon-derived epoxides [58]. Studies have shown that GSTP1 was present at high levels in many solid tumors and in a wide range of cancer cell lines [59], GSTP1 null mice disposed with carcinogen polycyclic aromatic hydrocarbons demonstrated highly significantly increased risk of cancer [60]. This signified the role of GSTP1 as an important determinant in cancer susceptibility.
Currently, relationship between GSTP1 polymorphisms and cancer is a major area of research focus. The GSTP1 313A > G polymorphism was shown to be a predisposing risk factor for a number of human malignancies, but small size of study is a common limitation of biomarker validation studies. In the present meta-analysis, our main focus was to establish a more conclusive association between the GSTP1 313A > G polymorphism and overall cancer susceptibility in Indian population. Meta-analysis increases statistical strength and precision in estimating effects by combining the results of previous studies, thus overcoming the problem of small sample size and the inadequate statistical strength of complex trait genetic studies [61].
To the best of our knowledge, this meta-analysis is the first study to investigate the association between the GSTP1 313 A > G polymorphism and susceptibility to overall cancer in a large number of Indian populations. After rigorous statistical analysis has been performed for overall comparison of pooled ORs, we found significant increased risk between the GSTP1 313 A > G polymorphism and susceptibility to cancer in allele, homozygous, heterozygous, dominant and recessive genetic models. This result suggested that the GSTP1 313 A > G polymorphism may be a possible susceptibility factor for cancer in the Indian population, especially in individuals with mutant allele and mutant homozygous genotype. Alteration of GSTP1 activity due to 313 A > G polymorphism may lead to increased cell vulnerability to oxidative DNA damage and the accumulation of DNA base adducts, which can precede other genetic alterations lead to carcinogenesis. Numerous studies supported that G allele of GSTP1 313 A > G polymorphism substantially reduced enzyme activity and increased the risk of DNA mutation, resulting in poor elimination of hydrophilic metabolites and consequently increasing the susceptibility to cancer when individuals are exposed to carcinogens [62].
Genetically complex diseases differ from simple Mendelian diseases and cancer etiology is polygenic, a single genetic variant is usually inadequate to predict the risk of this deadly disease. Though, we interpreted our findings with full caution, but, some limitation of our meta-analysis should be addressed. Heterogeneity is an important issue while interpreting the results of meta-analysis, although that can be minimized by applying random-effects model. In the present study we detected heterogeneity in the entire genetic model, which might be due to the control sources and mix of cancers. Most of the studies used hospital-based patients as controls, who were not strictly healthy individuals and could not represent the general population. Gene environment interaction and adjusted OR have not been performed due to the limited number of data.
In spite of these, our meta-analysis still has some advantages. First, this is the first large association study between GSTP1 313 A > G polymorphism demonstrating susceptibility to cancer, which dramatically increase the statistical power of the present analysis than single study. Second, all the eligible studies included in the current meta-analysis researched in Indian population. The participants have the same genetic background, which can reduce the effects of ethnicity on pooled ORs. Third, there was not any publication bias detected, which indicated that the entire pooled result is robust and authentic. Fourth, sensitivity analysis was carried out by deleting each single study involved in the meta-analysis each time and the results did not alter, suggesting that our meta-analysis results were robust and reliable. Moreover, we used strict data extraction criteria and statistical analysis to make satisfactory and consistent conclusion.
Conclusion
Our meta-analysis indicated that the GSTP1 313 A > G gene polymorphism is a strong contender as a genetic susceptibility to cancer in Indian population. This could be used as a biomarker for clinical application and early identification and prevention of cancer. Furthermore, larger scale studies and impact of gene–gene and gene-environment interactions on the GSTP1 313 A > G polymorphism and cancer risk is necessary for providing a better comprehensive understanding of the association.
References
Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol Biomark Prev. 2016;25:16–27.
Takiar R, Nadayil D, Nandakumar A. Projections of number of cancer cases in India (2010–2020) by cancer groups. Asian Pac J Cancer Prev. 2010;11:1045–9.
Bredberg A. Cancer: more of polygenic disease and less of multiple mutations? A quantitative viewpoint. Cancer. 2011;117:440–5.
Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85.
Brennan P, Wild CP. Genomics of cancer and a new era for cancer prevention. PLoS Genet. 2015;11(11):e1005522.
Hayes J, Flanagan J, Jowsey I, Hayes J, Flanagan J, Jowsey I. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88.
Cowell IG, Dixon KH, Pemble SE, Ketterer B, Taylor JB. The structure of the human glutathione S-transferase pi gene. Biochem J. 1988;255:79–83.
Hengstler JG, Arand M, Herrero ME, Oesch F. Polymorphisms of N-acetyltransferases, glutathione S-transferases, microsomal epoxide hydrolase and sulfotransferases: influence on cancer susceptibility. Eur PubMed Cent. 1998;154:47–85.
Ahmad H, Wilson DE, Fritz RR, Singh SV, Medh RD, Nagle GT, Awasthi YC, Kurosky A. Primary and secondary structural analyses of glutathione S-transferase pi from human placenta. Arch Biochem Biophys. 1990;278:398–408.
Hu X, Xia H, Srivastava SK, Pal A, Awasthi YC, Zimniak P, Singh SV. Catalytic efficiencies of allelic variants of human glutathione S-transferase P1-1 toward carcinogenic anti-diol epoxides of benzo[c]phenanthrene and benzo[g]chrysene. Cancer Res. 1998;58:5340–3.
Chung CC, Magalhaes WC, Gonzalez-Bosquet J, Chanock SJ. Genome-wide association studies in cancer—current and future directions. Carcinogenesis. 2010;31:111–20.
Satinder K, Sobti RC, Pushpinder K. Impact of single nucleotide polymorphism in chemical metabolizing genes and exposure to wood smoke on risk of cervical cancer in North-Indian women. Exp Oncol. 2017;39:69–74.
Ghatak S, Yadav RP, Lalrohlui F, Chakraborty P, Ghosh S, Ghosh S, Das M, Pautu JL, Zohmingthanga J, Senthil Kumar N. Xenobiotic pathway gene polymorphisms associated with gastric cancer in high risk Mizo-Mongoloid population, Northeast India. Helicobacter. 2016;21:523–35.
Ghosh S, Ghosh S, Bankura B, Saha ML, Maji S, Ghatak S, Pattanayak AK, Sadhukhan S, Guha M, Nachimuthu SK, Panda CK, Maity B, Das M. Association of DNA repair and xenobiotic pathway gene polymorphisms with genetic susceptibility to gastric cancer patients in West Bengal, India. Tumour Biol. 2016;37:9139–49.
Sharma N, Singh A, Singh N, Behera D, Sharma S. Genetic polymorphisms in GSTM1, GSTT1 and GSTP1 genes and risk of lung cancer in a North Indian population. Cancer Epidemiol. 2015;39:947–55.
Kimi L, Ghatak S, Yadav RP, Chhuani L, Lallawmzuali D, Pautu JL, Kumar NS. Relevance of GSTM1, GSTT1 and GSTP1 gene polymorphism to breast cancer susceptibility in mizoram population, Northeast India. Biochem Genet. 2016;54:41–9.
Moulik NR, Parveen F, Kumar A, Agrawal S. Glutathione-S-transferase polymorphism and acute lymphoblastic leukemia (ALL) in north Indian children: a case–control study and meta-analysis. J Hum Genet. 2014;59:529–35.
Pandith AA, Lateef A, Shahnawaz S, Hussain A, Malla TM, Azad N, Shehjar F, Salim M, Shah ZA. GSTP1 gene Ile105 Val polymorphism causes an elevated risk for bladder carcinogenesis in smokers. Asian Pac J Cancer Prev. 2013;14:6375–8.
Abbas M, Srivastava K, Imran M, Banerjee M. Association of Glutathione S-transferase (GSTM1, GSTT1 and GSTP1) polymorphisms and passive smoking in cervical cancer cases from North India. IJBR. 2013;4:12.
Sameer AS, Qadri Q, Siddiqi MA. GSTP1 I105 V polymorphism and susceptibility to colorectal cancer in Kashmiri population. DNA Cell Biol. 2012;31:74–9.
Dunna NR, Vuree S, Kagita S, Surekha D, Digumarti R, Rajappa S, Satti V. Association of GSTP1 gene (I105V) polymorphism with acute leukaemia. J Genet. 2012;91:e60–3.
Ahmad ST, Arjumand W, Seth A, Kumar Saini A, Sultana S. Impact of glutathione transferase M1, T1, and P1 gene polymorphisms in the genetic susceptibility of North Indian population to renal cell carcinoma. DNA Cell Biol. 2012;31:636–43.
Saxena A, Dhillon VS, Shahid M, Khalil HS, Rani M, Prasad Das T, Hedau S, Hussain A, Naqvi RA, Deo SV, Shukla NK, Das BC, Husain SA. GSTP1 methylation and polymorphism increase the risk of breast cancer and the effects of diet and lifestyle in breast cancer patients. Exp Ther Med. 2012;4:1097–103.
Chauhan PS, Ihsan R, Mishra AK, Yadav DS, Saluja S, Mittal V, Saxena S, Kapur S. High order interactions of xenobiotic metabolizing genes and P53 codon 72 polymorphisms in acute leukemia. Environ Mol Mutagen. 2012;53:619–30.
Qadri Q, Sameer AS, Shah ZA, Hamid A, Alam S, Manzoor S, Siddiqi MA. Genetic polymorphism of the glutathione-S-transferase P1 gene (GSTP1) and susceptibility to prostate cancer in the Kashmiri population. Genet Mol Res. 2011;10:3038–45.
Wang J, Jiang J, Zhao Y, Gajalakshmi V, Kuriki K, Suzuki S, Nagaya T, Nakamura S, Akasaka S, Ishikawa H, Tokudome S. Genetic polymorphisms of glutathione S-transferase genes and susceptibility to colorectal cancer: a case–control study in an Indian population. Cancer Epidemiol. 2011;35:66–72.
Ihsan R, Chauhan PS, Mishra AK, Yadav DS, Kaushal M, Sharma JD, Zomawia E, Verma Y, Kapur S, Saxena S. Multiple analytical approaches reveal distinct gene-environment interactions in smokers and non smokers in lung cancer. PLoS ONE. 2011;6:e29431.
Sailaja K, Surekha D, Rao DN, Rao DR, Vishnupriya S. Association of the GSTP1 gene (Ile105Val) polymorphism with chronic myeloid leukemia. Asian Pac J Cancer Prev. 2010;11:461–4.
Kaushal M, Mishra AK, Raju BS, Ihsan R, Chakraborty A, Sharma J, Zomawia E, Verma Y, Kataki A, Kapur S, Saxena S. Betel quid chewing as an environmental risk factor for breast cancer. Mutat Res. 2010;703:143–8.
Malik MA, Upadhyay R, Mittal RD, Zargar SA, Mittal B. Association of xenobiotic metabolizing enzymes genetic polymorphisms with esophageal cancer in Kashmir Valley and influence of environmental factors. Nutr Cancer. 2010;62:734–42.
Ruwali M, Pant MC, Shah PP, Mishra BN, Parmar D. Polymorphism in cytochrome P450 2A6 and glutathione S-transferase P1 modifies head and neck cancer risk and treatment outcome. Mutat Res. 2009;669:36–41.
Malik MA, Upadhyay R, Mittal RD, Zargar SA, Modi DR, Mittal B. Role of xenobiotic-metabolizing enzyme gene polymorphisms and interactions with environmental factors in susceptibility to gastric cancer in Kashmir Valley. J Gastrointest Cancer. 2009;40:26–32.
Saxena A, Dhillon VS, Raish M, Asim M, Rehman S, Shukla NK, Deo SV, Ara A, Husain SA. Detection and relevance of germline genetic polymorphisms in glutathione S-transferases (GSTs) in breast cancer patients from Northern Indian population. Breast Cancer Res Treat. 2009;115:537–43.
Kumar M, Agarwal SK, Goel SK. Lung cancer risk in north Indian population: role of genetic polymorphisms and smoking. Mol Cell Biochem. 2009;322:73–9.
Suneetha KJ, Nancy KN, Rajalekshmy KR, Sagar TG, Rajkumar T. Role of GSTM1 (Present/Null) and GSTP1 (Ile105Val) polymorphisms in susceptibility to acute lymphoblastic leukemia among the South Indian population. Asian Pac J Cancer Prev. 2008;9:733–6.
Syamala VS, Sreeja L, Syamala V, Raveendran PB, Balakrishnan R, Kuttan R, Ankathil R. Influence of germline polymorphisms of GSTT1, GSTM1, and GSTP1 in familial versus sporadic breast cancer susceptibility and survival. Fam Cancer. 2008;7:213–20.
Rajkumar T, Samson M, Rama R, Sridevi V, Mahji U, Swaminathan R, Nancy NK. TGFbeta1 (Leu10Pro), p53 (Arg72Pro) can predict for increased risk for breast cancer in south Indian women and TGFbeta1 Pro (Leu10Pro) allele predicts response to neo-adjuvant chemo-radiotherapy. Breast Cancer Res Treat. 2008;112:81–7.
Singh M, Shah PP, Singh AP, Ruwali M, Mathur N, Pant MC, Parmar D. Association of genetic polymorphisms in glutathione S-transferases and susceptibility to head and neck cancer. Mutat Res. 2008;638:184–94.
Sobti RC, Kaur P, Kaur S, Janmeja AK, Jindal SK, Kishan J, Raimondi S. Combined effect of GSTM1, GSTT1 and GSTP1 polymorphisms on histological subtypes of lung cancer. Biomarkers. 2008;13:282–95.
Tripathi S, Ghoshal U, Ghoshal UC, Mittal B, Krishnani N, Chourasia D, Agarwal AK, Singh K. Gastric carcinogenesis: possible role of polymorphisms of GSTM1, GSTT1, and GSTP1 genes. Scand J Gastroenterol. 2008;43:431–9.
Soya SS, Vinod T, Reddy KS, Gopalakrishnan S, Adithan C. Genetic polymorphisms of glutathione-S-transferase genes (GSTM1, GSTT1 and GSTP1) and upper aerodigestive tract cancer risk among smokers, tobacco chewers and alcoholics in an Indian population. Eur J Cancer. 2007;43:2698–706.
Samson M, Swaminathan R, Rama R, Sridevi V, Nancy KN, Rajkumar T. Role of GSTM1 (Null/Present), GSTP1 (Ile105Val) and P53 (Arg72Pro) genetic polymorphisms and the risk of breast cancer: a case control study from South India. Asian Pac J Cancer Prev. 2007;8:253–7.
Pandey SN, Jain M, Nigam P, Choudhuri G, Mittal B. Genetic polymorphisms in GSTM1, GSTT1, GSTP1, GSTM3 and the susceptibility to gallbladder cancer in North India. Biomarkers. 2006;11:250–61.
Jain M, Kumar S, Rastogi N, Lal P, Ghoshal UC, Tiwari A, Pant MC, Baiq MQ, Mittal B. GSTT1, GSTM1 and GSTP1 genetic polymorphisms and interaction with tobacco, alcohol and occupational exposure in esophageal cancer patients from North India. Cancer Lett. 2006;242:60–7.
Sobti RC, Kaur S, Kaur P, Singh J, Gupta I, Jain V, Nakahara A. Interaction of passive smoking with GST (GSTM1, GSTT1, and GSTP1) genotypes in the risk of cervical cancer in India. Cancer Genet Cytogenet. 2006;166:117–23.
Mittal RD, Mishra DK, Mandhani A. Evaluating polymorphic status of glutathione-S-transferase genes in blood and tissue samples of prostate cancer patients. Asian Pac J Cancer Prev. 2006;7:444–6.
Mittal RD, Srivastava DS. Genetic polymorphism of drug metabolizing enzymes (CYP2E1, GSTP1) and susceptibility to bladder cancer in North India. Asian Pac J Cancer Prev. 2005;6:6–9.
Srivastava DS, Mishra DK, Mandhani A, Mittal B, Kumar A, Mittal RD. Association of genetic polymorphism of glutathione S-transferase M1, T1, P1 and susceptibility to bladder cancer. Eur Urol. 2005;48:339–44.
Vijayalakshmi K, Vettriselvi V, Krishnan M, Shroff S, Vishwanathan KN, Jayanth VR, Paul SF. Polymorphisms at GSTM1 and GSTP1 gene loci and risk of prostate cancer in a South Indian population. Asian Pac J Cancer Prev. 2005;6:309–14.
Srivastava DS, Mandhani A, Mittal B, Mittal RD. Genetic polymorphism of glutathione S-transferase genes (GSTM1, GSTT1 and GSTP1) and susceptibility to prostate cancer in Northern India. BJU Int. 2005;95:170–3.
Mandal RK, Mittal RD. Genetic variant Arg399Gln G>A of XRCC1 DNA repair gene enhanced cancer risk among Indian Population: evidence from meta-analysis and trial sequence analyses. Indian J Clin Biochem. 2018;33:262–72.
Wu R, Li B. A multiplicative-epistatic model for analyzing interspecific differences in outcrossing species. Biometrics. 1999;2:355–65.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;4:719–48.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34.
Khoury MJ, Yang Q. The future of genetic studies of complex human diseases: an epidemiologic perspective. Epidemiology. 1998;9:350–4.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600.
Tew KD. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res. 1994;54:4313–20.
Henderson CJ, Smith AG, Ure J, Brown K, Bacon EJ, Wolf CR. Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proc Natl Acad Sci USA. 1998;95:5275–80.
Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–82.
Chasseaud LF. The role of glutathione and glutathione S-transferases in the metabolism of chemical carcinogens and other electrophilic agents. Adv Cancer Res. 1979;29:175–274.
Acknowledgements
The authors acknowledge the software related service support provided by Research and Scientific Studies Unit, College of Nursing and Allied Health Sciences, Jazan University, Saudi Arabia.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mandal, R.K., Mittal, R.D. Glutathione S-Transferase P1 313 (A > G) Ile105Val Polymorphism Contributes to Cancer Susceptibility in Indian Population: A Meta-analysis of 39 Case–Control Studies. Ind J Clin Biochem 35, 8–19 (2020). https://doi.org/10.1007/s12291-018-0787-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12291-018-0787-1