Abstract
The A/C transversion at 1166 of the angiotensin II Type 1 Receptor (AT1R) gene per se does not characterize any functional diversity but has been associated with expression of the AT1R, consequently molecular variants of the gene may modulate the possible risk of essential hypertension. The present study was performed to determine the genotypic frequency of the A1166C polymorphism of the AT1R gene in essential hypertensive patients with the aim to assess the effect of variants of this polymorphism in hypertension. AT1R gene amplification was performed by PCR and A1166C polymorphism was determined by enzyme digestion methodologies in 224 consecutively enrolled essential hypertensive patients and 257 controls. Suitable descriptive statistics was used for different variables. Results revealed that genotype and allele distribution of the A1166C variant differed significantly in hypertensives and normotensives. Allele frequency at the A1166C position was 61%A and 39%C for control and 52%A and 48%C for patients. Observed frequencies were compatible with HWE expected frequencies in cases as well as in controls. rs5186 was found to be associated with hypertension (95% CI 1.1453–2.7932, p: 0.0106). The difference remained statistically significant after the multivariate adjustment (p < 0.05), with C/C variant conferring a risk of 1.74-fold of essential hypertension. This association was confirmed by inter-genotypic variations in the mean systolic and diastolic blood pressure in patients. In conclusion, genetic variation at the AT1R gene influences the risk of hypertension stratification and might serve as a predictive marker for the susceptibility to hypertension among affected families.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Essential hypertension (EHT), a complex pleomorphic disorder, is often associated with development of co-morbidities with demanding health costs. The etiopathogenesis of EHT are likely to be emanating from underlying genetic factors and superimposing environmental entities. The complexity of the trait makes it difficult to identify the individuals who are at increased risk.
Over the last two decades or so, population based studies; adoption studies; twin studies and segregation analysis have suggested that inter-individual variability in human blood pressure (BP) is genetically determined [1] with a heritability factor of approximately 30–60% [1], elucidating and emphasizing the impact of genetic contribution. However, it does not follow the Mendelian mode of transmission [2], and a number of quantitative trait loci attributed to EHT across the genome have been proposed [2]. Among the many potential candidate components, genes that influence the renin–angiotensin–aldosterone system (RAAS) seem to be particularly important as the RAAS system plays an essential role in BP regulation by maintaining vascular tone and renal hemodynamics [3] and clinically relevant as most of the antihypertensive drugs are targeting this system [4]. Therefore, genes encoding for the components RAAS are one of the imperative contributing factors associated with both causation and progression of hypertension.
The effects of RAAS are predominantly mediated by angiotensin II, which has manifold effects, such as systemic vasoconstriction, cellular proliferation (on vascular smooth muscle and cardiomyocytes), vascular remodeling, aldosterone secretion, synthesis of the extracellular collagen matrix, and is tightly intertwined with the cascade of inflammatory, thrombotic, and fibrotic factors [5]. Most of these effects are mediated via the activation of angiotensin II type 1 receptor (AT1R) [6], which are expressed in vascular smooth muscle cells and adrenal glands, among others.
Because of the physiological role of the AT1R in the cardiovascular system, and substantiated by the findings that arterial BP can be effectively reduced by viral gene delivery of AT1R antisense in adult spontaneously hypertensive rat [7], the gene that encodes this mol-ecule represents an excellent potentially aetiological candidate and is reasonable to hypothesize that genetic variants, or polymorphisms, in AT1R gene modulates receptor function and may modify the risk of developing EHT.
The human AT1R gene spans more than 50 kb in length (05 exons and 04 introns), is located on chromosome 3q21-q25 and encodes a 360 amino acids peptide [8]. A polymorphism in the 3′ untranslated region of AT1R gene has been described with either an adenine (A) or a cytosine (C) base (A/C transversion) at the 1166 position [8] designated as AT1R A1166C (SNP ID: rs5186) [9] and a substantial number of association studies have addressed the role of this polymorphism in the complex etiology of EHT, albeit with equivocal results. However, there is a paucity of information on the trends of the A1166C polymorphisms in the in Indian population more so specifically in Guajarati hypertensive population. Hence, we opted to determine the genotypic frequency of the A1166C polymorphism of the AT1R gene in essential hypertensive patients with the aim to assess the effect of variants of this polymorphism in hypertension.
Material and Method
Ethical Consideration
The study was conducted in accordance with the ethical guidelines of the Declaration of Helsinki on biomedical research on humans, and was approved by the Institutional Human Research Ethical Committee. Informed consent was obtained from each participant in accordance with a study protocol after providing a detailed study overview.
Participants and Study Protocol
This case–control study was conducted from September 2014 to May 2016. Patient selected for this study included unrelated essential hypertensive adult Gujarati patients. The history of EHT was operationally based on patient self report of a prior physician diagnosis and use of prescribed antihypertensive medication for a minimum of one year and only patients with positive family history of hypertension, defined by at least one parent being hypertensive were selected. Patients showing clinical signs, symptoms, or laboratory findings suggestive of secondary hypertension were excluded. Other criteria for exclusion were, patients with a history of diabetes mellitus (DM) or renal disease; showing impaired glucose tolerance, current or history of tobacco or alcohol intake; patients with, any severe illness, impairment of speech, hearing, vision, or cognition, continuous or periodic use of corticosteroids; pregnant females or who had given birth within the preceding six weeks, or any significant medical illnesses that prevented participants from adhering to the protocol, lack of approval by physician and patients showing disinterest or refusal to sign the consent form. A total of 481 participants (224 hypertensive patients and 257 healthy controls) were recruited in the present study. It was based on consecutive sampling technique i.e. all the patients who fulfilled the criteria in the period October 2014 to March 2016 were included and it was ensured that 1:1 matching was done. Healthy controls were selected at random from outpatients, who were on routine health check-ups and was defined as participants not suffering from hypertension (as per the criteria formulated by Seventh Report of the Joint National Committee, JNC 7) [10], nor having any family history of hypertension in first-degree relatives, not suffering from DM or from any acute or chronic disease, nor taking any drugs believed to affect BP. All participants (patients and control) included in the study were ethnically matched, and we included only those individuals who had been born in and residents (without migration history) of Gujarat for at least three generations, including their own.
Participant’s Examination and Measurements
All participants were studied as outpatient. A detailed present and past history of each case was recorded including name, age, sex, address, religion, occupation, economic status, nutritional and personal habits, education, medication and history suggestive of any systemic illness. Each patient was then examined for various anthropometric parameters: Height was measured to the nearest centimeter and weight to the nearest 0.1 kg (using SECA 206 wall mounted metal tapes and Omron digital body weight scale HN-286 respectively) which was used for calculation of BMI (kg/m2). All anthropometric measures reflect the average of 3 measurements (measured by same person on same instrument to avoid inter-instrument and inter personal variation). BP was measured three times (on different days) in the seated position after 10 min of rest with a standard manual mercury sphygmomanometer (Diamond Deluxe Industrial Electronics and Products, Pune, India) and stethoscope (3 M Littmann, 3 M India Ltd, Banglore, India) by auscultatory method. The recorded pressure of the three measurements was averaged so as to exclude false values due to variability that may be caused by various factors at any one point in time. Age was defined as the age at the time of interview (though no documentary proof had been entertained) and the date of diagnosis of EHT was obtained from the patient.
Sampling and Biochemical Analysis
After an overnight fast of 12 h, venous sampling was done for biochemical determinations and for isolation of DNA. Serum and plasma was separated by centrifugation of blood sample and were subjected for analytical procedures. Glucose (Glucose oxidase–peroxidase method, CV %: 3.4), cholesterol (Cholesterol oxidase–peroxidase method, CV%: 3.9), triglycerides (Enzymatic method, CV%: 3.6), HDL-C (Phosphotungstic method, CV%: 4.7), LDL-C (CV%: 3.6) and creatinine (modified Jaffe’s method, CV%: 2.5) were measured in fully automated analyzer.
DNA Analysis: Determination of AT1R Gene A1166C Polymorphism
DNA Extraction
Genomic DNA was extracted from peripheral blood leukocytes using commercially available DNA extraction and purification based on standard proteinase K technique. Eluted DNA was either stored at −20 °C or amplified immediately. All DNA samples were amplified within three days following extraction. Before amplification, quantity of DNA in each sample was assessed by measuring the absorption at 260 nm (using molar extinction coefficient of double stranded DNA: 0.020 μg/ml/cm) in a standard spectrophotometer (UV–VIS double beam—2205, Systronics, Ahmedabad, India).
DNA Amplification
Two oligonucleotide primers, forward: 5′- ATAATGTAAGCTCATCCACC -3′ and reverse: 5′- GAGATTGCATTTCTGTCGGT -3′ based on the flanking sequences [8, 9] were used to amplify the corresponding DNA fragments by polymerase chain reaction (PCR). Amplification was carried out in a DNA Thermal Cycler (2720, Applied Biosystems) in a final reaction volume of 50 μl containing 100 ng genomic DNA, 25 pM of each primer, 200 μM of each deoxyribonucleotides triphosphate, 0.5 U of thermus aquaticus DNA polymerase, 2 mM magnesium chloride, 20 mM Tris-hydrocholric acid and 50 mM KCl. The thermocycling profile consisted of 5 min of initial denaturation at 94 °C followed by 30 cycles of amplification of denaturation at 94 °C for 30 s, annealing at 68 °C for 30 s and extension at 72 °C for 30 s, followed by a final extension at 72 °C for 5 min.
Restriction Endonuclease Digestion of PCR Products
The PCR amplified 359-bp products were analyzed for genotyping by restriction fragment length polymorphism (RFLP) using the restriction enzyme DdeI (Desulfovibrio desulfuricans I), whose recognition site is 5′CTNAG. The 359-bp PCR fragments {10 μL (1 μg/μL)} were submitted to restriction endonuclease digestion by addition of 2 U of DdeI endonucleases with restriction enzyme buffer, and incubated at 37 °C for 12 h. The restriction fragments were separated and distinguished on agarose gel stained in ethidium bromide using UV transillumination and DNA ladder as standard. After digestion, samples with no DdeI restriction site {DdeI (-); homozygous A/A} showed the single 359-bp band, those homozygous for the restriction site {DdeI (+); homozygous C/C} resulted in two bands (220 + 139 bp) after digestion, while heterozygous samples showed all the three bands.
Statistical Analysis
The statistical analyses were performed using a Statistical Package for Social Sciences for Windows version 15.0 (SPSS Inc, Chicago, Illinois, USA). Data were expressed as mean ± SD (continuous variables), or as percentages of total (categorical variables). Prior to hypothesis testing, histograms and normality plots were used for evaluating the normality of data. For those data with skewed distribution, log transformation was performed before analysis. Two-group comparisons were made using Chi square (χ2) for categorical variables and Student’s t tests or one-way ANOVA for continuous variables.
The distribution of alleles in studied groups was tested for fitting to the Hardy–Weinberg equilibrium (HWE) (using web base program: http://www.oege.org/software/hwe-mr-calc.shtml) through testing the difference between observed and expected frequencies of genetic variants using the χ2 goodness-of-fit test. Odds ratios (ORs) with 95% confidence intervals (CIs) were estimated to determine the risk levels for the associated genotypes. In addition, the strength of the association between EHT and the AT1R gene A1166C polymorphism was esti-mated using ORs (with the corresponding 95% CIs). The ORs were also performed for a dominant model [(DD% + ID%)vs II%], a co-dominant model [ID% vs (II% + DD%)] and a recessive model [DD% vs (II% + ID%)]. Multivariate logistic regression analysis was employed to determine the relations of gene polymorphisms and other risk factors with EHT. Associations were expressed as adjusted OR with 95% CI. For all analyses, two-sided probability values <0.05 were considered statistically significant.
Results
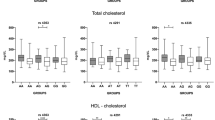
The mean acceptance rate of this protocol was 78.98% (n: 481) by eligible patients (n: 609) and there was no difference between hypertensive patients versus non-hypertensive groups in the number of participants who withdrew (p ≥ 0.74) after the initial consent. Table 1 shows the subgroup, anthropometric and clinical characteristics of the study participants. Among the study participants there was a preponder-ance of male individuals as compared with females (In cases: 63.83 vs 36.17%, p < 0.0001; in controls: 56.42 vs 43.58%, p: 0.0048) but the gender distribution i.e. male to female ratio (p: 0.1186) as well as mean age (p: 0.0618) of hypertensive participants and controls were similar. A comparison of clinico-biochemical characteristics revealed that the difference were statically significant between cases and controls with respect to glucose (p < 0.0001), BMI (p < 0.0001), and non-vegetarian dietary habit (p: 0.04), suggesting an elevated risk of the condition when associated with these parameters. Elevated body weight was a concomitant health disorder for most patients. Eight nine (88.83%) percent showed a body mass index equal to or greater than 25.0 kg/m2, and 55% (54.91%) of the patients were obese (body mass index ≥30.0 kg/m2). Barring Triglyceride, the other indices of lipid profile such as cholesterol, HDL-C and LDL-C were well controlled and comparable in patients and controls, most likely because of the high percentage of patients taking lipid lowering medications. However, detailed documentation of antihypertensive and lipid lowering medication could not be obtained for all patients and hence these data are not reported. Although patients were under antihypertensive medication, the mean systolic BP or diastolic BP of was apparently higher (p < 0.0001) than those of other participants.
The genotypic and allelic distribution of AT1R A1166C polymorphism in hypertensive patients and non-hypertensive individuals are presented in Table 2 and the observed genotype frequencies were compatible with frequencies predicted by Hardy–Weinberg equilibrium, both in the total sample group and segregated into hypertensive and normoten-sive groups. There was a significant difference between the hypertensive group and the controls as regard to C/C genotype (p: 0.0138). This may suggest that patients with C/C genotype are at high risk of developing hypertension (p: 0.0106, 95% CI 1.1453–2.7932; OR: 1.7886; Table 3). The genotypic level was also visible at the allelic front as C allele was found in a higher frequency in hypertensive patients than in the controls. (p: 0.0031, Table 2). The observed allele frequencies in patients were 0.48 and 0.52 for C and A alleles respectively, whereas 0.39 and 0.61 was observed in control groups, which resulted in significant differences in the allele (x2: 8.80, 2 d. f. p: 0.003) and genotype (x2: 8.35, 2 d. f. p: 0.0154) distribution among the case and control participants. The difference was due to significantly higher frequency of C/C homozygotes (25.89 vs. 16.34%, x2: 6.06, p: 0.0138), and lower frequency of wild type homozygotes (29.46 vs. 38.91%, x2: 4.32, p: 0.0377). Analogous results were obtained at all fronts when analyzed according to gender and age.
There was a significant association of mutant genotype with essential hypertension and AT1R C/C genotype conferred a 1.7886 fold risk of essential hypertension compared with the genotype A/C and A/A. After controlling for other risk factors, the CC genotype was still significantly associated with hypertension, conferring a 1.74 times higher risk (Table 4). To further evaluate the etiologic effects of AT1R polymorphisms, the data was stratified according to the three genotypes of AT1R A1166C polymorphism for the clinical characteristics of the patients and compared using univariate analysis. We found a statistically significant intergenotypic variation in the mean systolic and diastolic blood pressure with C/C genotypic patients having the highest values followed by A/C and A/A patients (Table 6).
The inheritance hypothesis for this polymorphism was tested according to three models: co-dominant, dominant and recessive. When comparison of homozygous carriers of C variant plus A/C heterozygote (CC + AC) versus homozygous carriers of A variant (AA) (dominant model) were made, the OR was 1.52 (p = 0.029). Whereas when A/C heterozygote versus homozygous carriers of C variant plus homozygous carriers of A variant (CC + AA) (Co-dominant model) were made, the OR was 1.43 (p = 0.004) and 1.79 (p = 0.010) under recessive mode [CC% vs (AA% + AC%)] (Table 5).
Discussion
Extensive analysis in spontaneously hypertensive rat had revealed an elevated level of AT1R gene expression in the brainstem [11] as well as a positive correlation between AT1R gene expression and systolic BP [12]. Such and similar findings from animal model as well as human studies [13, 14] supports a pivotal role of AT1R gene in BP homeostasis. Several lines of experimental clinical evidence also have alluded a strong evidence that development of hypertension is harboring the AT1R gene. Despite many positive find-ings of association studies, linkage and sib-pair linkage analysis in clearly linking rs5186 as one of the apt cause of hypertension, overwhelmingly negative results from preceding molecular genetic studies of humans also have appeared across various populations and ethnic groups. We therefore considered it worthwhile to unravel the functional significance of AT1R A1166C polymorphism with respect to disease and indeed found a significant interac-tion with the disease phenotype.
The primary finding from this study using a case control approach, involving individuals with EHT is that gene polymorphism in the 3′-untranslated region of the AT1R gene on chromosome 3 with transversion of an A to C at nucleotide 1166 is an independent risk factor for hypertension. Result presented here revealed significant heterogeneity under the allele (p: 0.003) and genotype (p: 0.0154) contrast in hypertensive patients than in normal controls. These differences might validate that the AT1R A1166C polymorphism is an important genetic determinant of hypertension. Conspicuously, subgroup analysis by gender or by age did not change this pattern of results. The frequency of 1166 A/A genotype among cases was found to be 29.46% and CC genotype frequency as 25.89%, which was higher and statically significant in regard to that in the control group (p: 0.83). Albeit the genotype distribution resulted in a higher frequency of the C allele in the cases than controls (p: 0.0031; Table: 2), testing genetic equilibrium between the observed and expected genotypes using Hardy–Weinberg equilibrium showed AT1R gene variants were confirming to the law. C allele of AT1R gene might confer a high risk of developing hypertension, as exemplified by an increased frequency of the rs5186 C allele in hypertensive individuals and its association with hypertension was highly compounded when C allele was present in homozygous state i.e. there was a statistically significant increase in odds of hypertension with the AT1R C/C genotype.
Multivariate meta-analysis after adjustment for the covariates, which included clinically significant variables along with variables with positive significant associations in univariate analysis, did not change the significance of the results. (p: 0.02, 95% CI 1.65–2.78, Table 5), implying a risk of approximately 1.74 times higher than for those homozygous for the A allele, indicating that the individuals with C/C genotypes may be at substantially higher risk of developing EHT. Furthermore, the AT1R C/C genotype was also associated with a higher systemic arterial pressure in patients than those with the other two genotypes at the locus in question. This reasserts that the AT1R gene C/C variant is associated with higher susceptibility to hypertension and is substantiated by intervention studies demonstrating greater benefits & physiological response by wild type of rs5186 AT1R gene polymorphism compare to mutant type to pharmacological maneuvers [15]. Therefore, the hypothesis purposed by researchers that polymor-phisms associated with increased RAAS activity may predispose to hypertension, as in AT1R C allele, which is associated with greater responses to angi-otensin II at lower concentrations [16] might all be more prev-alent within a hypertensive population is likely to be true for the studied population.
AT1R A1166C SNP has been studied in different populations with paradoxical results. Association of C/C variant with the EHT in our study has shown agreement with some studies [5, 17–19] and meta-analysis [9, 20]. Bonnardeaux et al. [8] were the first to report the linkage of the molecular variants A1166C with hypertension. Thereafter, several studies were performed in an effort to redefine the association between AT1R A1166C SNP and hypertension. However, many of the subsequent studies [21–25] are at variance with the finding of our study. This discrepancy could be due to methods of quantitation, study design, statistical power, environ-mental backgrounds, the heterogeneous phenotypic characterization of cases and controls or more importantly that might be due to particular genetic structure of studied population. It also clarifies a causality role to polymorphisms for the multifactorial nature of hypertension pathogenesis, in which many genetic variants could contribute, together with environmental and the interactions of other genes and a single gene or its polymorphism did not have a major effect on hypertension, at the most, it is capable of affecting, albeit weakly, the onset and progression of EHT. It is also probably one of the main reasons that no clear evidence for the precise mode of inheritance was observed (Table 5).
A1166C variant is a nonfunctional mutation [8] but has been associated with higher antiotensin II type 1 receptor responsiveness [25], defects in messenger RNA (mRNA) processing, receptor expression [26] or may well affect the function of regulatory elements [27], which could mechanistically contribute to its effects. Thus, it plays a pivotal role in the genomic context and may influence the expression levels, even being located within the 3′ untranslated regions of the gene. So, although, it do not lead to amino acid substitutions, positive associations observed in this study may be the result of linkage disequilibrium with another polymorphism or with an unidentified functional mutation in the AT1R gene or in another closely linked gene possibly located in regulatory regions and is supported by a study [28] that demonstrated that there is miR155 (from chromosome 21,), which down-regulates the expression of the 1166A allele but not of the 1166C. As a result 1166C allele is associated with hypertension just because miR155 cannot negatively control the expression levels of AT1R Table 6.
To exclude the genotyping error, we randomly regenotyped 20% of the samples by Tm-shift genotyping method [29] and the results were 100% concordant. To minimize the potential influence of mixed factors on EHT occurrence, we designed and performed this study in a population of similar socio-economical geographical background, so all participants had homogeneity of environments. Other potential strengths to our study includes the selection criteria of patient cohort as we excluded patients with features suggestive of secondary hypertension, thus confined our study to EHT; we excluded one of the main environmental influences (DM), thus likely most of the selected hypertensive case patients had a strong genetic predisposition to the disease.
Nonetheless, the present study provided only the statistical evidences of the association between the A1166C allele of AT1R with EHT at the gene level and do not imply causation, mechanism or the functionality of the variants. It remains possible that we have not necessarily identified the pathobiological genetic variant but potentially only a variant in the gene in linkage disequilibrium with the causal pathobiological variant. Hence, our data must be interpreted within the context of its precincts for any potential clinical implications. There are also other potential limitations. Firstly, hypertension is a polygenic disease, to which different physiologic pathways contribute. Therefore, a dichotomous classification—hypertension and normotension, may not be sensitive enough to detect genetic predispositions to individual pathways, especially when the gene under study can exert minor effects on hypertension status itself. Secondly, the number of cases for association study analysis is somewhat insufficient i.e. study involves a small sample size and may not be representative of the entire patient population. Nevertheless, still results of our study are particularly relevant, as our trial fulfils the criteria of a good genetic association study suggested by Hattersley et al. [30]. Finally, we did not make any adjustments for dietary factors (salts, electrolytes) and physical activity in the covariate list of regression analysis, which seem to affect BP variance. In spite of these limitations, we believe that AT1R gene A1166C SNP can be use-ful for outcome predictions during diagnostic processes and can be helpful in finding new treatment strategies.
To conclude, our findings suggest that the AT1 gene A1166C polymorphism appeared to be an important genetic determinant in etiopathogenesis of HTN and genetic variants of AT1R might serve as a predictor for the susceptibility to hypertension among affected families. This could in the future become an important part of the clinical process of HTN risk stratification. This possibility now needs to be confirmed by prospective studies. Additional studies in this regard in other popu-lations along with the effects of other candidate genes of the RAAS are required to consider the joint effects of several candidate genes to dissect the genetic framework of essential hypertension which we envisage will open a plethora of options like timing, type and doses of anti-hypertensive therapy.
References
Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–56.
Butler MG. Genetics of hypertension: current status. J Med Liban. 2010;58:175–8.
Navar LG, Kobori H, Prieto MC, Gonzalez-Villalobos RA. Intratubular renin-angiotensin system in hypertension. Hypertension. 2011;57:355–62.
van Vark LC, Bertrand M, Akkerhuis KM, Brugts JJ, Fox K, Mourad JJ, et al. Angiotensin-converting enzyme inhibitors reduce mortality in hypertension: a meta-analysis of randomized clinical trials of renin-angiotensin-aldosterone system inhibitors involving 158,998 patients. Eur Heart J. 2012;33:2088–97.
Jiang ZN, Zhao WS, Yu F, Xu G. Association of angiotensin II type 1 receptor gene polymorphism with essential hypertension. Chin Med J. 2001;114:1249–51.
Chiu AT, Herblin WF, McCall DE, Ardecky RJ, Carini DJ, Duncia JV, et al. Identification of angioensin II receptor subtypes. Biochem Biophys Res Commun. 1989;165:196–203.
Katovich MJ, Gelband CH, Reaves P, Wang HW, Raizada MK. Reversal of hypertension by angiotensin II type 1 receptor antisense gene therapy in the adult SHR. Am J Physiol. 1999;277:H1260–4.
Bonnardeaux A, Davies E, Jeunemaitre X, Féry I, Charru A, Clauser E, et al. Angiotensın II Type 1 receptor gene polymorphism in human essential hypertension. Hypertension. 1994;24:63–9.
Wang JL, Xue Li, Hao PP, Feng Xu, Chen YG, Zhang Y. Angiotensin II type 1 receptor gene A1166C polymorphism and essential hypertension in Chinese: a meta-analysis. J Renin Angiotensin Aldosterone Syst. 2010;11:127–35
Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL Jr, et al. The seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure: the JNC 7 report. JAMA. 2003;289:2560–72.
Raizada MK, Sumners C, Lu D. Angiotensin II type 1 receptor mRNA levels in the brains of normotensive and spontaneously hypertensive rats. J Neurochem. 1993;60:1949–52.
Reja V, Goodchild AK, Phillips JK, Pilowsky PM. Upregulation of angiotensin AT1 receptor and intracellular kinase gene expression in hypertensive rats. Clin Exp Pharmacol Physiol. 2006;33:690–5.
Chandra S, Narang R, Sreenivas V, Bhatia J, Saluja D, Srivastava K. Association of angiotensin II type 1 receptor (A1166C) gene polymorphism and its increased expression in essential hypertension: a case-control study. PLoS ONE. 2014;9(7):e101502.
Ceolotto G, Papparella I, Bortoluzzi A, Strapazzon G, Ragazzo F, Bratti P, et al. Interplay between miR-155, AT1R A1166C polymorphism, and AT1R expression in young untreated hypertensives. Am J Hypertens. 2011;24:241–6.
Brugts JJ, Isaacs A, de Maat MP, Boersma E, van Duijn CM, Akkerhuis KM, et al. A pharmacogenetic analysis of determinants of hypertension and blood pressure response to angiotensin-converting enzyme inhibitor therapy in patients with vascular disease and healthy individuals. J Hypertens. 2011;29:509–19.
Spiering W, Kroon AA, Fuss-Lejeune MM, Daemen MJ, De Leeuw PW. Angiotensin II sensitivity is associated with the angiotensin II type 1 receptor A(1166)C polymorphism in essential hypertensives on a high sodium diet. Hypertension. 2000;36:411–6.
Bayramoglu A, Kurt H, Gunes HV, Ata N, Birdane A, Dikmen M, et al. Angiotensin II type 1 receptor (AT1) gene A1166C is associated with the risk of hypertension. Genet Test Mol Biomark. 2015;19:14–7.
Patnaik M, Pati P, Swain SN, Mohapatra MK, Dwibedi B, Kar SK, Ranjit M. Aldosterone synthase C-344T, angiotensin II type 1 receptor A1166C and 11-β hydroxysteroid dehydrogenase G534A gene polymorphisms and essential hypertension in the population of Odisha. India J Genet. 2014;93:799–808.
Chung WK, Deng L, Carroll JS, Mallory N, Diamond B, Rosenzweig EB, et al. Polymorphism in the angiotensin II Type 1 receptor (AGTR1) is associated with age at diagnosis in pulmonary arterial hypertension. J Heart Lung Transplant. 2009;28:373–9.
Niu W, Qi Y. Association of the angiotensin II type I receptor gene +1166 A>C polymorphism with hypertension risk: evidence from a meta-analysis of 16474 subjects. Hypertens Res. 2010;33:1137–43.
Shahin DS, Irshaid YM, Saleh AA. The A(1166)C26 polymorphism of the AGTR1 gene is associated with an early onset of hypertension and high waist circumference in Jordanian males attending the Jordan University Hospital. Clin Exp Hypertens. 2014;36:333–9.
Kooffreh ME, Anumudu CI, Duke R, Okpako EC, Kumar PL. Angiotensin II type 1 receptor A1166C gene polymorphism and essential hypertension in Calabar and Uyo cities, Nigeria. Indian J Hum Genet. 2013;19:213–8.
Jiang X, Sheng H, Li J, Xun P, Cheng Y, Huang J, Xiao H, Zhan Y. Association between renin–angiotensin system gene polymorphism and essential hypertension: a community-based study. J Hum Hypertens. 2009;23:176–81.
Sugimoto K, Katsuya T, Ohkubo T, Hozawa A, Yamamoto K, Matsuo A, et al. Association between angiotensin II type 1 receptor gene polymorphism and essential hypertension: the Ohasama study. Hypertens Res. 2004;27:551–6.
Van Geel PP, Pinto YM, Voors AA, Buikema H, Oosterga M, Crijns HJ, et al. Angiotensin II type 1 receptor A1166C gene polymorphism is associated with an increased response to angiotensin II in human arteries. Hypertension. 2000;35:717–21.
Thekkumkara TJ, Linas SL. Evidence for involvement of 3′-untranslated region in determining angiotensin II receptor coupling specificity to G-protein. Biochem J. 2003;370:631–9.
Atkinson TJ, Halfon MS. Regulation of gene expression in the genomic context. Comput Struct Biotechnol J. 2014;9:e201401001.
Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–13.
Wang J, Chuang K, Ahluwalia M, Patel S, Umblas N, Mirel D, et al. High-throughput SNP genotyping by single-tube PCR with Tm-shift primers. Biotechniques. 2005;39:885–93.
Hattersley AT, McCarthy MI. What makes a good genetic association study? Lancet. 2005;366:1315–23.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Parchwani, D.N., Patel, D.D., Rawtani, J. et al. Analysis of Association of Angiotensin II Type 1 Receptor Gene A1166C Gene Polymorphism with Essential Hypertension. Ind J Clin Biochem 33, 53–60 (2018). https://doi.org/10.1007/s12291-017-0644-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12291-017-0644-7