Abstract
The global public health burden of bacterial antimicrobial resistance (AMR) is intensified by Gram-negative bacteria, which have an additional membrane, the outer membrane (OM), outside of the peptidoglycan (PG) cell wall. Bacterial two-component systems (TCSs) aid in maintaining envelope integrity through a phosphorylation cascade by controlling gene expression through sensor kinases and response regulators. In Escherichia coli, the major TCSs defending cells from envelope stress and adaptation are Rcs and Cpx, which are aided by OM lipoproteins RcsF and NlpE as sensors, respectively. In this review, we focus on these two OM sensors. β-Barrel assembly machinery (BAM) inserts transmembrane OM proteins (OMPs) into the OM. BAM co-assembles RcsF, the Rcs sensor, with OMPs, forming the RcsF-OMP complex. Researchers have presented two models for stress sensing in the Rcs pathway. The first model suggests that LPS perturbation stress disassembles the RcsF-OMP complex, freeing RcsF to activate Rcs. The second model proposes that BAM cannot assemble RcsF into OMPs when the OM or PG is under specific stresses, and thus, the unassembled RcsF activates Rcs. These two models may not be mutually exclusive. Here, we evaluate these two models critically in order to elucidate the stress sensing mechanism. NlpE, the Cpx sensor, has an N-terminal (NTD) and a C-terminal domain (CTD). A defect in lipoprotein trafficking results in NlpE retention in the inner membrane, provoking the Cpx response. Signaling requires the NlpE NTD, but not the NlpE CTD; however, OM-anchored NlpE senses adherence to a hydrophobic surface, with the NlpE CTD playing a key role in this function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Globally, bacterial antimicrobial resistance (AMR) is becoming a major public health burden, with antibiotic overuse being one of the main causes (UNEP, 2023). At the same time, the development of new antibiotics has slowed significantly. It has been estimated that AMR may result in a mortality rate of 10 million people per year by 2050 (O'Neill, 2016). In 2017, the World Health Organization issued a list of twelve bacterial strains that present the greatest risk to human health, most of which are highly virulent and cause AMR. Among these twelve strains, three strains, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae, are resistant to the antibiotic carbapenem and have been recognized as critical-priority pathogens that require the urgent development of new antibiotics (Tacconelli et al., 2018). Importantly, these three pathogens are Gram-negative. Moreover, a recent report presented surprising statistical data about the death rate attributable to and associated with AMR, showing that Gram-negative bacteria, including Escherichia coli, are the leading pathogens (Antimicrobial Resistance Collaborators, 2022). Compared with Gram-positive bacteria, Gram-negative bacteria show a higher association with AMR (Breijyeh et al., 2020; Tacconelli et al., 2018) because these bacteria have an additional permeability barrier, the outer membrane (OM). This additional barrier comprises a unique lipid bilayer and can restrict many types of chemicals, including antibiotics (Nikaido, 2003).
The bacterial cell wall consists of polysaccharide with peptide stems called peptidoglycan (PG), located outside the cytoplasmic membrane, which is also known as the inner membrane (IM). By establishing peptide bonds between polysaccharide chains, PG becomes mesh-like in structure, with a high physical strength to withstand turgor pressure (Höltje, 1998; Typas et al., 2012). In Gram-negative bacteria, the aforementioned OM exists outside the PG layer and, unlike the IM, forms an asymmetric lipid bilayer composed of phospholipid in the inner leaflet and lipopolysaccharide (LPS) in the outer leaflet (Nikaido, 2003). The IM is a symmetric lipid bilayer comprised of phospholipid. Because of the presence of polysaccharide chains in the LPS layer, the OM has the unique ability to serve a barrier against hydrophobic molecules. In addition to acting as a chemical barrier, the OM also serves as a physical barrier to protect cells (Rojas et al., 2018; Sun et al., 2022). The region between the OM and IM, which contains the PG layer, is called the periplasm. Because this space is closed, this region forms a compartment outside of the cytoplasm. Figure 1 illustrates these structural components of the envelope of Gram-negative bacteria.
Gram-negative envelope architecture and lipoprotein processing and trafficking. The cytoplasmic membrane (inner membrane [IM]) in Gram-negative bacteria is surrounded by two additional layers—a peptidoglycan (PG) layer and the outer membrane (OM). The space between the IM and OM is closed, forming a compartment called the periplasm. Lipoprotein processing occurs as follows. After a lipoprotein is secreted, it can remain in the IM via its signal peptide (1). Lgt recognizes a lipobox in a lipoprotein and adds diacylglycerol to a cysteine residue in the lipobox (2). LspA removes the signal peptide (3). Finally, Lnt acylates the N-terminus of the new N-terminal cysteine (4). Depending on the lipobox signal, triacylated lipoproteins can be targeted to the OM. LolCDE, an ATPase complex, extracts a lipoprotein from the IM and transfers it to soluble LolA. An OM-anchored lipoprotein, LolB, receives the lipoprotein and facilitates its insertion into the OM
While transmembrane α-helical proteins are present in the IM, transmembrane β-barrel proteins (termed OM proteins [OMPs]) are found in the OM. Lipoproteins are anchored to both of the periplasmic faces of the IM and OM, and soluble proteins are present in the periplasm. Lpp, the most abundant OM lipoprotein in E. coli, connects the PG layer and OM (Braun & Rehn, 1969).
Growth of PG occurs during cellular division and elongation. PG growth requires the synthesis of new PG chains, cleaving of existing PG, and crosslinking of new PG chains with existing chains. Penicillin-binding proteins (PBPs) are synthases that synthesize a new PG chain and crosslink it with existing PG chains. β-Lactams such as penicillin inhibit the crosslinking step (Höltje, 1998; Typas et al., 2012).
OM biogenesis requires the biosynthesis and translocation of LPS, phospholipid, β-barrel transmembrane proteins, and lipoproteins (Konovalova et al., 2017; Ruiz et al., 2006; Tokuda, 2009). After synthesis in the cytoplasm and IM, LPS can be translocated onto the outer leaflet of the OM. LptBFG ATPase extracts LPS from the IM, and LptA forms a bridge to transfer LPS to LptDE, the OM translocon, which finally inserts LPS into the outer leaflet of the OM. It remains unclear how phospholipids move from the IM to the OM, but recent reports show that dedicated machinery may enable this process (Douglass et al., 2022; Grimm et al., 2020; Ruiz et al., 2021). β-Barrel assembly machinery (BAM) mediates the insertion of OMPs into the OM (Wu et al., 2005) (see details in the following section). OM lipoproteins are processed via Lgt, LspA, and Lnt and translocated via the Lol system (Okuda & Tokuda, 2011) (Fig. 1; see details in the following sections).
For Gram-negative bacteria to survive and protect themselves from a physically and chemically changing environment or from an attack by their host, the envelope must maintain its structural integrity. Remodeling structural integrity for adaptation is also imperative. The envelope stress response system, RpoE (σE), is responsible for maintaining structural integrity, with the Rcs and Cpx two-component systems (TCSs) being major components. Interestingly, a ribosome profiling study identified the Rcs and Cpx systems as the second and third most abundant TCSs in E. coli (Li et al., 2014). While the sensing mechanism of the σE system is well understood, the mechanisms for the Rcs and Cpx TCSs have only recently begun to be explored (Barchinger & Ades, 2013; Lima et al., 2013). Our review focuses on the mechanisms by which stress signals in the envelope can be converted for transcription in the cytoplasm by means of the IM signal cascade component. In this context, the sensing mechanisms of the sensor proteins, RcsF for Rcs and NlpE for Cpx, are especially relevant. Recently, many interesting observations and hypotheses for the sensing mechanism of RcsF have been reported; however, some details remain under debate. Thus, we will also describe and evaluate these issues in detail.
Rcs Pathway
The Rcs signaling pathway was first identified and named for its role in the regulation of capsular polysaccharide synthesis (Majdalani & Gottesman, 2005; Wall et al., 2018). In E. coli, mucoid colonies resulting from the production of colanic acid (capsular polysaccharide; wza and rcsA) and non-motile cells caused by the inhibition of flagella genes (flhDC) are the two strongest phenotypes that arise from Rcs activation (Francez-Charlot et al., 2003; Gottesman et al., 1985). Thus, Rcs is critical to the formation of biofilms in pathogenic bacteria as well as their virulence (Danese et al., 2000; Majdalani & Gottesman, 2005; Wall et al., 2018). Other genes induced by Rcs also appear to be necessary for maintenance and remodeling of the envelope. For example, rprA encodes a small RNA, RprA, which activates the translation of the stationary phase sigma factor-encoding rpoS (σS) mRNA, ftsZ encodes a protein that forms a ring at the future division site, osmB encodes an unknown osmotically induced lipoprotein, and osmC encodes a periplasmic peroxidase (Boulanger et al., 2005; Davalos-Garcia et al., 2001; Majdalani et al., 2002). The Rcs response can be activated by high osmolarity during growth as well as more direct stresses against envelope integrity (Sledjeski & Gottesman, 1996). As an example of a direct stress, mutations in LPS biosynthesis genes or treatment with the cationic antimicrobial peptide polymixin B (PMB) lead to a defect in OM integrity (Farris et al., 2010; Konovalova et al., 2016; Parker et al., 1992). PG elongation is arrested by treatment with mecillinam, a β-lactam inhibitor of PBP2, or S-(3,4-dichlorobenzyl) isothiourea (A22), an inhibitor of MreB, which forms the cytoskeleton for the elongasome of PG growth (Laubacher & Ades, 2008).
Composition of the Rcs System
As indicated by its name, a TCS consists of two components, a sensor kinase and a response regulator. Each TCS usually involves a unique pair. Environmental or stress signals are sensed by the sensor kinase, which then phosphorylates itself, and the phosphate can be transferred to the response regulator. The phosphorylated response regulator transcriptionally induces or represses the expression of corresponding genes (regulon).
RcsC is a sensor kinase and phosphorylates RcsB, a response regulator. The homodimer of modified RcsB binds to the promoters of the regulons (ftsZ, rprA, osmB, osmC, etc.) (Fig. 2A). Phosphorylated RcsB can also form a heterodimer with RcsA to control capsule synthesis and motility. Additionally, non-phosphorylated RcsB can form a heterodimer with other proteins to regulate specific gene expression (Wall et al., 2018). This process is independent of Rcs phosphorylation signaling. Rcs signaling leads to a more complicated situation. RcsD, structurally resembling RcsC, transfers phosphate between RcsC and RcsB (Fig. 2A). IgaA acts as a negative regulator of Rcs activation by interacting with RcsD (Cano et al., 2002; Wall et al., 2020). Finally, an OM stress sensing lipoprotein, RcsF, serves to control the system. The manner in which these multiple components contribute to Rcs activation is an interesting topic. In particular, researchers have proposed that RcsF acts as a sensor. The mechanism by which RcsF senses stress is one of the most interesting questions in Rcs research because RcsF is located exactly where stress is generated.
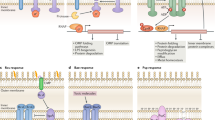
Rcs signaling pathway. A RcsC is a sensor kinase and phosphorylates RcsD, a phosphorelay component. RcsD phosphorylates RcsB, a response regulator. IgaA acts as a negative regulator for Rcs signaling by inhibiting RcsD function. RcsF, an OM lipoprotein, can activate the Rcs response by relieving the inhibition of IgaA. B RcsF-OMP sensor model. OMPs are assembled in the OM by BAM, which also funnels RcsF into OMPs. The flexible N-terminal linker (NTL) of RcsF can be threaded through the lumens of OmpC/F when they form complexes. LPS stresses can destabilize the complexes to allow RcsF to interact with the Rcs regulation system. C BAM sensor model. RcsF funneling into OmpC/F is mediated by BAM, as in the RcsF-OMP model. However, in the BAM sensor model, BAM function plays a vital role in the process, because the assembly of OMPs and the loading of RcsF into BamA are both dependent on BAM function. The unloaded RcsF in the periplasm interacts with the C-terminal domain (CTD) of OmpA or IgaA. This fraction increases under stressed conditions, and then IgaA, which has a much stronger affinity for RcsF, takes up more RcsF, activating Rcs. D Ribbon diagram of structure of the BamA–RcsF complex in side view. (PDB: 6T1W). BamA adopts an inward-open conformation, with the lateral gate to the membrane closed. RcsF is lodged deep within the lumen of the BamA barrel. Five POTRA (P1-P5) domains are present as NTDs. BamA, yellow; RcsF, magenta. The lateral gate is indicated by an arrow
RcsF
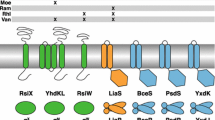
RcsF is an OM lipoprotein with a long flexible linker domain at the N-terminus and a C-terminal globular domain (Leverrier et al., 2011; Rogov et al., 2011; Umekawa et al., 2013). The C-terminal domain (CTD) interacts directly with the negative regulator, IgaA, to relieve Rcs repression in stress conditions (Fig. 2A) (Cho et al., 2014; Hussein et al., 2018). In E. coli, the OM and PG are covalently connected by the most abundant protein, Lpp, which forms an α-helical trimer with coiled-coiled motifs and a rigid pillar-like structure dictating a relatively constant height between the OM and PG (Asmar & Collet, 2018; Braun & Rehn, 1969). When Lpp is artificially elongated by the addition of its own amino-acid residues (+ 14, + 21, + 42, and + 63), the distance between the PG and OM can also be increased (Asmar et al., 2017; Cohen et al., 2017). Under these conditions, RcsF signaling is blocked because RcsF cannot reach IgaA in the IM. However, by elongating the N-terminal linker (NTL) of RcsF, Rcs stress sensing is rescued, demonstrating that the OM-targeted RcsF directly reaches the IM component, IgaA (Asmar et al., 2017). Recent genetic analysis and structural simulation showed that the periplasmic domain of IgaA is quite extended toward the OM and that its tip can interact with the N-terminal helix of RcsF CTD (Lach et al., 2023).
Studies have shown that the length and flexibility of the RcsF NTL are critical in RcsF trafficking to the OM (El Rayes et al., 2021). Many other OM-targeting lipoproteins also share this property. Furthermore, the Lol trafficking system appears to select the lipoprotein NTL with this property (Fig. 1). LolCDE, an ATPase complex, extracts lipoproteins from the IM and transfers them to LolA. LolB receives lipoproteins from LolA and inserts them into the OM (Okuda & Tokuda, 2011). The structures of LolCDE have been solved, and studies have shown that some of these structures can carry passenger lipoproteins, including RcsF inside the cavity formed by LolC and LolE (Bei et al., 2022; Sharma et al., 2021; Tang et al., 2021). RcsF NTL or N-terminal unstructured residues of Lpp with the triacyl chains could be visualized, but not the globular domain or α-helical structure of Lpp, which may explain why the long, flexible NTL is essential for lipoprotein trafficking.
Stress Sensing Mechanism of RcsF (RcsF-OMP Sensor Model vs. BAM Sensor Model)
The RcsF stress sensing mechanism has received much attention because it clearly explains how envelope stresses are sensed at a molecular level (Meng et al., 2021; Narita & Tokuda, 2017; Wall et al., 2018). There is agreement on the involvement of multiple OMPs in the sensing mechanism via RcsF, but currently, two different models have been proposed for the mechanistic details. These models may not be mutually exclusive. We provide a point-by-point discussion of these details and express our opinions about the mechanisms. As our group proposed the “BAM sensor model,” we aim to give insight into this elegant, but very complicated signaling system.
RcsF-OMP Sensor Model
BAM mediates the formation of a complex between RcsF and OMPs such as OmpA, OmpC, and OmpF, the most abundant proteins (see details in the next section) (Cho et al., 2014; Konovalova et al., 2014). The structurally flexible NTL of RcsF is threaded through the lumen of OmpC and OmpF (OmpC/F) (Konovalova et al., 2014), and it has been proposed that the N-terminal triacyls are embedded in the LPS layer. It is thought that this interaction allows the RcsF NTL to be detected on the surface of bacterial cells while the CTD is facing the periplasm (Fig. 2B). This result is quite surprising, as no lipoprotein has been previously shown to have such a topology in E. coli. Yet, in another study using a specific antibody against the RcsF CTD, RcsF was detected on the surface of bacterial cells (Cho et al., 2014). This topology suggests that the N-terminal triacyls are embedded in the inner leaflet of the OM. Thus, the two proposed topologies are contradictory and need to be resolved by further biochemical analyses.
As described above, Konovalova et al. (2014) claimed that the RcsF NTL is surface-exposed by being complexed with OmpC/F. While the RcsF NTL has many positively charged residues, the LPS molecule is negatively charged by multiple phosphorylations. Therefore, Konovalova et al. (2016) proposed that the electrostatic charge interaction between LPS and the RcsF NTL plays a crucial role in sensing. Furthermore, it has also been hypothesized that when this interaction is impaired by LPS stress, the RcsF-OMP complex can be disassembled, thus freeing RcsF and inducing the Rcs response (Fig. 2B). Herein, we refer to this hypothesis as the “RcsF-OMP sensor model.” This hypothesis is supported by the following evidence. (1) When the OMP flux is limited by ompA deletion (OmpA is one of the most abundant proteins in E. coli) or when the RcsF NTL is mutated to cause a defect in the interaction with OmpA, Rcs activation is reduced because the formation of the sensory component, RcsF-OMP, decreases. (2) Several LPS variants with reduced negative charges induce the Rcs response. (3) Changing the positively charged amino acids into alanines in the RcsF NTL dampens the Rcs response when LPS stress is applied. We have concerns about this model due to the indirect nature of its supporting evidence, leading to potential misinterpretations or incorrect conclusions. For example, the RcsF NTL may influence the availability of RcsF for IgaA by increasing the interaction with other proteins or by impacting the flexibility of the NTL for the RcsF CTD to reach IgaA (see section entitled “RcsF”). A reduction of negative charges in LPS can also influence the activity of the BAM complex, which is surrounded by LPS (see section entitled “BAM Sensor Model”). The topological state of the RcsF-OMP complex is controversial, but the presence of the RcsF NTL on the surface is a key feature of this model. Hence, biochemical experiments, such as determining the complex structure or analyzing crosslinking patterns before and after stress in vivo and in vitro, are necessary to validate the “RcsF-OMP sensor model”.
There is also a critical need to clarify how RcsF and OmpA interact. The Konovalova group has claimed that the interacting mode is similar to that between RcsF and OmpC/F, but our work has shown that RcsF interacts with the OmpA CTD, as shown in Fig. 2B and C (Dekoninck et al., 2020; Tata & Konovalova, 2019; Tata et al., 2021). Major porins in E. coli have distinctive conformations—OmpA has an 8-β-strand barrel in the N-terminal domain (NTD) and a soluble CTD while OmpC/F have 16-β-strand barrels. RcsF forms complexes with these porins (Cho et al., 2014; Dekoninck et al., 2020; Konovalova et al., 2014). OmpC/F has a luminal space that can accommodate a flexible peptide, such as an unstructured region of a bacteriocin, and, most likely, the RcsF NTL (Housden et al., 2013). It has been proposed that a very small fraction of OmpA exists as a large transmembrane β-barrel comprised of the β-barrel NTD and the CTD, which is normally soluble (Reusch, 2012). However, only indirect evidence has been provided to support this conformation. The formation of this large barrel would be required for OmpA to function as OmpC/F for RcsF. Thus far, no such data have been reported. We have sought to obtain evidence of the large barrel of OmpA, but have been unsuccessful. Instead, we have found that RcsF interacts with the OmpA CTD, not the β-barrel, supporting the two-domain model of OmpA (Dekoninck et al., 2020). The affinity between RcsF and the OmpA CTD is very weak; this finding is perplexing because the interaction occurs in the periplasm, where the periplasmic domain of IgaA can strongly interact with RcsF for Rcs activation (Dekoninck et al., 2020). However, the abundance of these two proteins (OmpA vs. IgaA: 300,000 vs. 200) (Li et al., 2014) may be an important factor in these interactions. We have proposed that a large number of OmpA CTDs can reduce Rcs activation by competing with IgaA, acting as a “buffer.” Overactivation of Rcs can kill cells, even under stressed conditions (Cano et al., 2002; Cho et al., 2014). Of note, this hypothesis is supported by the finding that an absence of OmpA induces the Rcs response (Cho et al., 2014; Dekoninck et al., 2020). However, Konovalova et al. (2016) observed the opposite result in the ompA strain. This discrepancy needs to be addressed.
It is risky to assume that the RcsF-OmpA complex has the same structure as the RcsF-OmpC/F complex. If the “RcsF-OMP sensor model” turns out to be valid, it would be likely that RcsF-OmpC/F, but not RcsF-OmpA, acts as the sensor in this process.
BAM Sensor Model
The BAM complex serves as machinery to assemble OMPs into the OM. BamA itself is an OMP and is the central component of the BAM complex, containing BamB, BamC, BamD, and BamE (BamBCDE), four OM lipoproteins, as subunits (Fig. 2B and C). When RcsF is translocated into the OM, it interacts with BamA. Although the BAM complex assembles OMPs, it can also co-assemble RcsF with OmpC/F (Fig. 2B and C; we denote this ability as the funneling function of BAM for RcsF), which has currently been proposed to govern the formation of RcsF-OmpC/F (Cho et al., 2014; Dekoninck et al., 2020; Konovalova et al., 2014; Rodriguez-Alonso et al., 2020). However, as described above, we suggest that some fraction of RcsF can escape this process, be exposed to the periplasm, and interact with the OmpA CTD. We proposed that under envelope-stressed conditions, BAM function is impaired, and the BAM complex fails to assemble OmpC/F and bind to RcsF, which leads to increased free RcsF and activation of Rcs (Fig. 2C). We denote this model as the “BAM sensor model” in this review. This model is supported by the following evidence (Cho et al., 2014; Rodriguez-Alonso et al., 2020). (1) BamA can be chemically crosslinked (which demonstrates protein–protein interactions) with RcsF under non-stressed conditions but not under diverse envelope-stressed conditions, clearly showing the stress sensitivity of the interaction of RcsF and BamA. (2) RcsF can be purified with the BamABCDE holocomplex as well as the BamAB complex. We have proposed that this complex is the intermediate state of the RcsF-Bam complex prior to the formation of the RcsF-OMP complex. (3) The crystal structure of RcsF-BamA clearly demonstrates how RcsF senses stress (Fig. 2D). RcsF is lodged in the luminal space of the BamA barrel, where the gate for OMP assembly is located, which is essential for BamA function (Noinaj et al., 2017). In this structure, BamA displays an inward-open conformation and the gate is in a closed conformation. RcsF binding regions have been proposed to undergo outward and lateral opening during OMP insertion. (4) The closed-gate conformation, but not the open-gate form, of BamA allows RcsF to bind (Fig. 2D). This mode of binding not only fits well with the funneling function of BamA for RcsF and OMPs, but also suggests that stress signals may impact the gate conformation of BamA.
The recent research by the Konovalova group challenges the validity of “BAM sensor model” (Tata et al., 2021). However, in the following section, we present arguments in support of the “BAM sensor model.” Before presenting our rebuttal argument, we first clarify the apparent opposite interacting behavior of BamA with RcsF depending on the type of stress, to help avoid confusion regarding the mechanism. Rigel et al. (2012, 2013) have shown that deletion of bamE causes a defect in BAM function, where BamA takes on a conformation that is more sensitive to proteolytic digestion. It is possible that this conformation allows RcsF to stall more on BamA, thus dampening the Rcs response (Konovalova et al., 2016; Tata & Konovalova, 2019). BamA alone is not functional. When BamA is overexpressed, more RcsF stalls on BamA, resulting in dampened Rcs activation, as in bamE deletion (Cho et al., 2014; Tata et al., 2021). In this state, BamA must be in the closed-gate conformation (Fig. 2D), which we refer to as the “locked-up state of BamA for RcsF.” In contrast, RcsF does not interact with BamA upon treatment with Rcs-inducing stresses, such as stress induced by mecillinam, A22, PMB, or mutations in LPS biosynthesis genes (Cho et al., 2014). We have attributed this defect to the impairment of BamA function. Because the latter two stresses target the OM, they are not difficult to explain; however, the former two stresses require confirmation. In these stress conditions, perhaps BamA is in another conformation and cannot interact with RcsF. We call this conformation the “non-binding state of BamA for RcsF.” Thus, BamA can remain in at least two compromised conformations, either with or without RcsF binding, which can result in different Rcs responses. Perhaps the “BAM sensor model” needs to be expanded to explain why the Rcs response is weaker in some cases. In fact, it may be possible for the “locked-up state of BamA for RcsF” to occur without genetic manipulation under certain stress conditions.
Here, we provide point-by-point rebuttal arguments to support the “BAM sensor model” in light of arguments by the Konovalova group. (i) It is possible to isolate rcsF mutants and bamA mutants, which show a weaker interaction between RcsF and BamA but the same Rcs response to stress observed for the wild type (WT). Accordingly, Tata et al. (2021) have claimed that RcsF does not monitor BAM function. However, in such isolated mutants, it must be considered that RcsF funneling into OMPs still depends on BAM activity (BamA function). If BamA function is compromised, then Rcs activation should occur, which would verify the “BAM sensor model.” Regarding the RcsF-BamA crosslinking results, these isolated mutant proteins are likely to have a lower affinity for each other, which could affect the kinetics of the funneling process, but not the function itself. Otherwise, a high level of Rcs activation should be observed. Yet, stresses that negatively impact RcsF-BamA complex formation have an effect on BamA function for OMP assembly as well as RcsF funneling, thus inducing the Rcs response. (ii) BamAE373K is not functional at 37 °C (Ricci et al., 2012). However, RcsF is similarly trapped in both BamAE373K and WT BamA. Because BamAE373K does not allow RcsF funneling, excess RcsF should be released to induce the Rcs response. Thus, in this context, the “BAM sensor model” is still valid. (iii) Darobactin is a direct inhibitor of BamA function (Imai et al., 2019). Therefore, its treatment causes misfolded OMPs to accumulate and the sensory machinery, the σE system, to be activated (Barchinger & Ades, 2013). Darobactin treatment can also induce the Rcs response. At higher concentrations of magnesium ions, the LPS layer can be stabilized further, but it cannot reverse the effect of darobactin on BamA function; thus, the σE response is still induced, but not the Rcs response. This may provide strong evidence against RcsF monitoring of BAM function. However, recent reports have revealed that darobactin-bound BamA is in a closed-gate conformation (Imai et al., 2019; Kaur et al., 2021). Moreover, to grow crystals of darobactin-bound BamA, magnesium ions were added. Therefore, as discussed above, it may be possible for darobactin to stimulate a shift in the BamA conformation into the “locked-up state of BamA for RcsF” in the presence of magnesium ions. (iv) Two groups have found that when bamE is deleted, BamA retains a high level of RcsF, causing a defective function of BAM (Hart et al., 2019a, 2019b; Tata and Konovalova, 2019). This defect can be suppressed by deleting rcsF. Tata et al. (2021) observed that when BamE was depleted, the crosslinked quantity of RcsF-BamA increased and that of RcsF-OmpA decreased. BamE induction did not overturn this effect. Therefore, they concluded that RcsF-BamA is a dead-end product, not an intermediate for RcsF-OMPs. We disagree and present two pieces of evidence that contradict this conclusion (Rodriguez-Alonso et al., 2020). First, RcsF can be purified with the BamABCDE holocomplex. Second, RcsF-BamA crosslinking is always observed in WT cells, but overexpression of BamCDE reduces this crosslinking. This suggests that a small fraction of RcsF-BamA without BamCDE exists as an intermediate, where binding of BamCDE causes instability in RcsF, either by itself or by OMP flux.
In sum, we believe that the “BAM sensor model” is a reasonable model for explaining how RcsF monitors BamA activity to control Rcs activation. Future work will need to show that RcsF indeed interacts in the same way with BamA(B) when complexed with BamABCDE. Further research should also focus on obtaining detailed structural information for the “locked-up state of BamA for RcsF” and “non-binding state of BamA for RcsF.” The manner in which PG stresses can be translated into a defect in BamA and Rcs activation is also an outstanding issue. A recent report on the differential interaction between BamA and new or old PG may be relevant to this topic (Mamou et al., 2022).
Cpx Pathway
Misfolding of IM proteins or periplasmic proteins activates the Cpx TCS. However, Cpx sensing appears to be inactive for misfolded OMPs in the periplasm, which are mainly sensed by the σE system (Mitchell & Silhavy, 2019; Raivio, 2014). The specific signals for this kind of Cpx activation include elevated pH, altered membrane phospholipid composition, overexpression of misfolded pilus subunits, perturbation in lipoprotein production, high osmolarity, and copper (Danese & Silhavy, 1998; Hung et al., 2001; Itou et al., 2012; Jubelin et al., 2005; Yamamoto & Ishihama, 2006). Recent work has shown that PG perturbation can also induce a signal (Bernal-Cabas et al., 2015; Delhaye et al., 2016; Evans et al., 2013). Protein misfolding due to PG perturbation has also been implicated in Cpx activation, but no evidence has been provided (Grabowicz & Silhavy, 2017). When bacteria attach to a hydrophobic surface, the adhesion signal activates the Cpx system (Otto & Silhavy, 2002; Shimizu et al., 2016). Many proteins expressed by the Cpx activation signals play a role in protein homeostasis, including a periplasmic protease chaperone, DegP; a protein disulfide oxidase, DsbA; peptidyl prolyl isomerases, PpiA and PpiD; chaperones CpxP and Spy; and YccA, an adaptor for degradation (Danese & Silhavy, 1997, 1998; Danese et al., 1995; Pogliano et al., 1997; Yamamoto & Ishihama, 2006). A PG l,d-transpeptidase (LdtD), a PG-lytic transglycosylase (Slt), and two PG amidases (AmiA and AmiC) can also be expressed to remodel the perturbed PG (Bernal-Cabas et al., 2015; Delhaye et al., 2016; Weatherspoon-Griffin et al., 2011). The Cpx response is also critical for biofilm formation. One can reduce the motility by inhibiting the induction of genes for parts of the flagella motors (motAB), but the production of curli proteins increases with increasing expression of a regulator for curli assembly (csgD) (De Wulf et al., 1999; Jubelin et al., 2005). The Cpx response induces the expression of small RNAs, RprA, CpxQ, and CyaR (Chao & Vogel, 2016; Vogt et al., 2014). rprA is also a Rcs regulon. CpxQ is derived from cpxP mRNA and negatively regulates the translation of skp, encoding a periplasmic chaperone to prevent mistargeting of OMPs into the IM (Grabowicz et al., 2016).
Composition of the Cpx System
The Cpx TCS consists of a sensor kinase CpxA and a response regulator CpxR (Fig. 3A). cpxP is a regulon of the Cpx TCS, and CpxP is a direct negative regulator of CpxA; thus, the Cpx response is controlled by feedback inhibition (Fleischer et al., 2007; Raivio et al., 1999). CpxP is also a chaperon and can thus interact with misfolded proteins. Interestingly, DegP can degrade CpxP when it interacts with misfolded proteins, relieving inhibition of the Cpx response (Buelow & Raivio, 2005; DiGiuseppe & Silhavy, 2003; Isaac et al., 2005), which provides a mechanism for sensing misfolded proteins. NlpE, an OM lipoprotein, is also a signaling module for the Cpx system. NlpE overexpression can activate the Cpx response (Snyder et al., 1995).
Cpx signaling pathway. A Misfolding of IM proteins or periplasmic proteins activates the Cpx system. CpxA is a sensor kinase and phosphorylates CpxR, a response regulator. Two auxiliary proteins, CpxP and NlpE, control the Cpx response. CpxP acts as a negative regulator of Cpx signaling by inhibiting CpxA. NlpE, an OM lipoprotein, can activate the Cpx response. CpxA contains a periplasmic domain that interacts with CpxP and NlpE. B NlpE is a sensor for OM lipoprotein trafficking. Under stress conditions, retention in the IM causes Cpx activation, which is crucial to cell survival. The N-terminal domain (NTD) of NlpE interacts directly with the periplasmic domain of CpxA. NlpE homologs containing only the NTD appear to be highly conserved outside of Enterobacterales (Fig. 1A) and have longer NTLs than E. coli NlpE. C The NlpE CTD is implicated in Cpx signaling due to redox perturbation or adhesion to a hydrophobic surface. The NlpE CTD has a disulfide bond, but when this bond cannot be formed, a Cpx response can be induced. Both NlpE and OmpA are required for the activation of Cpx via hydrophobic surface adhesion. The OmpA CTD interacts with the NlpE NTD. Sensing is dependent on OmpA CTD binding to PG. Moreover, the NlpE CTD is indispensable for this sensing. However, in two recent studies (Cho et al., 2022; Delhaye et al., 2019), there was no evidence to show a direct interaction between the NlpE CTD and CpxA, suggesting that indirect activation may occur
As two auxiliary components are present, a negative regulator and an OM sensor, the Cpx system appears to have substantial similarity to the Rcs system. Recently, many excellent studies have elucidated the molecular mechanisms of NlpE function, as described in the following sections.
Stress Sensing Mechanism via NlpE
In its crystal structure, NlpE has two soluble β-barrel domains (NTD and CTD) and forms a dimer (Hirano et al., 2007). The two monomers form a domain-swapped dimer in which the two NTDs exchange the last β-sheet at the C-terminus. It has been proposed that the NTD is unstable after unfolding and extending, which allows the CTD to reach CpxA in the IM across the periplasm to activate Cpx. However, this does not appear to be the case, since the NTD alone, but not the CTD, can directly activate the Cpx system (Delhaye et al., 2019; May et al., 2019). Thus, the significance of the NTD swap remains unclear.
Two studies have recently demonstrated that NlpE senses lipoprotein trafficking from the IM to the OM, where failure of this trafficking leads to the induction of Cpx in an NlpE-dependent manner (Fig. 3B) (Delhaye et al., 2019; May et al., 2019). Importantly, Cpx activation is crucial for cell survival under conditions of lipoprotein trafficking stress. NlpE itself is a lipoprotein; thus, by being retained in the IM, it can activate Cpx by interacting with CpxA in the IM. Copper stress activates Cpx, allowing cells to adapt, and research has shown that NlpE confers copper tolerance (Gupta et al., 1995; Yamamoto & Ishihama, 2006). However, the molecular mechanism remains unclear. In their study, the Grabowicz group explained the Cpx induction mechanism in the presence of copper stress as follows (May et al., 2019). A lipoprotein has a lipobox containing cysteine at the C-terminus of the signal peptide (Fig. 1). The thiol of the cysteine can be di-acylated by the Lgt enzyme; subsequently, the signal peptide is digested just before the cysteine, and the new N-terminal cysteine is acylated. The copper ion can oxidize the thiol of the cysteine in the lipobox, preventing the acylation and therefore the trafficking of lipoproteins. Thus, copper stress causes a malfunction in lipoprotein trafficking.
Moreover, two studies have shown that perturbation of oxidative protein folding can activate Cpx. This is not surprising, since dsbA, which encodes the periplasmic protein thiol oxidase DsbA, is a Cpx regulon. One group showed that the Cpx response can be induced in a dsbA-null mutant in an NlpE-dependent manner (Fig. 3C) (Delhaye et al., 2019). NlpE has four cysteines forming two pairs of disulfide bonds, and each pair is present in the NTD and CTD. Their formation depends on DsbA. However, the NlpE NTD alone cannot activate Cpx in the dsbA strain. Furthermore, the variant containing alanines instead of the two cysteines in the CTD, which cannot have a disulfide bond, induces the Cpx response in the presence of DsbA, but not the NTD variant. This finding suggests that misfolding of the CTD can trigger the Cpx response. However, in the second study, the Cpx response was activated independent of NlpE (Jaswal et al., 2021). Long-chain fatty acids can inhibit the oxidation of quinone in the IM, which is critical for the recycling of DsbA by DsbB, a DsbA:quinone oxido-reductase (Bader et al., 1999). Consequently, oxidative folding in the periplasm was perturbed in the second study. This stress can indeed activate the Cpx response but is independent of NlpE. Further study is needed to clarify this discrepancy.
As described above, the NlpE NTD alone is sufficient to transduce Cpx activation signals. However, as some evidence indicates that the CTD plays a vital role in activation, the activation mechanism may not be as simple as previously thought. Misfolding of the NlpE CTD can activate Cpx as shown above. This result could be due to direct or indirect communication of the CTD with the Cpx signaling molecules. Alternatively, this result could be due to a defect in NTD trafficking caused by misfolding of the CTD. These possibilities must be tested.
NlpE also senses hydrophobic surface adhesion (Otto & Silhavy, 2002), and a recent study revealed the importance of the NlpE CTD in this function (Cho et al., 2022). Overexpression of OmpA and hydrophobic surface adhesion via OmpA are sensed by NlpE, which triggers the Cpx response. The NlpE NTD interacts with the OmpA CTD, which can bind to the PG (Fig. 3C) (Samsudin et al., 2016). PG binding determines the Cpx response. Interestingly, the NlpE CTD alone is sufficient to activate the Cpx response. From these results, Cho et al. (2022) concluded that NlpE in the OM senses the surface adhesion status via OmpA, in contrast to the mechanism for lipoprotein mislocalization stress. OmpA plays at least two roles in its functionality: the NTD has surface-exposed domains as well as an OM-embedded β-barrel, which can be used for phage adsorption, while the CTD connects the OM and PG (Bertozzi Silva et al., 2016). Therefore, it could be interesting to investigate how NlpE exploits these features of OmpA and transduces the signal of adherence to a hydrophobic surface to the Cpx system.
NlpE is composed of an NTD and CTD. However, the Pfam database reveals that NlpE homologs carrying only an NTD are more widely conserved (El-Gebali et al., 2019). Approximately 6000 sequences containing the NlpE NTD have been deposited (https://www.ebi.ac.uk/interpro/entry/pfam/PF04170/domain_architecture/; December 26, 2022). Overall, 1542 NlpE homologs contain both domains whereas 3777 NlpE homologs contain only the NTD. The two-domain NlpE architecture is primarily found in Enterobacterales (Fig. S1A). NlpE homologs without the CTD are more widely conserved in Proteobacteria and Bacteroidetes. This trend may align with the finding that the NlpE NTD interacts directly with CpxA. The addition of the NlpE CTD may have evolved to modulate signaling parameters such as surface adhesion. Bacteroides fragilis, Vibrio cholera, and Acinetobacter baumannii are the three representative bacteria that have only the NTD. Interestingly, all three of these bacteria have a longer NTL than the E. coli NlpE (Fig. S1B). As mentioned in the previous section, the length of the RcsF NTL is critical for Rcs signaling. Perhaps this long linker in the OM allows the NlpE NTD to reach Cpx components or their own targets in the IM for signaling purposes, similar to RcsF (Fig. 3B).
The molecular mechanism of NlpE is just beginning to be understood. Exciting future work awaits. We conclude this section by suggesting two interesting lines of further research. The NlpE NTD has two conserved cysteines forming Cys-Xaa-Xaa-Cys (CXXC) (Fig. S1B). Although the two cysteines have been shown to be dispensable for copper stress sensing, it may be interesting to investigate the function of CXXC because it is conserved and may be redox-active (May et al., 2019). As another outstanding area of research, future work could focus on how the Cpx response (which may involve a regulon) suppresses the toxicity caused by the malfunction in lipoprotein trafficking.
Concluding Remarks
The Rcs and Cpx systems are two major TCSs that protect cells from envelope stress. This review focused on the mechanisms by which the two OM lipoproteins RcsF and NlpE monitor stress and transduce these signals to downstream components. Here, we conclude by discussing three perspectives, including a biomedical application based on a mechanical understanding of these two sensors. (1) In the “BAM sensor model” proposed for stress sensing by RcsF, BAM activity is a key feature. Together with Lpt machinery, BAM is critical to OM biogenesis. Gram-negative bacteria are more problematic in AMR than Gram-positive bacteria. Therefore, BAM and Lpt machinery are good targets for developing novel antimicrobials because they are located in the OM and are thus more accessible to antimicrobials (Hart et al., 2019a, 2019b; Imai et al., 2019; Luther et al., 2019). Recently, the concept of the “BAM sensor model” was exploited to identify BAM inhibitors (Steenhuis et al., 2021). Darobactin, a BamA inhibitor, can also induce the Rcs response (Tata et al., 2021). (2) Other TCSs in E. coli or other Gram-negative bacteria may also deploy an OM lipoprotein as a sensor. Therefore, pioneering studies of RcsF and NlpE may aid in identifying and characterizing such sensors. (3) Rcs and Cpx TCSs play a critical role in modulating virulence in some pathogenic bacteria. Revisiting these cases with an updated mechanistic understanding may help us to better understand their roles.
Data availability
The statistics for NlpE NTD (PF04170) architecture from the website for the Pfam database and supplementary figures are available upon request.
References
Antimicrobial Resistance Collaborators. (2022). Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. The Lancet, 399, 629–655.
Asmar, A. T., & Collet, J. F. (2018). Lpp, the braun lipoprotein, turns 50-major achievements and remaining issues. FEMS Microbiology Letters, 365, fny199.
Asmar, A. T., Ferreira, J. L., Cohen, E. J., Cho, S. H., Beeby, M., Hughes, K. T., & Collet, J. F. (2017). Communication across the bacterial cell envelope depends on the size of the periplasm. PLoS Biology, 15, e2004303.
Bader, M., Muse, W., Ballou, D. P., Gassner, C., & Bardwell, J. C. (1999). Oxidative protein folding is driven by the electron transport system. Cell, 98, 217–227.
Barchinger, S. E., & Ades, S. E. (2013). Regulated proteolysis: Control of the Escherichia coli σE-dependent cell envelope stress response. In D. Dougan (Ed.), Regulated proteolysis in microorganisms. Subcellular biochemistry (Vol. 66, pp. 129–160). Springer.
Bei, W., Luo, Q., Shi, H., Zhou, H., Zhou, M., Zhang, X., & Huang, Y. (2022). Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli. PLoS Biology, 20, e3001823.
Bernal-Cabas, M., Ayala, J. A., & Raivio, T. L. (2015). The Cpx envelope stress response modifies peptidoglycan cross-linking via the L, D-transpeptidase LdtD and the novel protein YgaU. Journal of Bacteriology, 197, 603–614.
Bertozzi Silva, J., Storms, Z., & Sauvageau, D. (2016). Host receptors for bacteriophage adsorption. FEMS Microbiology Letters, 363, fnw002.
Boulanger, A., Francez-Charlot, A., Conter, A., Castanie-Cornet, M. P., Cam, K., & Gutierrez, C. (2005). Multistress regulation in Escherichia coli: Expression of osmB involves two independent promoters responding either to σS or to the RcsCDB His-Asp phosphorelay. Journal of Bacteriology, 187, 3282–3286.
Braun, V., & Rehn, K. (1969). Chemical characterization, spatial distribution and function of a lipoprotein (Murein-lipoprotein) of the E. coli cell wall. European Journal of Biochemistry, 10, 426–438.
Breijyeh, Z., Jubeh, B., & Karaman, R. (2020). Resistance of Gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules, 25, 1340.
Buelow, D. R., & Raivio, T. L. (2005). Cpx signal transduction is influenced by a conserved N-terminal domain in the novel inhibitor CpxP and the periplasmic protease DegP. Journal of Bacteriology, 187, 6622–6630.
Cano, D. A., Domínguez-Bernal, G., Tierrez, A., Garcia-Del Portillo, F., & Casadesus, J. (2002). Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics, 162, 1513–1523.
Chao, Y., & Vogel, J. (2016). A 3’ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Molecular Cell, 61, 352–363.
Cho, S. H., Szewczyk, J., Pesavento, C., Zietek, M., Banzhaf, M., Roszczenko, P., Asmar, A., Laloux, G., Hov, A. K., Leverrier, P., et al. (2014). Detecting envelope stress by monitoring β-barrel assembly. Cell, 159, 1652–1664.
Cho, T. H. S., Wang, J., & Raivio, T. L. (2022). NlpE is an OmpA-associated outer membrane sensor of the Cpx envelope stress response. bioRxiv. https://doi.org/10.1101/2022.10.18.512811
Cohen, E. J., Ferreira, J. L., Ladinsky, M. S., Beeby, M., & Hughes, K. T. (2017). Nanoscale-length control of the flagellar driveshaft requires hitting the tethered outer membrane. Science, 356, 197–200.
Danese, P. N., Pratt, L. A., & Kolter, R. (2000). Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. Journal of Bacteriology, 182, 3593–3596.
Danese, P. N., & Silhavy, T. J. (1997). The σE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes and Development, 11, 1183–1193.
Danese, P. N., & Silhavy, T. J. (1998). CpxP, a stress-combative member of the Cpx regulon. Journal of Bacteriology, 180, 831–839.
Danese, P. N., Snyder, W. B., Cosma, C. L., Davis, L. J., & Silhavy, T. J. (1995). The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes and Development, 9, 387–398.
Davalos-Garcia, M., Conter, A., Toesca, I., Gutierrez, C., & Cam, K. (2001). Regulation of osmC gene expression by the two-component system rcsB-rcsC in Escherichia coli. Journal of Bacteriology, 183, 5870–5876.
De Wulf, P., Kwon, O., & Lin, E. C. (1999). The cpxra signal transduction system of Escherichia coli: Growth-related autoactivation and control of unanticipated target operons. Journal of Bacteriology, 181, 6772–6778.
Dekoninck, K., Létoquart, J., Laguri, C., Demange, P., Bevernaegie, R., Simorre, J. P., Dehu, O., Iorga, B. I., Elias, B., Cho, S. H., & Collet, J. F. (2020). Defining the function of OmpA in the Rcs stress response. eLife, 9, e60861.
Delhaye, A., Collet, J. F., & Laloux, G. (2016). Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. mBio, 7, e00047-16.
Delhaye, A., Laloux, G., & Collet, J. F. (2019). The lipoprotein NlpE is a cpx sensor that serves as a sentinel for protein sorting and folding defects in the Escherichia coli envelope. Journal of Bacteriology, 201, e00611-e618.
DiGiuseppe, P. A., & Silhavy, T. J. (2003). Signal detection and target gene induction by the CpxRA two-component system. Journal of Bacteriology, 185, 2432–2440.
Douglass, M. V., McLean, A. B., & Trent, M. S. (2022). Absence of YhdP, TamB, and YdbH leads to defects in glycerophospholipid transport and cell morphology in Gram-negative bacteria. PLoS Genetics, 18, e1010096.
El Rayes, J., Szewczyk, J., Deghelt, M., Csoma, N., Matagne, A., Iorga, B. I., Cho, S. H., & Collet, J. F. (2021). Disorder is a critical component of lipoprotein sorting in Gram-negative bacteria. Nature Chemical Biology, 17, 1093–1100.
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., Qureshi, M., Richardson, L. J., Salazar, G. A., Smart, A., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Research, 47, D427–D432.
Evans, K. L., Kannan, S., Li, G., de Pedro, M. A., & Young, K. D. (2013). Eliminating a set of four penicillin binding proteins triggers the Rcs phosphorelay and Cpx stress responses in Escherichia coli. Journal of Bacteriology, 195, 4415–4424.
Farris, C., Sanowar, S., Bader, M. W., Pfuetzner, R., & Miller, S. I. (2010). Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. Journal of Bacteriology, 192, 4894–4903.
Fleischer, R., Heermann, R., Jung, K., & Hunke, S. (2007). Purification, reconstitution, and characterization of the CpxRAP envelope stress system of Escherichia coli. The Journal of Biological Chemistry, 282, 8583–8593.
Francez-Charlot, A., Laugel, B., Van Gemert, A., Dubarry, N., Wiorowski, F., Castanié-Cornet, M. P., Gutierrez, C., & Cam, K. (2003). RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Molecular Microbiology, 49, 823–832.
Gottesman, S., Trisler, P., & Torres-Cabassa, A. (1985). Regulation of capsular polysaccharide synthesis in Escherichia coli k-12: Characterization of three regulatory genes. Journal of Bacteriology, 162, 1111–1119.
Grabowicz, M., Koren, D., & Silhavy, T. J. (2016). The CpxQ sRNA negatively regulates Skp to prevent mistargeting of β-barrel outer membrane proteins into the cytoplasmic membrane. mBio, 7, e00312-16.
Grabowicz, M., & Silhavy, T. J. (2017). Envelope stress responses: An interconnected safety net. Trends in Biochemical Sciences, 42, 232–242.
Grimm, J., Shi, H., Wang, W., Mitchell, A. M., Wingreen, N. S., Huang, K. C., & Silhavy, T. J. (2020). The inner membrane protein YhdP modulates the rate of anterograde phospholipid flow in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 117, 26907–26914.
Gupta, S. D., Lee, B. T., Camakaris, J., & Wu, H. C. (1995). Identification of cutC and cutF (nlpE) genes involved in copper tolerance in Escherichia coli. Journal of Bacteriology, 177, 4207–4215.
Hart, E. M., Gupta, M., Wühr, M., & Silhavy, T. J. (2019a). The synthetic phenotype of ΔbamB ΔbamE double mutants results from a lethal jamming of the Bam complex by the lipoprotein RcsF. mBio, 10, e00662-19.
Hart, E. M., Mitchell, A. M., Konovalova, A., Grabowicz, M., Sheng, J., Han, X., Rodriguez-Rivera, F. P., Schwaid, A. G., Malinverni, J. C., Balibar, C. J., et al. (2019b). A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proceedings of the National Academy of Sciences of the United States of America, 116, 21748–21757.
Hirano, Y., Hossain, M. M., Takeda, K., Tokuda, H., & Miki, K. (2007). Structural studies of the Cpx pathway activator NlpE on the outer membrane of Escherichia coli. Structure, 15, 963–976.
Höltje, J. V. (1998). Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiology and Molecular Biology Reviews, 62, 181–203.
Housden, N. G., Hopper, J. T., Lukoyanova, N., Rodriguez-Larrea, D., Wojdyla, J. A., Klein, A., Kaminska, R., Bayley, H., Saibil, H. R., Robinson, C. V., & Kleanthous, C. (2013). Intrinsically disordered protein threads through the bacterial outer-membrane porin OmpF. Science, 340, 1570–1574.
Hung, D. L., Raivio, T. L., Jones, C. H., Silhavy, T. J., & Hultgren, S. J. (2001). Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. The EMBO Journal, 20, 1508–1518.
Hussein, N. A., Cho, S. H., Laloux, G., Siam, R., & Collet, J. F. (2018). Distinct domains of Escherichia coli IgaA connect envelope stress sensing and down-regulation of the rcs phosphorelay across subcellular compartments. PLoS Genetics, 14, e1007398.
Imai, Y., Meyer, K. J., Iinishi, A., Favre-Godal, Q., Green, R., Manuse, S., Caboni, M., Mori, M., Niles, S., Ghiglieri, M., et al. (2019). A new antibiotic selectively kills Gram-negative pathogens. Nature, 576, 459–464.
Isaac, D. D., Pinkner, J. S., Hultgren, S. J., & Silhavy, T. J. (2005). The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. Proceedings of the National Academy of Sciences of the United States of America, 102, 17775–17779.
Itou, A., Matsumoto, K., & Hara, H. (2012). Activation of the Cpx phosphorelay signal transduction system in acidic phospholipid-deficient pgsA mutant cells of Escherichia coli. Biochemical and Biophysical Research Communications, 421, 296–300.
Jaswal, K., Shrivastava, M., & Chaba, R. (2021). Revisiting long-chain fatty acid metabolism in Escherichia coli: Integration with stress responses. Current Genetics, 67, 573–582.
Jubelin, G., Vianney, A., Beloin, C., Ghigo, J. M., Lazzaroni, J. C., Lejeune, P., & Dorel, C. (2005). CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. Journal of Bacteriology, 187, 2038–2049.
Kaur, H., Jakob, R. P., Marzinek, J. K., Green, R., Imai, Y., Bolla, J. R., Agustoni, E., Robinson, C. V., Bond, P. J., Lewis, K., et al. (2021). The antibiotic darobactin mimics a β-strand to inhibit outer membrane insertase. Nature, 593, 125–129.
Konovalova, A., Kahne, D. E., & Silhavy, T. J. (2017). Outer membrane biogenesis. Annual Review of Microbiology, 71, 539–556.
Konovalova, A., Mitchell, A. M., & Silhavy, T. J. (2016). A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. eLife, 5, e15276.
Konovalova, A., Perlman, D. H., Cowles, C. E., & Silhavy, T. J. (2014). Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proceedings of the National Academy of Sciences of the United States of America, 111, E4350–E4358.
Lach, S. R., Kumar, S., Kim, S., Im, W., & Konovalova, A. (2023). Conformational rearrangements in the sensory RcsF/OMP complex mediate signal transduction across the bacterial cell envelope. PLoS Genetics, 19, e1010601.
Laubacher, M. E., & Ades, S. E. (2008). The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. Journal of Bacteriology, 190, 2065–2074.
Leverrier, P., Declercq, J. P., Denoncin, K., Vertommen, D., Hiniker, A., Cho, S. H., & Collet, J. F. (2011). Crystal structure of the outer membrane protein RcsF, a new substrate for the periplasmic protein-disulfide isomerase DsbC. Journal of Biological Chemistry, 286, 16734–16742.
Li, G. W., Burkhardt, D., Gross, C., & Weissman, J. S. (2014). Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell, 157, 624–635.
Lima, S., Guo, M. S., Chaba, R., Gross, C. A., & Sauer, R. T. (2013). Dual molecular signals mediate the bacterial response to outer-membrane stress. Science, 340, 837–841.
Luther, A., Urfer, M., Zahn, M., Muller, M., Wang, S. Y., Mondal, M., Vitale, A., Hartmann, J. B., Sharpe, T., Monte, F. L., et al. (2019). Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature, 576, 452–458.
Majdalani, N., & Gottesman, S. (2005). The Rcs phosphorelay: A complex signal transduction system. Annual Review of Microbiology, 59, 379–405.
Majdalani, N., Hernandez, D., & Gottesman, S. (2002). Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Molecular Microbiology, 46, 813–826.
Mamou, G., Corona, F., Cohen-Khait, R., Housden, N. G., Yeung, V., Sun, D., Sridhar, P., Pazos, M., Knowles, T. J., Kleanthous, C., et al. (2022). Peptidoglycan maturation controls outer membrane protein assembly. Nature, 606, 953–959.
May, K. L., Lehman, K. M., Mitchell, A. M., & Grabowicz, M. (2019). A stress response monitoring lipoprotein trafficking to the outer membrane. mBio, 10, e00618-19.
Meng, J., Young, G., & Chen, J. (2021). The Rcs system in Enterobacteriaceae: Envelope stress responses and virulence regulation. Frontiers in Microbiology, 12, 627104.
Mitchell, A. M., & Silhavy, T. J. (2019). Envelope stress responses: Balancing damage repair and toxicity. Nature Reviews Microbiology, 17, 417–428.
Narita, S. I., & Tokuda, H. (2017). Bacterial lipoproteins; biogenesis, sorting and quality control. BBA Molecular and Cell Biology of Lipids, 1862, 1414–1423.
Nikaido, H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiology and Molecular Biology Reviews, 67, 593–656.
Noinaj, N., Gumbart, J. C., & Buchanan, S. K. (2017). The beta-barrel assembly machinery in motion. Nature Reviews Microbiology, 15, 197–204
Okuda, S., & Tokuda, H. (2011). Lipoprotein sorting in bacteria. Annual Review of Microbiology, 65, 239–259.
O'Neill, J. (2016). Tackling drug-resistant infections globally: Final report and recommendations. Review on Antimicrobial Resistance.
Otto, K., & Silhavy, T. J. (2002). Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proceedings of the National Academy of Sciences of the United States of America, 99, 2287–2292.
Parker, C. T., Kloser, A. W., Schnaitman, C. A., Stein, M. A., Gottesman, S., & Gibson, B. W. (1992). Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. Journal of Bacteriology, 174, 2525–2538.
Pogliano, J., Lynch, A. S., Belin, D., Lin, E. C., & Beckwith, J. (1997). Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes and Development, 11, 1169–1182.
Raivio, T. L. (2014). Everything old is new again: An update on current research on the Cpx envelope stress response. BBA Molecular Cell Research, 1843, 1529–1541.
Raivio, T. L., Popkin, D. L., & Silhavy, T. J. (1999). The Cpx envelope stress response is controlled by amplification and feedback inhibition. Journal of Bacteriology, 181, 5263–5272.
Reusch, R. N. (2012). Insights into the structure and assembly of Escherichia coli outer membrane protein A. The FEBS Journal., 279, 894–909.
Ricci, D. P., Hagan, C. L., Kahne, D., & Silhavy, T. J. (2012). Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proceedings of the National Academy of Sciences of the United States of America, 109, 3487–3491.
Rigel, N. W., Ricci, D. P., & Silhavy, T. J. (2013). Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 110, 5151–5156.
Rigel, N. W., Schwalm, J., Ricci, D. P., & Silhavy, T. J. (2012). BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. Journal of Bacteriology, 194, 1002–1008.
Rodriguez-Alonso, R., Létoquart, J., Nguyen, V. S., Louis, G., Calabrese, A. N., Iorga, B. I., Radford, S. E., Cho, S. H., Remaut, H., & Collet, J. F. (2020). Structural insight into the formation of lipoprotein-β-barrel complexes. Nature Chemical Biology, 16, 1019–1025.
Rogov, V. V., Rogova, N. Y., Bernhard, F., Lohr, F., & Dotsch, V. (2011). A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RcsF. The Journal of Biological Chemistry, 286, 18775–18783.
Rojas, E. R., Billings, G., Odermatt, P. D., Auer, G. K., Zhu, L., Miguel, A., Chang, F., Weibel, D. B., Theriot, J. A., & Huang, K. C. (2018). The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature, 559, 617–621.
Ruiz, N., Davis, R. M., & Kumar, S. (2021). Yhdp, TamB, and YdbH are redundant but essential for growth and lipid homeostasis of the Gram-negative outer membrane. mBio, 12, e0271421.
Ruiz, N., Kahne, D., & Silhavy, T. J. (2006). Advances in understanding bacterial outer-membrane biogenesis. Nature Reviews Microbiology, 4, 57–66.
Samsudin, F., Ortiz-Suarez, M. L., Piggot, T. J., Bond, P. J., & Khalid, S. (2016). OmpA: A flexible clamp for bacterial cell wall attachment. Structure, 24, 2227–2235.
Sharma, S., Zhou, R., Wan, L., Feng, S., Song, K., Xu, C., Li, Y., & Liao, M. (2021). Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins. Nature Communications, 12, 4687.
Shimizu, T., Ichimura, K., & Noda, M. (2016). The surface sensor NlpE of enterohemorrhagic Escherichia coli contributes to regulation of the type III secretion system and Flagella by the Cpx response to adhesion. Infection and Immunity, 84, 537–549.
Sledjeski, D. D., & Gottesman, S. (1996). Osmotic shock induction of capsule synthesis in Escherichia coli K-12. Journal of Bacteriology, 178, 1204–1206.
Snyder, W. B., Davis, L. J., Danese, P. N., Cosma, C. L., & Silhavy, T. J. (1995). Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. Journal of Bacteriology, 177, 4216–4223.
Steenhuis, M., Corona, F., Ten Hagen-Jongman, C. M., Vollmer, W., Lambin, D., Selhorst, P., Klaassen, H., Versele, M., Chaltin, P., & Luirink, J. (2021). Combining cell envelope stress reporter assays in a screening approach to identify BAM complex inhibitors. ACS Infectious Diseases, 7, 2250–2263.
Sun, J., Rutherford, S. T., Silhavy, T. J., & Huang, K. C. (2022). Physical properties of the bacterial outer membrane. Nature Reviews Microbiology, 20, 236–248.
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., Pulcini, C., Kahlmeter, G., Kluytmans, J., Carmeli, Y., et al. (2018). Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. The Lancet Infectious Diseases, 18, 318–327.
Tang, X., Chang, S., Zhang, K., Luo, Q., Zhang, Z., Wang, T., Qiao, W., Wang, C., Shen, C., Zhang, Z., et al. (2021). Structural basis for bacterial lipoprotein relocation by the transporter LolCDE. Nature Structural & Molecular Biology, 28, 347–355.
Tata, M., & Konovalova, A. (2019). Improper coordination of BamA and BamD results in bam complex jamming by a lipoprotein substrate. mBio, 10, e00660-19.
Tata, M., Kumar, S., Lach, S. R., Saha, S., Hart, E. M., & Konovalova, A. (2021). High-throughput suppressor screen demonstrates that RcsF monitors outer membrane integrity and not Bam complex function. Proceedings of the National Academy of Sciences of the United States of America, 118, e2100369118.
Tokuda, H. (2009). Biogenesis of outer membranes in Gram-negative bacteria. Bioscience, Biotechnology, Biochemistry, 73, 465–473.
Typas, A., Banzhaf, M., Gross, C. A., & Vollmer, W. (2012). From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nature Reviews Microbiology, 10, 123–136.
Umekawa, M., Miyagawa, H., Kondo, D., Matsuoka, S., Matsumoto, K., & Hara, H. (2013). Importance of the proline-rich region for the regulatory function of RcsF, an outer membrane lipoprotein component of the Escherichia coli Rcs signal transduction system. Microbiology, 159, 1818–1827.
United Nations Environment Programme, UNEP. (2023). Bracing for Superbugs: Strengthening environmental action in the One Health response to antimicrobial resistance.
Vogt, S. L., Evans, A. D., Guest, R. L., & Raivio, T. L. (2014). The Cpx envelope stress response regulates and is regulated by small noncoding RNAs. Journal of Bacteriology, 196, 4229–4238.
Wall, E., Majdalani, N., & Gottesman, S. (2018). The complex Rcs regulatory cascade. Annual Review of Microbiology, 72, 111–139.
Wall, E. A., Majdalani, N., & Gottesman, S. (2020). IgaA negatively regulates the Rcs phosphorelay via contact with the RcsD phosphotransfer protein. PLoS Genetics, 16, e1008610.
Weatherspoon-Griffin, N., Zhao, G., Kong, W., Kong, Y., Morigen, Andrews-Polymenis, H., McClelland, M., & Shi, Y. (2011). The CpxR/CpxA two-component system up-regulates two tat-dependent peptidoglycan amidases to confer bacterial resistance to antimicrobial peptide. The Journal of Biological Chemistry, 286, 5529–5539.
Wu, T., Malinverni, J., Ruiz, N., Kim, S., Silhavy, T. J., & Kahne, D. (2005). Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell, 121, 235–245.
Yamamoto, K., & Ishihama, A. (2006). Characterization of copper-inducible promoters regulated by Cpxa/Cpxr in Escherichia coli. Bioscience, Biotechnology, and Biochemistry, 70, 1688–1695.
Acknowledgements
This work was funded by WELBIO (Grant no. WELBIO-CR-20190-03).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to report.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cho, SH., Dekoninck, K. & Collet, JF. Envelope-Stress Sensing Mechanism of Rcs and Cpx Signaling Pathways in Gram-Negative Bacteria. J Microbiol. 61, 317–329 (2023). https://doi.org/10.1007/s12275-023-00030-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-023-00030-y