Abstract
The G-quadruplex (G4) formed in single-stranded DNAs or RNAs plays a key role in diverse biological processes and is considered as a potential antiviral target. In the genome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), 25 putative G4-forming sequences are predicted; however, the effects of G4-binding ligands on SARS-CoV-2 replication have not been studied in the context of viral infection. In this study, we investigated whether G4-ligands suppressed SARS-CoV-2 replication and whether their antiviral activity involved stabilization of viral RNA G4s and suppression of viral gene expression. We found that pyridostatin (PDS) suppressed viral gene expression and genome replication as effectively as the RNA polymerase inhibitor remdesivir. Biophysical analyses revealed that the 25 predicted G4s in the SARS-CoV-2 genome formed a parallel G4 structure. In particular, G4-644 and G4-3467 located in the 5′ region of ORF1a, formed a G4 structure that could be effectively stabilized by PDS. We also showed that PDS significantly suppressed translation of the reporter genes containing these G4s. Taken together, our results demonstrate that stabilization of RNA G4s by PDS in the SARS-CoV-2 genome inhibits viral infection via translational suppression, highlighting the therapeutic potential of G4-ligands in SARS-CoV-2 infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has resulted in unprecedented global health and economic crises. SARS-CoV-2 is a single-stranded ( +) RNA virus belonging to the family Coronaviridae (Andersen et al. 2020; Dhama et al. 2020; Wu et al. 2020). It is the seventh coronavirus known to infect humans, with the six previously identified being human coronavirus (HCoV)-229E, HCoV-HKU1, HCoV-OC43, HCoV-NL63, SARS-CoV and Middle East respiratory syndrome (MERS)-CoV (Andersen et al. 2020). Among these, SARS-CoV-2 shares the highest nucleotide sequence identity (79.7%) with SARS-CoV (Zhou et al. 2020). The SARS-CoV-2 genome is ~ 30 kb, which encodes 16 non-structural proteins (Nsps1 − 16), four conserved structural proteins including the spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins and strain specific accessory proteins (Gussow et al. 2020; V'Kovski et al. 2021). During the COVID-19 pandemic, various genetic variants of SARS-CoV-2 have been emerging and spreading around the world. Considering their phenotypic characteristics such as transmissibility, disease severity, risk of reinfection and impacts on diagnostics and vaccine performance, WHO has classified SARS-CoV-2 strains as variants of concern (VOCs) or variants of interest (VOIs). As of March 2023, circulating VOCs are the omicron variants since the end of 2021, while there have been four previously circulating VOCs (alpha, beta, gamma and delta) and eight VOIs (Epsilon, Zeta, Eta, Theta, Iota, Kappa, Lambda and Mu) (Fernandes et al. 2022; Thakur et al. 2022).
Various pharmacological interventions including antiviral drugs, monoclonal antibodies (Wang et al. 2020; Kumar et al. 2021), non-steroidal anti-inflammatory drugs (Chen et al. 2021) and immunomodulators (Berry and Fontana 2020) have been evaluated and some have become available in the markets for the management of COVID-19, such as remdesivir, molnupiravir, paxlovid and ritonavir (Rubin et al. 2020; Burki 2022; Fischer et al. 2022). In addition, vaccine candidates including mRNA-based vaccines, viral vector-based vaccines, and recombinant protein-based vaccines have been investigated, many of which have been approved to control SARS-CoV-2 infection (Creech et al. 2021; Dong et al. 2021). Despite widespread availability of vaccines and antiviral therapeutics, the COVID-19 pandemic situation is still unpredictable, due to continuous emergence of clinical variants. While antiviral drugs that target viral proteins are conventional routes to combat against viral infection, targeting specific viral genomic sites that are highly conserved may provide a novel opportunity to expand the scope of antiviral therapeutics.
These novel targets include G-quadruplexes (G4s), which are non-canonical nucleic acid secondary structures formed by guanine-rich sequences and have been studied extensively in the human genome (Hansel-Hertsch et al. 2017; Ravichandran et al. 2019; Ravichandran et al. 2021; Shamim et al. 2020). In the past few years, a number of studies reported G4 structures in the genomes of both DNA and RNA viruses, such as human immunodeficiency virus-1 (HIV-1), Zika virus (ZIKV), hepatitis C virus (HCV), rhinovirus, Ebola virus (EBOV), influenza virus, human papillomavirus (HPV), herpes simplex virus 1 (HSV-1), Epstein-Barr virus (EBV) and human cytomegalovirus (HCMV) (Lavezzo et al. 2018; Ruggiero and Richter 2020; Abiri et al. 2021). Viral G4s are known to play key roles in controlling genome replication, maintenance of genome integrity, transcription, and translation (Ruggiero and Richter 2020), thereby becoming potential targets for antiviral therapeutics. Several small molecules can bind to G4s with high binding affinity because of π–π stacking or cationic- π or electronic interactions (Monchaud and Teulade-Fichou 2008; Sun et al. 2019). In previous studies, G4-binding ligands have been screened against many human viruses as potential therapeutic candidates. Recently, G4s have been identified in the genome of SARS-CoV-2 and proposed to be therapeutic targets (Cui and Zhang 2020; Panera et al. 2020; Xie et al. 2020b; Bezzi et al. 2021; Zhao et al. 2021). However, most of these studies focused on understanding the role of individual few viral G4s, and until now there have been no studies evaluating G4 formation in the entire SARS-CoV-2 genome or bona-fide antiviral activity of G4-binding ligands in SARS-CoV-2-infected cells. Furthermore, no extensive studies of those ligands on each G4 in the viral genome and their correlation to the antiviral effect have been explored. Since G4s have been shown to play a key role in the regulation of viral virulence genes (Tuesuwan et al. 2008; Ravichandran et al. 2018; Bohálová et al. 2021), a genome-wide analysis is critical to understand the biological significance of SARS-CoV-2 G4s. Importantly, the effects of G4-ligands on the SARS-CoV-2 genome should be examined in the context of viral infection.
In this study, we screened the antiviral effects of several G4-binding ligands on SARS-CoV-2 replication in infected cells and identified pyridostatin (PDS) as the most potent inhibitor. Following a genome-wide characterization of viral G4s, we investigated whether the antiviral effect of G4-ligands involves their ability to stabilize viral G4s and regulate protein translation using mutational analyses of G4-forming sequences. Our study sheds light on the prospects of targeting viral RNA G4s with G4-binding ligands as a potential therapeutic strategy against SARS-CoV-2.
Materials and methods
Cells, viruses and plaque assays
Vero cells were obtained from the ATCC (ATCC-CCL81) and maintained in high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. The cells were incubated at 37 °C in a humidified incubator under 5% CO2. The original SARS-CoV-2 wild-type (WT) stock (hCoV-19/Korea/KCDC03/2020) was provided by the Korea Disease Control and Prevention Agency (KDCA; Osong, Republic of Korea). An additional SARS-CoV-2 virus expressing the Nano-luciferase reporter protein (SARS-CoV-2-Nluc) was kindly provided by Dr. Pei-Yong Shi (University of Texas Medical Branch, Galveston, TX) (Xie et al. 2020a, b). SARS-CoV-2 viruses were propagated in Vero cells, and the virus titers were determined by plaque assays. Briefly, Vero cells were prepared one day prior to the assays and inoculated with ten-fold serially diluted stock of SARS-CoV-2. After one hour of infection, the inoculum was discarded, and the cells were overlaid with DMEM-F12 (Sigma-Aldrich) containing final concentration of 0.6% oxoid agarose, also supplemented with 0.01% DEAE-dextran (Sigma-Aldrich) and 2 μg/ml TPCK-trypsin (Thermo Fisher Scientific). After three to four days, the cells were stained with 0.2% crystal violet solution (20% ethanol) overnight. Plaques were counted in duplicates of each sample, and the virus titer was calculated.
Chemicals
The chemicals used were as follows: chloroquine diphosphate (C6628, Sigma-Aldrich), lopinavir (S1380, SelleckChem), remdesivir (HY-104077, MedChemExpress), NMM (N-methyl mesoporphyrin IX; SC-396879, Santa Cruz Biotechnology), TMPyP4 (5,10,15,20-tetra-(N-methyl-4-pyridyl)porphyrin; GC12092, GLPBIO), TMPyP2 (5,10,15,20-tetra-(N-methyl-2-pyridyl)porphyrin; T40846, Frontier Scientific), BRACO19 (N,N’-(9-(4-(dimethylamino)phenylamino)acridine-3,6-diyl)bis(3-(pyrrolidin-1-yl)propanamide); GC50140, GLPBIO), PDS (pyridostatin; 18,013, Cayman Chemical), PhenDC3 (3,3′-[1,10-Phenanthroline-2,9-diylbis(carbonylimino)]bis[1-methylquinolinium] 1,1,1-trifluoromethanesulfonate; CS-7711, Chemscene), Thioflavin T (2390-54-7, MedChemExpress) and CX3543 (A12380, Adooq).
Antiviral analysis of G4-ligands against SARS-CoV-2
As previously described (Corman et al. 2020; Bae et al. 2021), Vero cells were seeded into a 24-well plate at a density of 1 × 105 cells/well one day prior to infection. On the day of infection, plates were transferred to a BSL3 facility, and the cells were pre-treated for 1 h with the G4-ligands in 10 twofold serial dilutions to achieve in-well concentrations ranging from 0.08 to 50 µM. Cells were either mock-infected or infected with SARS-CoV-2-Nluc or SARS-CoV-2 (WT) at MOI of 0.01 for 1 h. The virus inoculum was removed and the cells were washed once with PBS before the medium was replaced with 2% FBS DMEM containing the G4-ligands at the same concentrations used for pre-treatment. For SARS-CoV-2-Nluc infection, at 24 h post-infection, cells were washed once with PBS and collected using a passive lysis buffer (Promega). Cell lysates were frozen and thawed for complete lysis. Afterward, 20 μl Nano-Luc® luciferase substrate (Promega) was added to 20 μl cell lysate in a white opaque 96-well plate and incubated for at least three minutes before measurement of luciferase signals using a Hidex Sense Microplate Reader (Hidex). The luciferase activity of G4-ligand treated groups was normalized to DMSO or DW controls. For SARS-CoV-2 (WT) infection, at 24- and 48 h post-infection, virus culture supernatants were collected, and automated nucleic acid purification using a Maxwell® RSC 48 instrument (Promega) was performed to extract total RNA from virions in the supernatants. The RT-qPCR assay was performed using primers specific for the SARS-CoV-2 RNA polymerase gene using forward (5′-GTGARATGGTCATGTGTGGCGG-3′) and reverse (5′-CARATGTTAAASACACTATTAGCATA-3′) primers and a probe (5′-FAMCAGGTGGAACCTCATCAGGAGATGC– BHQI-3′) in duplicate for each sample using a QuantStudio™ 3 Real-Time PCR System (Thermo) (Corman et al. 2020). Transcript levels were analyzed using the delta-delta Ct method, and relative inhibition of infection was calculated by normalizing the G4-ligand-treated groups to DMSO or distilled water (DW) controls.
The IC50 for each compound was calculated using Graphpad Prism 5.0 software and the following formula: Y = Bottom + (Top–Bottom)/(1 + 10((LogIC50−X)*HillSlope)). Three independent experiments were conducted to calculate IC50 and CC50 values from the dose–response curves.
Cytotoxicity assays
The cytotoxicity of G4-ligands was assessed via the 3-(4,5- dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays, as previously described (Cho et al. 2013). Briefly, Vero cells (104 cells/well) were seeded in 96-well plates one day prior to the treatments and incubated with the G4-ligands in 10 two-fold serial dilutions (0.08–50 µM) for 24 or 48 h. After treatment, MTT solution (10 µl of 5 mg/ml) was added to each well and the mixtures were incubated for additional 3–4 h at 37 °C. The plates were then centrifuged, the supernatants were discarded, and DMSO (100 µl) was added to each well. After the crystals were dissolved, the amount of reduced MTT was measured at 540 nm.
Circular dichroism (CD) spectra measurement
For measurement of the CD spectra, RNA oligonucleotides (15 µM) were prepared in a conventional G4 buffer containing 10 mM Tris–HCl (pH 7.5) and 100 mM KCl (Katsuda et al. 2016; Hu et al. 2017). Annealing of G4 samples was performed in a thermoblock (denaturation at 95 °C for 10 min followed by cooling at 0.5 °C/min to 4 °C). After annealing, samples were stored overnight at 4 °C and equilibrated at room temperature for 15 min before performing experiments. The CD spectra were measured at 20 °C at the wavelengths from 220 to 320 nm with a scan speed of 100 nm/min, data pitch of 0.2 nm, and bandwidth of 0.2 nm using a 1 mm quartz cuvette with 200 µl sample volume. All CD experiments were performed using a JASCO J-810 CD spectropolarimeter equipped with a JASCO CDC-426F Peltier temperature controller.
CD thermal melting analysis
For CD melting, samples were heated from 20 to 90 °C at a ramp rate of 2 °C/min and data pitch of 0.2 °C. The CD melting analysis was performed in the absence or presence G4-binding ligands at a 1:2 RNA:ligand molar ratio. The G4 RNA (15 µM) samples were incubated at room temperature for 30 min in buffer containing 10 mM Tris–HCl (pH 7.5) and 100 mM KCl with or without G4 binding ligand (30 µM). The ellipticity was recorded at the wavelength showing maximum ellipticity. The data were performed in two trials and curve fitting was performed using a sigmoidal four-parameter equation in SigmaPlot 12.5. The melting temperature (Tm) was calculated at the point where 50% of the G4s were folded and 50% were unfolded. Error bars indicate ± SEM.
Thermal difference spectra (TDS)
UV/VIS spectra were obtained on a dual beam V-750 UV–Visible Spectrophotometer (JASCO) using WT and Mut-G4s (15 μM) preformed in G4 buffer (10 mM Tris, pH 7.5, 100 mM KCl). The spectra were measured at 20 °C and 90 °C in triplicate and blanked by subtracting G4 spectra with only G4 buffer. TDS were generated by subtracting buffer-corrected spectra at 20 °C from those at 90 °C (Ariyo et al. 2015). The spectra were normalized using the maximum TDS value to yield the normalized TDS. Single-stranded RNA (20mer of poly-A RNA) was used as a negative control.
ThT fluorescence turn-on assay
The experiments were carried out with 96-well microplates from CORNING (Flat Bottom Black) on a Biotek Synergy Neo multiplate reader (TECAN). The preformed RNA G4s in 100 mM KCl or LiCl G4 buffer conditions were mixed with Thioflavin T (ThT) at 2 μM or 4 μM final concentrations, respectively. The total reaction volume was 100 µl and plate was incubated at room temperature for 30 min before taking measurement. Fluorescence emission spectra was recorded in two trials from 440 to 700 nm after excitation at 425 nm. Error bars represent ± SEM. All the data were analyzed with Microsoft Excel.
In vitro transcription and coupled transcription/translation assays of G4-containing reporter genes
The peYFP-N1 reporter plasmids (Promega) containing the viral G4 sequence were generated by cloning the target G4-containing DNA fragment in a frame upstream of the eYFP reporter coding region. For this, DNA fragments containing G4-644 (284 bp), or G4-3467 (580 bp) were synthesized by Cosmogenetech, cloned into a pUC-GW-Amp donor plasmid, and then subcloned into a peYFP-N1 recipient plasmid between the Xho1/BamH1 restriction sites; this resulted in pN1-WT-G4-644, and pN1-WT-G4-3467 plasmids, respectively. The primers used for PCR amplification are listed in (Table S2). To introduce mutations that disrupted G4 formation in these plasmids, PCR reactions were performed according to the Stratagene QuikChange® site-directed mutagenesis protocol. Primers used for site-directed mutagenesis are listed in (Table S2). The resulting Mut-G4-eYFP plasmids were named pN1-Mut-G4-644 and pN1-Mut-G4-3467. To prepare the T7-G4-eYFP templates for in vitro transcription (IVT) or in vitro coupled transcription/translation (TNT) reactions, pN1-WT/Mut-G4-eYFP plasmids were used for PCR amplification using primers covering the T7 promoter site and 3′ end of eYFP (Table S2). These amplified templates were named T7-WT/Mut-G4-644 and T7-WT/Mut-G4-3467, and their sizes were 1044 and 1341 bp, respectively.
For IVT reactions, PCR amplicons containing the T7 promoter site, G4 sequence, and eYFP gene were used as templates. Transcripts containing WT or Mut-G4s followed by the eYFP gene were prepared using an mMESSAGE mMACHINE™ T7 Transcription Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, the PCR amplicons (0.125 µg) were incubated in a 20 μl reaction mixture at 30 °C for 90 min with DW or increasing concentration of PDS (15 and 50 µM). After incubation, the DNA template was removed by DNase treatment for 15 min at 37 °C and the remaining RNA transcripts were precipitated by addition of lithium chloride. The RNA concentrations were measured using a Colibri microvolume spectrometer (Titertek-Berthold) and RNA qualities were analyzed in 0.8% agarose gel with ethidium bromide staining.
For in vitro coupled transcription/translation (TNT) reactions, the T7-G4-eYFP templates were transcribed and translated using a TNT® Quick Coupled Transcription/Translation System (Promega) containing a standard rabbit reticulocyte lysate system. For the in vitro TNT reaction, PCR template (0.5 µg) was mixed in a 50 μl reaction mixture containing 1 mM methionine, TNT® Quick Master Mix and incubated at 30 °C for 90 min. To test the effect of G4-ligands, G4-binding ligands were added to the reaction mixtures at various concentrations.
Western blot analyses
To assess the expression of viral proteins in infected cells, cells were lysed in 1x-RIPA buffer or passive lysis buffer, and then lysates were mixed with 5 × sample buffer containing 62.5 mM Tris–HCl (pH 6.8), 20% glycerol, 5% β-mercaptoethanol, 2% SDS and 0.025% bromophenol blue. Cell lysates or in vitro translated samples were resolved by SDS-PAGE, electroblotted onto polyvinylidene fluoride (PVDF) membranes. The membranes of cell lysates were probed with primary antibodies against the SARS-CoV-2 spike (1:2000 dilution; Abcam, ab272504) and nucleocapsid (1:2000; SinoBiological, 40143-R019) proteins overnight at 4 °C. After three 5 min washes in Tris-buffered saline (TBS) containing 0.05% Tween-20 (TBST), blots were incubated with horseradish peroxide (HRP)-conjugated secondary mouse anti-rabbit (1:10,000; Santa Cruz, sc-2357) and goat anti-rabbit (1:10,000; Invitrogen, 31460) antibodies, respectively, for 1 h at room temperature. The membranes of in vitro translated samples were probed with mouse anti-eYFP monoclonal antibody (1:5000; MAB 8759, Abnova) at 4 °C for 1 h, followed by incubation with an HRP-conjugated secondary goat anti-mouse antibody (1:5000; SC-2031, Santa Cruz Biotechnology) for 1 h at room temperature. Blots were then washed, and the protein bands were detected using enhanced chemiluminescence (ECL) detection reagents (ELPIS, Daejeon, Korea) and analyzed using an LAS3000 or LAS4000 luminescence image analyzer (Fujifilm). The band intensity was quantified using ImageJ software (Ma-Lauer et al. 2016). GAPDH is used as a sample loading control. The eYFP band intensity value of the drug untreated (DW- or DMSO-treated) sample was set as 100% in both WT and Mut-G4 constructs. For drug treated samples, the expression change (%) was quantified as (Treated/Untreated) × 100 to get relative values. Therefore, each WT and Mut values were independently measured and calculated.
Statistical analysis
Data were analyzed using GraphPad Prism software version 8.3.0. One-way ANOVA tests were used to determine significant differences between untreated and ligand-treated samples with respect to in vitro eYFP transcription and coupled translation. Data represent either the mean ± standard errors of the mean (SEM) or the mean ± standard deviations. A p value less than or equal to 0.05 was considered statistically significant.
Results
Identification of G4-ligands with antiviral activity against SARS-CoV-2
To test whether G4-binding ligands can suppress SARS-CoV-2 infection, primary antiviral screening assays employing detection of nucleocapsid (N) protein expression was conducted as previously described (Jeon et al. 2020). Briefly, Vero cells were infected with SARS-CoV-2 (WT) in the presence of a G4-binding ligand for 24 h and subjected to immunofluorescence assays (IFAs) to detect N protein expression. In our screening, seven commercially available and widely used G4-ligands with various chemical structures (PDS, CX3543, NMM, PhenDC3, TMPyP2, TMPyP4 and BRACO19) were chosen to be tested and their chemical structures are illustrated in Figs. 1A and S1. Remdesivir, chloroquine and lopinavir, with known antiviral activity against SARS-CoV-2, were included as positive controls. In these assays, PDS was the most potent inhibitor of SARS-CoV-2 infection among the tested G4-ligands, with an IC50 of 13.64 µM (Fig. S2). The IC50 value of PDS was comparable to those of the positive controls, remdesivir (8.79 µM), chloroquine (9.61 µM), and lopinavir (11.58 µM). While PhenDC3, NMM and BRACO19 showed moderate antiviral effects at higher concentrations, CX3543, TMPyP2 and TMPyP4 showed no or little antiviral activity at the tested concentrations. These results suggest that PDS may have a potent antiviral activity against SARS-CoV-2.
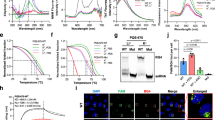
Antiviral effects of G4-ligands on SARS-CoV-2-Nluc reporter virus infection. A The chemical structures of G4-ligands, PDS and CX3543, are shown. B–D Vero cells cultured in 24-well plates were infected with a recombinant SARS-CoV-2 expressing Nano-luciferase (SARS-CoV-2-Nluc) at MOI of 0.01 with PDS or CX3543 at 10 concentrations ranging from 50 to 0.08 μM (two-fold serial dilutions). B At 24 h after infection, cell lysates were harvested and Nano-luciferase activity of each sample was measured and normalized to that of distilled water (DW) and DMSO vehicle control for PDS and CX3543, respectively. Antiviral activity against SARS-CoV-2 is shown as ‘inhibition of infection (%)’ and the IC50 was calculated as described in the text. Representative results of 10-point concentration–response curves are shown with estimated IC50 values. C The mean estimated IC50 values with standard deviations (n = 3) are summarized in a table. D The cytotoxicity of PDS and CX3543 was measured by MTT assays in uninfected Vero cells at the concentrations above-mentioned for 24 h. The mean values of cell viability were calculated from three independent experiments and shown with standard errors of the mean (± SEM)
To validate the primary screening results, we took advantage of a recombinant SARS-CoV-2 expressing the Nano-luciferase protein (SARS-CoV-2-Nluc), a model that has been employed as an efficient antiviral screening assay with high sensitivity (Xie et al. 2020b). We selected PDS to study as a G4-binding ligand candidate with strong antiviral activity and CX3543 with no or little antiviral activity against SARS-CoV-2. We included CX3543 as a control, because CX3543 is the only first-in-class G4-interacting drug enrolled in phase II clinical trials (Drygin et al. 2009). When Nano-luciferase activity was assessed in Vero cells infected with SARS-CoV-2-Nluc, the IC50 of PDS was estimated to be 2.44 ± 0.7 µM and the IC50 of CX3543 was higher than 50 µM (Fig. 1B, C). When the cytotoxicity was measured by MTT assays in uninfected Vero cells treated with PDS or CX3543 (0.08 µM to 50 µM) for 24 h, PDS showed no cytotoxicity, but CX3543 was cytotoxic at higher concentrations (Fig. 1D). These results suggest that PDS is a highly potent inhibitor of SARS-CoV-2 replication, compared to CX3543.
Antiviral effects of PDS on SARS-CoV-2 replication
To further confirm the inhibitory effect of PDS on SARS-CoV-2 replication, Vero cells were infected with SARS-CoV-2 with PDS, CX3543, or remdesivir (as a control) for 24- and 48 h, and the levels of progeny viral RNA genome in the culture supernatants were measured by RT-qPCR (Corman et al. 2020). As shown in representative results, PDS exhibited strong antiviral activity comparable to that of remdesivir (Fig. 2A, B). From the results of three independent experiments, the IC50 values of PDS were estimated to be 1.5 ± 0.1 µM and 3.5 ± 1.0 µM at 24- and 48 h post-infection, respectively, which was comparable to those of remdesivir, 2.6 ± 0.7 µM and 3.5 ± 1.3 µM at 24- and 48 h post-infection, respectively (Fig. 2C). Under the same experimental conditions, CX3543 moderately inhibited the progeny viral RNA genome levels, possibly due to cytotoxicity at higher concentrations and IC50 values were 13.0 ± 2.8 µM and 25.5 ± 11.6 µM at 24- and 48 h post-infection, respectively (Fig. 2A–C). Western blot analyses of infected cell lysates also showed that PDS more potently inhibited the expression of viral proteins such as viral spike (S) and nucleocapsid (N) than CX3543 and that this inhibitory activity of PDS was similar to that of remdesivir (Fig. 2D). When the effects of PDS, CX3543 or remdesivir on virus titers were measured by plaque assays, PDS more effectively reduced the progeny virus titer than CX3543 (Fig. 2E). The cytotoxicity of PDS and CX3543 in cells treated for 48 h was similar to that for 24 h; PDS exhibited no cytotoxicity, but CX3543 showed cytotoxicity at higher concentrations (Fig. S3). Taken together, our results demonstrate that PDS suppresses SARS-CoV-2 replication by inhibiting the expression of viral proteins as effectively as remdesivir.
Antiviral effects of PDS on SARS-CoV-2 replication. Vero cells cultured in 24-well plates were infected with SARS-CoV-2 at MOI of 0.01 and incubated with PDS or CX3543 at 10 concentrations from 50 to 0.08 μM (two-fold serial dilutions). Remdesivir was included as a positive control. (A and B) Culture supernatants were collected at 24- (A) or 48 h (B) post-infection and viral RNA genome levels in the supernatants were measured by RT-qPCR with primers specific to the viral RNA polymerase gene. The amount of viral RNA genome in each sample was normalized to that of distilled water (DW) and the DMSO vehicle control for PDS and CX3543, respectively. Antiviral activity against SARS-CoV-2 is shown as ‘inhibition of infection (%)’ and the IC50 was calculated as described in the text. Representative results of 10-point concentration–response curves with calculated IC50 values are shown. C IC50 values of PDS, CX3543 and remdesivir on SARS-CoV-2 replication at 24- and 48 h post-infection. The mean IC50 values of tested G4-ligands from three independent experiments are shown with standard deviations. D At 24 h post-infection, the cell lysates from the samples at the indicated concentrations (μM) of each compound were prepared and subjected to western blot analysis to determine the expression of viral proteins such as spike and nucleocapsid. Cellular α-tubulin was used as a loading control. The concentrations of compounds were chosen based on their IC50 values to examine dose-dependency. E At 24 h post-infection in the presence of PDS, CX3543 or remdesivir at the indicated concentrations (μM), the culture supernatants of SARS-CoV-2 infected cells were harvested and subjected to plaque assays to measure infectious virion production. ‘Inhibition of infection (%)’ was calculated as the virus titer in each sample was normalized to that of the DW and the DMSO vehicle control for PDS and CX3543, respectively. The experiments were repeated twice, and the bar graphs are shown as the mean values with error ranges
PDS has been shown to bind RNA G4s in the cytoplasm as well as G4s in the nucleus (Dolinnaya et al. 2016; Bao et al. 2017; Neckles et al. 2019) and increase cytoplasmic G4 staining by BG4 antibody. To test whether PDS primarily targets cytoplasmic G4s, which are thought to be largely viral G4s, in SARS-CoV-2-infected cells, we examined the effects of PDS on cytoplasmic G4 stabilization in SARS-CoV-2 infected cells by conducting IFAs using G4-specific BG4 antibody. Vero cells were infected with a recombinant SARS-CoV-2 expressing the mNeonGreen protein (SARS-CoV-2-mNG) at MOI of 0.01 in the presence or absence of PDS and CX3543 (10 μM) and subjected to IFAs at 24 h post-infection (Fig. S4). In DMSO-treated cells, the BG4 signals of uninfected cells were found mostly in the nucleus, while the signals of SARS-CoV-2-mNG-infected cells were detected in the cytoplasm, suggesting the increased G4 formation in the cytoplasm during virus infection. Upon PDS treatment, the BG4 signals became apparent even in the cytoplasm of uninfected cells, suggesting that PDS stabilized cytoplasmic G4s. Moreover, PDS treatment enhanced the intensity of the cytoplasmic BG4 signals in virus-infected cells. Similar results were obtained in CX3543-treated, virus-infected cells, albeit to a lesser extent. These results indicate that cytoplasmic G4s are highly produced in SARS-CoV-2 infected cells, which are further stabilized by G4-ligands.
Biophysical analysis of the putative SARS-CoV-2 G4-forming sequences
We identified 25 putative G4-forming sequences by applying QGRS Mapper to the SARS-CoV-2 genome (Accession no: NC_045512.2) with the following criteria: maximum length 30 bp, minimum G group length 2 and loop size range of 0–36 (Table S1). Each putative G4 was named by a starting sequence number of each G4 in the genome. These sequences were identical to those predicted in previous studies (Panera et al. 2020; Ji et al. 2021) and found throughout the genome, with a relatively greater density at the 5′ and 3′ regions of the viral genome (Fig. S5). The average density of G4-forming sequences in the viral genome is approximately one G4 per kb. Based on the location of G4s in the genome, we divided them into three groups, Group A at the 5′ region of ORF1a (n = 8), Group B at the center of the genome (n = 7), and Group C at the 3′ region of the genome (n = 10) (Fig. S5). The G4s in SARS-CoV-2 consist of two G-quartets, and G4s with three (GGG) or four (GGGG) contiguous G-runs were not found.
To validate G4 formation from the predicted sequences, we performed biophysical analyses for each of the 25 predicted G4-forming sequences using the corresponding synthetic RNA oligonucleotides (Table S1). The folding topologies of G4s were confirmed by CD spectroscopy since CD is the primary tool to characterize G4 formation and evaluate the thermal stability of G4s. The CD spectra of all SARS-CoV-2 G4s were characterized by a strong positive peak at 265 nm and a negative peak at 240 nm in the presence of 100 mM KCl (Fig. S6), which is the typical signature of parallel G4 structures (Majee et al. 2020, 2021; Zhao et al. 2021). In addition to CD spectra, thermal melting curves were obtained by monitoring the change in ellipticity at the wavelength showing the highest ellipticity (Fig. S7). The SARS-CoV-2 G4s showed Tm values ranging from 40.1 ± 0.2 to 58.5 ± 0.3 with an average of 49.3 °C (Table S3). We also carried out the fluorescence turn-on assay for all SARS-CoV-2 G4s to differentiate an a-type RNA duplex from the parallel type RNA G4s using a G4 specific ligand, Thioflavin T (ThT). ThT acts as an efficient fluorescence probe for selectively targeting RNA G4s among other RNA structures (Xu et al. 2016). Substantial fluorescence enhancement was observed at 490 nm in 100 mM KCl, compared to 100 mM LiCl G4 buffer condition for all SARS-CoV-2 G4s, while RNA controls such as duplex and single strand RNAs had no noteworthy effect on emission (Figs. S8 and S9).
Effects of PDS on the stability of the SARS-CoV-2 G4s
Since PDS but not CX3543 effectively suppressed SARS-CoV-2 infection, we hypothesized that the effect of these ligands on the topology or stability of G4s in the SARS-CoV-2 genome might be different. To test this hypothesis, we first investigated how these two G4-binding ligands affect the topology of SARS-CoV-2 G4s. The CD spectra of G4s treated with either G4-binding ligand in a molar ratio 2:1 (Ligand:RNA) were not significantly altered, indicating that the effect of ligands on G4 topology was limited (Fig. S10). However, in some cases such as G4-29123, alteration in the maximum wavelength upon chemical treatment was observed.
We further evaluated the effect of G4-binding ligands on the stability of SARS-CoV-2 G4s by measuring difference in melting temperature (∆Tm) in the absence and presence of ligands (Fig. S11). The melting curve analyses indicated that CX3543 did not induce any appreciable change in ∆Tm for most G4s, while PDS tended to induce significant changes in ∆Tm and stabilize many G4s (Fig. 3A and Table S3). Among the 25 predicted G4s in SARS-CoV-2, PDS increased ∆Tm of nine G4s by > 8 °C, whereas CX3543 increased ∆Tm by > 8 °C for only one G4 (Fig. 3B). Notably, G4-644 (42.0 ± 0.5 to 61.2 ± 1.0 °C), G4-3467 (54.8 ± 0.9 to 74.5 ± 0.6 °C) and G4-29123 (48.3 ± 0.2 to 69.6 ± 2.3 °C), showed the significant Tm increase (Fig. 3B and Table S3). These results suggest that PDS can be a more potent candidate for modulating SARS-CoV-2 G4s than CX3543 in terms of the stabilizing effect. The differential effects of PDS and CX3543 on Tm of G4s were correlated with the effects of these ligands on viral growth in cultured cells.
Comparison of the effects of PDS and CX3543 on Tm changes of SARS-CoV-2 RNA G4s. A Tm values of G4s in the absence or presence of CX3543 and PDS. For ligand treatment, preformed G4s were incubated with CX3543 or PDS in a molar ratio of 2:1 (Ligand:RNA). Error bars represent mean ± SEM (n = 2). B The Tm change (ΔTm) ranges (°C) of corresponding G4s after PDS (red circles) and CX3543 (green circles) treatment. The Tm changes of all G4s are shown in Table S3
Mutational analyses of SARS-CoV-2 G4-644 and G4-3467 for G4 stabilization by PDS
Among 25 viral G4s, we selected G4-644 located in Nsp1 for further mutational and functional analyses. We also targeted G4-3467 in Nsp3 which is involved in polyprotein processing and impedes the type 1 interferon (IFN) antiviral activities of the host cells (Hossain et al. 2021). Since these G4s are located at the 5′ end of ORF1ab, when stabilized by G4-binding ligands, they were expected to have great impact on the expression of nonstructural proteins that regulate viral genome replication, RNA processing, proofreading to maintain genome integrity, and immune evasion (Hossain et al. 2021; V'Kovski et al. 2021). Furthermore, all these G4s exhibited significantly higher ∆Tm (> 10 °C) with PDS treatment as compared to CX3543 (Table S3). G4-29123 was not chosen for the intensive analysis due to the large CD spectra change upon chemical treatment (Fig. S10) and its position in the genome (Fig. S5).
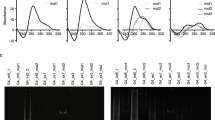
To evaluate the impact of targeting G4-644 and G4-3467, we synthesized mutant G4 sequences (Mut-G4s) that abrogate G4 formation from wild-type G4 sequences (WT-G4s; Fig. S5B), and compared the structural features of the WT- and Mut-G4s. Compared with the parallel CD spectra of WT-G4s, Mut-G4-3467 exhibited a shift of the peak from 265 to 270 nm, indicating disruption of G4 topology; however, the peak shift in Mut-G4-644 was not distinguishable (Fig. 4A). We further validated Mut-G4s by CD melting analysis, thermal difference spectra (TDS), and ThT fluorescence turn-on assay. In CD melting analysis, Mut-G4s had lower Tm values than their WT counterparts, indicating destabilization of G4s by the introduced mutations (Fig. 4B). In TDS, WT-G4s exhibited two positive peaks near 243 and 276 nm and a negative peak at 297 nm, which are characteristic G4 spectra (Mergny et al. 2005; Ariyo et al. 2015). Compared to those of WT-G4s, TDS spectra of Mut-G4s did not show G4 signature spectra but were more similar to that of the single-stranded RNA (ssRNA), confirming the dissociation of Hoogsteen base pairs and disruption of G4 configuration in the mutant G4s (Fig. 4C). In ThT fluorescence turn-on assay, Mut-G4s showed severely decreased fluorescence enhancement compared to WT-G4s, which also confirms the disruption of the G4 structure (Fig. 4D). Collectively, these results support that G4-644 and G4-3467 form stable parallel intramolecular G4s, while their mutants lose the G4 structural properties.
Biophysical analyses of the wild-type and mutant SARS-CoV-2 G4-644 and G4-3467. A CD spectra of wild-type (WT) and mutant (Mut) G4 sequences for G4-644 and G4-3467 were obtained as in Fig. S10 and compared. B The CD thermal melting curve for wild-type and mutant G4 sequences were obtained as described in Fig. S11. The melting temperature (Tm) for preformed WT- and Mut-G4s (G4-644 and G4-3467) is shown as the mean ± SEM (n = 2). C Thermal difference spectra (TDS) of preformed WT- and Mut-G4s (G4-644 and G4-3467) in 100 mM KCl. ssRNA was used as a negative control. D ThT fluorescence emission spectra at 490 nm of preformed WT-G4s and Mut-G4s for G4-644 and G4-3467
PDS interacts with SARS-CoV-2 G4-3467 via end-stacking binding mode
To investigate the binding of PDS to SARS-CoV-2 G4s, we conducted a Thioflavin T-fluorescent intercalator displacement assay (ThT-FID) specifically targeting G4-3467. ThT-FID has proven effective in identifying G4-binding ligands and elucidating their binding modes (Tse and Boger 2005). This assay involves the displacement of the fluorescence probe ThT binding to a G4 by a G4-ligand, resulting in reduced fluorescence intensity. Considering that ThT typically interacts with RNA G4s through the end-stacking mode (Mohanty et al. 2013), it is anticipated that G4-ligands displacing ThT exhibit a similar binding mode to ThT. Upon increasing concentrations of PDS (0.5–8.0 µM), we observed a significant decrease in ThT fluorescence emission at 492 nm, indicating the displacement of ThT by PDS from G4-3467 (Fig. S12). These findings suggest that PDS likely interacts with RNA G4s through the end-stacking mode on the top G-tetrad plane, as previously reported (Xu et al. 2016).
To gain further insights into the binding mode of PDS to G4-3467, we conducted molecular dynamics simulations (Fig. S13). In the presence of PDS, the G4 structure of WT-G4-3467 was observed to be stabilized through π–π stacking interactions between the planar surface of PDS and the top G-quartet, along with electrostatic interactions between the phosphate backbone of RNA and the amines in PDS (Fig. S13A–B). On the other hand, the Mut-G4-3467, which lacks the G-tetrad tract required for G4 formation (Fig. S13C), did not form a stable G4 structure either in the absence or presence of PDS (Fig. S13D). While highly unstable conformation of Mut-G4-3467 was evident as shown in the substantial fluctuations of RMSD (Root Mean Square Deviation) values in the presence of PDS, stabilization of WT-G4-3467 structure by PDS was indicated as the lower RMSD values in comparison to those in the absence of PDS (Fig. S13E).
Effects of G4 stabilization by PDS on SARS-CoV-2 protein translation
To investigate the effects of G4 stabilization at G4-644 and G4-3467 by PDS on the translation process, we performed T7 polymerase-driven in vitro coupled transcription/translation (TNT) reactions of G4 containing reporters with increasing doses of PDS. First, we constructed reporter plasmids in which the synthetic DNA fragments containing G4-forming sequences (WT-G4s or Mut-G4s) were placed at in-frame locations upstream of the enhanced yellow fluorescent protein (eYFP) gene (Fig. 5A). We cloned the Nsp1 fragment (284 bp, genomic location: nt 518–801) containing G4-644 alone and the Nsp3-MAC1 domain fragment (580 bp, genomic location: nt 3305–3884) containing G4-3467 for T7-WT-G4-644 and T7-WT-G4-3467, respectively (Fig. S14). Then, through PCR amplification of these reporter plasmids, we produced the corresponding DNA templates (T7-WT/Mut-G4-644 and T7-WT/Mut-G4-3467) (Fig. 5B), which were subjected to in vitro transcription (IVT) or in vitro coupled TNT reactions. Due to the smaller size of the Nsp1 fragment, the PCR amplicon of T7-G4-644-eYFP is shorter than that of T7-G4-3467-eYFP (Fig. 5B). When the effects of PDS were examined on in vitro transcription (IVT) reactions as a control experiment, PDS did not alter the G4-eYFP mRNA expression, except for WT-G4-644 (Fig. 5C). These results indicated that G4 formation did not significantly affect T7 polymerase-driven transcription of these templates. The template DNAs were subjected to in vitro coupled TNT reactions, and the production of G4 peptide-eYFP fusion proteins was analyzed by western blot using anti-eYFP antibody (Fig. 5D). The results showed that PDS treatment led to a significant reduction of the expression of G4 peptide-eYFP proteins from both templates containing G4-644 and G4-3467 in a concentration-dependent manner but did not affect protein production from the Mut-G4 templates (Figs. 5D and S15). These results demonstrated that PDS treatment suppressed the translation of mRNA containing G4-644 and G4-3467 sequences by stabilizing the G4 structures. In control experiments, the effect of CX3543 on the production of G4 peptide-eYFP proteins was marginal except in the case of G4-644, where CX-3543 treatment showed translational attenuation (Fig. S16).
The effect of PDS on SARS-CoV-2 G4-containing reporter gene expression from in vitro transcription and translation assays. A Structures of the T7-WT/Mut-G4-eYFP templates used for in vitro transcription (IVT) or coupled transcription/translation (TNT) reactions are shown. The sizes of the G4-containing DNA fragments synthesized and the T7-G4-eYFP templates PCR-amplified are summarized in the table. B Agarose gels showing the T7-G4-eYFP templates amplified by PCR. C IVT with the T7-WT/Mut-G4-eYFP templates shown in B with DW or increasing concentrations of PDS (15 and 50 μM). The relative levels of transcription were analyzed based on the total amount of RNA transcripts and normalized to that of DW-treated sample for each sample. The graphs represent the mean relative transcription levels with standard errors from three independent experiments. No statistically significant differences were found among samples in a one-way ANOVA. D In vitro coupled TNT reactions were carried out with the T7-WT/Mut-G4-eYFP templates shown in B with DW or increasing concentrations (5 to 50 μM) of PDS. The levels of translated proteins were analyzed by western blotting and quantitated using ImageJ software. The graphs indicate the relative mean protein levels with (± SEM) from three independent experiments. One-way ANOVA was used to determine significant differences in the band intensities between DW-treated and ligand-treated samples. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 are indicated
In addition to PDS and CX3543, we evaluated the effects of two additional G4-binding ligands, PhenDC3 and NMM, on translational suppression by stabilizing individual G4-644 and G4-3467. Initially, we examined the CD spectra and ∆Tm resulting from the interaction of PhenDC3 and NMM. Both ligands did not alter the parallel G4 topology of G4-644 and G4-3467 (Fig. S17A). Interestingly, PhenDC3 exhibited a significantly higher increase in ∆Tm than PDS and NMM, although the accurate determination of Tm values following PhenDC3 treatment was challenging due to the lack of the inflection points till 90 °C (Fig. S17B–C). Consistent with the CD Tm analysis, PhenDC3 effectively suppressed eYFP reporter gene expression for both G4-644 and G4-3467, while NMM did not impact eYFP expression for any G4 fragments (Figs. S18, S19). Despite the dramatic increase in ∆Tm and suppression of protein translation for G4-644 and G4-3467 by PhenDC3, PhenDC3 demonstrated a low antiviral efficacy compared to PDS, as indicated by its high IC50 value (Fig. S2). The discrepancy between in vitro translation suppression and in cellulo antiviral activity of these G4 ligands could be attributed to various factors, in addition to the affinity to targets within the complex cellular environments, such as drug stability, binding interactions with other cellular factors, etc. Nevertheless, our findings with these G4 ligands consistently suggest that the change in Tm values may correlate with in vitro activity to inhibit translation, highlighting the impact of G4 stability on translational activity. Taken together, among the G4-binding ligands tested, PDS potently suppresses the translation of ORF1a and ORF1ab mRNAs by stabilizing the G4s present in the 5′ terminus of viral mRNA. This mechanism, at least in part, contributes to the antiviral activity of PDS against SARS-CoV-2.
Discussion
Targeting of G4s with G4-binding ligands has been reported as an antiviral strategy to control virus infection in many RNA viruses including ZIKV, HCV, EBOV, and HIV-1 (Ruggiero and Richter 2018). This strategy has also been proposed for the development of therapeutic interventions against SARS-CoV-2 (Cui and Zhang 2020; Panera et al. 2020; Bezzi et al. 2021; Zhao et al. 2021). However, antiviral effects of G4-ligands targeting viral genomes have not been examined in SARS-CoV-2-infected cells. Here we explored a novel approach for identifying G4-targeting drugs in SARS-CoV-2. We found that, among various G4-binding ligands tested, PDS exhibited the most potent antiviral activity against SARS-CoV-2 by inhibiting both viral protein expression and genome replication in the SARS-CoV-2 infected Vero cells without cytotoxicity (Figs. 1, 2, S2 and S3). To our best knowledge, this is the first to demonstrate a potent antiviral activity of PDS in SARS-CoV-2 infected cells.
In the following genome-wide study, we found that all 25 predicted G4s in SARS-CoV-2 genome showed the characteristic CD spectra of the parallel G4 topology (Figs. S5–S7). The ThT fluorescence turn-on assay further supported that they adopted the G4 conformation (Figs. S8, S9). Our analysis for the effects of PDS and CX3543 on the stability of 25 SARS-CoV-2 G4s revealed that PDS stabilized SARS-CoV-2 G4s better than did CX3543 (Fig. 3 and Table S3), suggesting that there could be a potential molecular mechanism by which PDS is more capable of suppressing protein translation, thereby better inhibiting SARS-CoV-2 replication than CX3543. The results of in vitro coupled TNT assays were consistent with this notion; PDS was more potent in inhibiting protein translation than CX3543 in vitro (Fig. 5). Given that PDS exhibited stronger antiviral activity in SARS-CoV-2 infected cells than CX3543 (Figs. 1, 2 and S2), these results support our hypothesis that stabilization of viral G4s may inhibit SARS-CoV-2 virus replication via translational suppression. A recent study reported the antiviral activity of another G4-ligand, TMPyP4 in SARS-CoV-2 infected cells (Qin et al. 2022), which was not seen in our study, possibly due to different drug concentrations and infection conditions used in the study.
We found that PDS stabilized the SARS-CoV-2 G4s present in the mRNA encoding pp1a and pp1ab polyproteins, spike and nucleocapsid proteins (Fig. 3). Notably, two G4s (G4-644 and G4-3467) present in the 5′ region of ORF1a showed Tm values (42.0 ± 0.5 and 54.8 ± 0.9 °C), which were further enhanced by PDS treatment by (19.2ºC ± 1.5 and 19.7ºC ± 1.4 °C), respectively; they also exhibited well-defined G4 TDS (Table S3 and Fig. 4C). Given their genomic locations, these G4s might have greater impact on the translation of pp1a and pp1ab than other G4s found in ORF1a and ORF1b. Moreover, these G4s can affect the expression of other ORFs including spike and nucleocapsid, which are dependent on Nsps including RNA-dependent RNA polymerase (RdRp) produced from pp1a and pp1ab, thereby efficiently regulating SARS-CoV-2 replication. Our CD analysis with mutant sequences and in vitro coupled TNT analyses with the reporter constructs demonstrated that PDS-mediated stabilization of these G4s effectively suppressed the translation of the corresponding WT-G4 containing mRNAs compared to Mut-G4 mRNAs (Figs. 5D and S15). Accordingly, we propose a model to explain how the stabilization of G4 structures by PDS can effectively contribute to the regulation of translation during virus infection (Fig. S20). When PDS binds to the SARS-CoV-2 G4, the stabilized G4 structure blocks the progression of the translational machinery, which leads to the subsequent translational inhibition. However, in the case of the Mut-G4 where PDS is unable to bind, the destabilization of the G4 structure allows the translational machinery to progress through the G4 formation site, facilitating protein synthesis. These results confirm that stabilization of viral RNA G4 structures is highly correlated with suppression of protein translation. The effects of G4-binding ligands might be attributed to targeting not only a few G4s, but also multiple G4s. Indeed, PDS treatment also highly increased the Tm of several other G4s with relatively moderate Tm (40–50 °C) (Table S3). Thus, compared to those in in vitro TNT assay where we tested the effect of PDS on protein translation individually for a single G4 sequence of SARS-CoV-2, the effects of PDS might be more prominent on viral protein expression in infected cells with multiple viral RNA G4 sequences. Our results have improved our current understanding of SARS-CoV-2 G4s and underscored the importance of genome-wide analysis as a strategy for developing G4-targeting drugs.
This work stands as a proof of concept for using G4-binding ligands as anti-SARS-CoV-2 drugs. In addition, because many G4s are found in the host genome and can be formed in cellular RNAs (De Magis et al. 2019; Moruno-Manchon et al. 2020; Kosiol et al. 2021), stabilization of cellular G4s by G4-binding ligands may also contribute to their antiviral activity via regulating critical cellular genes to induce host innate immunity such as interferon and cytokine responses (Miglietta et al. 2021; Sun et al. 2022) or to affect viral permissiveness and replication in the host cells (Shen et al. 2020; Liu et al. 2022). Consistent with this notion, a recent study reported that targeting of TMPRSS2 mRNA G4s compromised SARS-CoV-2 replication by reducing the viral entry process (Liu et al. 2022). However, considering the amount of viral RNA in the infected cells, the antiviral effects by G4-ligands can be largely dependent on their targeting to viral RNAs. In support of this notion, the transcriptome analysis of SARS-CoV-2 infection revealed that the majority (65.4%) of the reads mapped to SARS-CoV-2 viral genome, indicating that viral transcripts dominate the transcriptome while the host gene expression is strongly suppressed during viral infection (Kim et al. 2020). Under this condition, there would be more viral RNA G4 targets available for PDS in SARS-CoV-2 infected cells than cellular G4 targets although PDS may affect both cellular DNA/RNA G4s and viral RNA G4 targets. G4-ligands including PDS have been shown to target DNA and RNA G4s in the cytoplasm as well as in the nucleus (Biffi et al. 2013, 2014; Bao et al. 2017; Neckles et al. 2019; Su et al. 2021). Although no G4-ligand specific to viral G4s has been developed yet, the use of G4-ligands as anti-SARS-CoV-2 measures is promising because G4-ligands may directly target to abundant viral G4s and treatment of G4- ligands can increase antiviral innate immune responses in cells (Miglietta et al. 2021).
There have been several cell- and in vivo-based studies where PDS has been safely used as a therapeutic agent for various diseases, including viral infections and cancers (Ruggiero et al. 2021). In line with these findings, our cytotoxicity assay demonstrated a high CC50 value (> 50 μM) for PDS, indicating its safe utilization in cellular systems (Figs. 1D and S3). However, conflicting results have been reported, depending on cell types and conditions; PDS was shown to induce neurotoxicity in cultured neurons by downregulating the BRCA1 gene (Moruno-Manchon et al. 2017), while PDS also induced DNA damages in p53-knockout, hTERT-immortalized RPE1 cell line by promoting topoisomerase II (TOP2) trapping on DNA (Olivieri et al. 2020). PDS treatment was cytotoxic in BRCA2-deficient DLD1 cells, suggesting its potential use as a cancer therapeutic with minimal toxicity to normal cells (Groelly et al. 2022). Nonetheless, further animal studies are necessary to investigate any potential adverse effects of PDS as an antiviral agent in physiological contexts.
Targeting viral proteins such as polymerases and proteases has been a conventional approach for developing antiviral agents. However, a major challenge with targeting key viral proteins is the inevitable emergence of drug-resistant strains due to the high mutation rates of viruses (Nijhuis et al. 2009). Thus, there is a pressing need to identify new antiviral targets with distinct mechanisms of actions to expand the repertoire of antiviral drugs, particularly against drug resistant strains. In this regard, G4s are worth considering as potential antiviral targets. Our analysis of mutation rates in the regions of SARS-CoV-2 G4 sequences found in ten clinical variants of SARS-CoV-2 (α, β, γ, δ, η, ι, κ, λ, μ and ο) showed that most identified G4 sequences are highly conserved and less susceptible to mutations, while few sites are prone to mutations (Fig. S21). For recently emerging SARS-CoV-2 Omicron (ο) variants, G4 site variations were quite lower than those in other variants, despite its highly variable mutations reported in Spike protein (based on sequences available at GISAID as of May 11, 2023); only one G4 at G4-4162 in SARS-CoV-2 Omicron variants shows a meaningful mutation rate (70.2%), while most G4s are not mutated. G4-4162 mutations were introduced in the later Omicron variants starting from BA.2 (> 99%), but not found in an earlier variant, BA.1.
In recent years, there has been a significant increase in research focusing on G4s in viruses, providing strong evidence for the regulatory role of G4s in viral growth such as genome replication, gene transcription and protein translation (Ruggiero et al. 2021). Analysis of G4-forming sequences in the genomes of all known viruses available in the NCBI database has revealed a statistically significant distribution of G4s in viral genomes, suggesting crucial biological roles of G4s in virus life cycle (Lavezzo et al. 2018; Bohálová et al. 2021). Although the G4-644 and G4-3467 sequences of SARS-CoV-2 identified in our current study are not conserved in other RNA and DNA viruses, numerous studies have demonstrated the presence, functionality, and conservation of G4s in viral infections such as HIV-1, HCV and EBOV (Ruggiero and Richter 2020). Therefore, targeting G4s using G4-ligands can potentially block transcription, translation, and replication of viral genomes, exhibiting antiviral activity (Ruggiero and Richter 2023). Moreover, multiple G4 sequences in viral genome may attribute to the minimal emergence of resistant strains. These findings collectively establish G4s in viral genomes as effective targets for antiviral intervention. G4-binding ligands possess druggable characteristics, including a wide range of antiviral activities and properties akin to traditional drugs (e.g., small size and favorable pharmacokinetics). Consequently, the use of G4-ligands opens up possibilities for innovative strategies in combating against viral infections. Our study certainly has limitations that presence of G4s in the host cell genome should be considered as they can also be affected by the G4-ligands. Because G4s are ubiquitous in nature, the challenge in developing antiviral G4-ligands is to ensure the selectivity toward viral G4s. A major limitation of the described G4 ligands is their large flat aromatic core that stacks on the G tetrad, which reduces the chances to discriminate among G4 topologies (Ruggiero and Richter 2018). However, interestingly, we found that the in vitro G4 stabilization effect and in cellulo antiviral activity of PDS are specific for SARS-CoV-2 under our experimental conditions compared to those of CX3543, another G4-binding ligand tested in this study. These results indicate that the antiviral effect of G4-ligands can be selective, depending on the structure of ligands and the nature of G4s in the RNA genome. Further structural studies should be accompanied to explain the detailed interaction specificity and stability changes.
Taken together, our results demonstrate the potential use of G4-binding ligands as an effective antiviral drug: targeting and further stabilizing multiple viral G4s attribute to suppression of protein translation, leading to antiviral activity. Considering the frequent emergence of SARS-CoV-2 variants that could limit the use of current vaccines and antiviral drugs, a novel strategy targeting viral G4s may serve as an alternative therapeutic option, so that a combinatorial therapy with multiple mechanism of actions can be developed to control SARS-CoV-2 infection with increased efficacy and decreased frequency of drug-resistant strains. Thus, in our efforts to combat against current COVID-19 and further prepare for the future outbreaks of human CoVs, the availability of genome-wide studies of G4 formation along with screening of G4-binding ligands can help provide an effective platform for development of antiviral drugs based on G4 targeting.
Data availability
All the data published in this paper will be availlable upon request.
References
Abiri A, Lavigne M, Rezaei M et al (2021) Unlocking G-quadruplexes as antiviral targets. Pharmacol Rev 73(3):897–923. https://doi.org/10.1124/pharmrev.120.000230
Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF (2020) The proximal origin of SARS-CoV-2. Nat Med 26(4):450–452. https://doi.org/10.1038/s41591-020-0820-9
Ariyo EO, Booy EP, Patel TR et al (2015) Biophysical characterization of G-quadruplex recognition in the PITX1 mRNA by the specificity domain of the helicase RHAU. PLoS ONE 10(12):e0144510. https://doi.org/10.1371/journal.pone.0144510
Bae JY, Lee GE, Park H et al (2021) Antiviral efficacy of pralatrexate against SARS-CoV-2. Biomol Ther (seoul) 29(3):268–272. https://doi.org/10.4062/biomolther.2021.032
Bao HL, Ishizuka T, Iwanami A, Oyoshi T, Xu Y (2017) A simple and sensitive 19F NMR approach for studying the interaction of RNA G-quadruplex with ligand molecule and protein. ChemistrySelect 2(15):4170–4175. https://doi.org/10.1002/slct.201700711
Berry SK, Fontana RJ (2020) Potential treatments for SARS-CoV-2 infection. Clin Liver Dis 15(5):181–186. https://doi.org/10.1002/cld.969
Bezzi G, Piga EJ, Binolfi A, Armas P (2021) CNBP binds and unfolds in vitro G-quadruplexes formed in the SARS-CoV-2 positive and negative genome strands. Int J Mol Sci 22(5):2614. https://doi.org/10.3390/ijms22052614
Biffi G, Tannahill D, McCafferty J, Balasubramanian S (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem 5(3):182–186. https://doi.org/10.1038/nchem.1548
Biffi G, Di Antonio M, Tannahill D, Balasubramanian S (2014) Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat Chem 6(1):75–80. https://doi.org/10.1038/nchem.1805
Bohálová N, Cantara A, Bartas M et al (2021) Analyses of viral genomes for G-quadruplex forming sequences reveal their correlation with the type of infection. Biochimie 186:13–27. https://doi.org/10.1016/j.biochi.2021.03.017
Burki TK (2022) The role of antiviral treatment in the COVID-19 pandemic. Lancet Respir Med 10(2):e18. https://doi.org/10.1016/S2213-2600(22)00011-X
Chen JS, Alfajaro MM, Chow RD et al (2021) Nonsteroidal anti-inflammatory drugs dampen the cytokine and antibody response to SARS-CoV-2 infection. J Virol 95(7):e00014-21. https://doi.org/10.1128/JVI.00014-21
Cho HJ, Jeong SG, Park JE et al (2013) Antiviral activity of angelicin against gammaherpesviruses. Antiviral Res 100(1):75–83. https://doi.org/10.1016/j.antiviral.2013.07.009
Corman VM, Landt O, Kaiser M et al (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25(3):2000045. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045
Creech CB, Walker SC, Samuels RJ (2021) SARS-CoV-2 vaccines. JAMA 325(13):1318–1320. https://doi.org/10.1001/jama.2021.3199
Cui H, Zhang L (2020) G-quadruplexes are present in human coronaviruses including SARS-CoV-2. Front Microbiol 11(2570):567317. https://doi.org/10.3389/fmicb.2020.567317
De Magis A, Manzo SG, Russo M et al (2019) DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc Natl Acad Sci USA 116(3):816–825. https://doi.org/10.1073/pnas.1810409116
Dhama K, Khan S, Tiwari R et al (2020) Coronavirus disease 2019-COVID-19. Clin Microbiol Rev 33(4):e00028-e120. https://doi.org/10.1128/CMR.00028-20
Dolinnaya NG, Ogloblina AM, Yakubovskaya MG (2016) Structure, properties, and biological relevance of the DNA and RNA G-quadruplexes: overview 50 years after their discovery. Biochem Mosc 81(13):1602–1649. https://doi.org/10.1134/S0006297916130034
Dong Y, Dai T, Wang B et al (2021) The way of SARS-CoV-2 vaccine development: success and challenges. Signal Transduct Target Ther 6(1):387. https://doi.org/10.1038/s41392-021-00796-w
Drygin D, Siddiqui-Jain A, O’Brien S et al (2009) Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Can Res 69(19):7653–7661. https://doi.org/10.1158/0008-5472.can-09-1304
Fernandes Q, Inchakalody VP, Merhi M et al (2022) Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann Med 54(1):524–540. https://doi.org/10.1080/07853890.2022.2031274
Fischer WA, Eron JJ, Holman W et al (2022) A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci Transl Med. https://doi.org/10.1126/scitranslmed.abl7430
Groelly FJ, Porru M, Zimmer J et al (2022) Anti-tumoural activity of the G-quadruplex ligand pyridostatin against BRCA1/2-deficient tumours. EMBO Mol Med 14(3):e14501. https://doi.org/10.15252/emmm.202114501
Gussow AB, Auslander N, Faure G, Wolf YI, Zhang F, Koonin EV (2020) Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc Natl Acad Sci 117(26):15193–15199. https://doi.org/10.1073/pnas.2008176117
Hansel-Hertsch R, Di Antonio M, Balasubramanian S (2017) DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol 18(5):279–284. https://doi.org/10.1038/nrm.2017.3
Hossain MU, Bhattacharjee A, Emon MTH et al (2021) Novel mutations in NSP-1 and PLPro of SARS-CoV-2 NIB-1 genome mount for effective therapeutics. J Genet Eng Biotechnol 19(1):52. https://doi.org/10.1186/s43141-021-00152-z
Hu MH, Chen SB, Wang B et al (2017) Specific targeting of telomeric multimeric G-quadruplexes by a new triaryl-substituted imidazole. Nucleic Acids Res 45(4):1606–1618. https://doi.org/10.1093/nar/gkw1195
Jeon S, Ko M, Lee J et al (2020) Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.00819-20
Ji D, Juhas M, Tsang CM, Kwok CK, Li Y, Zhang Y (2021) Discovery of G-quadruplex-forming sequences in SARS-CoV-2. Brief Bioinform 22(2):1150–1160. https://doi.org/10.1093/bib/bbaa114
Katsuda Y, Sato SI, Asano L et al (2016) A small molecule that represses translation of G-quadruplex-containing mRNA. J Am Chem Soc 138(29):9037–9040. https://doi.org/10.1021/jacs.6b04506
Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H (2020) The architecture of SARS-CoV-2 transcriptome. Cell 181(4):914-921.e10. https://doi.org/10.1016/j.cell.2020.04.011
Kosiol N, Juranek S, Brossart P, Heine A, Paeschke K (2021) G-quadruplexes: a promising target for cancer therapy. Mol Cancer 20(1):40. https://doi.org/10.1186/s12943-021-01328-4
Kumar S, Chandele A, Sharma A (2021) Current status of therapeutic monoclonal antibodies against SARS-CoV-2. PLOS Pathog 17(9):e1009885. https://doi.org/10.1371/journal.ppat.1009885
Lavezzo E, Berselli M, Frasson I et al (2018) G-quadruplex forming sequences in the genome of all known human viruses: a comprehensive guide. PLOS Comput Biol 14(12):e1006675. https://doi.org/10.1371/journal.pcbi.1006675
Liu G, Du W, Sang X et al (2022) RNA G-quadruplex in TMPRSS2 reduces SARS-CoV-2 infection. Nat Commun 13(1):1444. https://doi.org/10.1038/s41467-022-29135-5
Majee P, Kumar Mishra S, Pandya N et al (2020) Identification and characterization of two conserved G-quadruplex forming motifs in the Nipah virus genome and their interaction with G-quadruplex specific ligands. Sci Rep 10(1):1477. https://doi.org/10.1038/s41598-020-58406-8
Majee P, Pattnaik A, Sahoo BR et al (2021) Inhibition of Zika virus replication by G-quadruplex-binding ligands. Mol Ther Nucleic Acids 23:691–701. https://doi.org/10.1016/j.omtn.2020.12.030
Ma-Lauer Y, Carbajo-Lozoya J, Hein MY et al (2016) p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc Natl Acad Sci U S A 113(35):E5192–E5201. https://doi.org/10.1073/pnas.1603435113
Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB (2005) Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res 33:e138–e138
Miglietta G, Russo M, Duardo RC, Capranico G (2021) G-quadruplex binders as cytostatic modulators of innate immune genes in cancer cells. Nucleic Acids Res 49(12):6673–6686. https://doi.org/10.1093/nar/gkab500
Mohanty J, Barooah N, Dhamodharan V, Harikrishna S, Pradeepkumar PI, Bhasikuttan AC (2013) Thioflavin T as an efficient inducer and selective fluorescent sensor for the human telomeric G-quadruplex DNA. J Am Chem Soc 135(1):367–376. https://doi.org/10.1021/ja309588h
Monchaud D, Teulade-Fichou MP (2008) A hitchhiker’s guide to G-quadruplex ligands. Org Biomol Chem 6(4):627–636. https://doi.org/10.1039/b714772b
Moruno-Manchon JF, Koellhoffer EC, Gopakumar J et al (2017) The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging 9(9):1957–1970. https://doi.org/10.18632/aging.101282
Moruno-Manchon JF, Lejault P, Wang Y et al (2020) Small-molecule G-quadruplex stabilizers reveal a novel pathway of autophagy regulation in neurons. Elife 9:e52283. https://doi.org/10.7554/eLife.52283
Neckles C, Boer RE, Aboreden N et al (2019) HNRNPH1-dependent splicing of a fusion oncogene reveals a targetable RNA G-quadruplex interaction. RNA (new York, NY) 25(12):1731–1750. https://doi.org/10.1261/rna.072454.119
Nijhuis M, van Maarseveen NM, Boucher CAB (2009) Antiviral resistance and impact on viral replication capacity: evolution of viruses under antiviral pressure occurs in three phases. In: Kräusslich HG, Bartenschlager R (eds) Antiviral strategies. Springer, Berlin, pp 299–320
Olivieri M, Cho T, Álvarez-Quilón A et al (2020) A genetic map of the response to DNA damage in human cells. Cell 182(2):481-496.e21. https://doi.org/10.1016/j.cell.2020.05.040
Panera N, Tozzi AE, Alisi A (2020) The G-quadruplex/helicase world as a potential antiviral approach against COVID-19. Drugs 80(10):941–946. https://doi.org/10.1007/s40265-020-01321-z
Qin G, Zhao C, Liu Y et al (2022) RNA G-quadruplex formed in SARS-CoV-2 used for COVID-19 treatment in animal models. Cell Discov 8(1):86. https://doi.org/10.1038/s41421-022-00450-x
Ravichandran S, Kim YE, Bansal V et al (2018) Genome-wide analysis of regulatory G-quadruplexes affecting gene expression in human cytomegalovirus. PLoS Pathog 14(9):e1007334. https://doi.org/10.1371/journal.ppat.1007334
Ravichandran S, Ahn JH, Kim KK (2019) Unraveling the regulatory G-quadruplex puzzle: lessons from genome and transcriptome-wide studies. Front Genet 10(1002):1002. https://doi.org/10.3389/fgene.2019.01002
Ravichandran S, Razzaq M, Parveen N, Ghosh A, Kim KK (2021) The effect of hairpin loop on the structure and gene expression activity of the long-loop G-quadruplex. Nucleic Acids Res 49(18):10689–10706. https://doi.org/10.1093/nar/gkab739
Rubin D, Chan-Tack K, Farley J, Sherwat A (2020) FDA Approval of remdesivir—a step in the right direction. N Engl J Med 383(27):2598–2600. https://doi.org/10.1056/NEJMp2032369
Ruggiero E, Richter SN (2018) G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res 46(7):3270–3283. https://doi.org/10.1093/nar/gky187
Ruggiero E, Richter SN (2020) Viral G-quadruplexes: new frontiers in virus pathogenesis and antiviral therapy. Annu Rep Med Chem 54:101–131. https://doi.org/10.1016/bs.armc.2020.04.001
Ruggiero E, Richter SN (2023) Targeting G-quadruplexes to achieve antiviral activity. Bioorg Med Chem Lett 79:129085. https://doi.org/10.1016/j.bmcl.2022.129085
Ruggiero E, Zanin I, Terreri M, Richter SN (2021) G-quadruplex targeting in the fight against viruses: an update. Int J Mol Sci 22(20):10984
Shamim A, Razzaq M, Kim KK (2020) MD-TSPC4: computational method for predicting the thermal stability of I-motif. Int J Mol Sci. https://doi.org/10.3390/ijms22010061
Shen LW, Qian MQ, Yu K et al (2020) Inhibition of Influenza A virus propagation by benzoselenoxanthenes stabilizing TMPRSS2 Gene G-quadruplex and hence down-regulating TMPRSS2 expression. Sci Rep 10(1):7635. https://doi.org/10.1038/s41598-020-64368-8
Su H, Xu J, Chen Y et al (2021) Photoactive G-quadruplex ligand identifies multiple G-quadruplex-related proteins with extensive sequence tolerance in the cellular environment. J Am Chem Soc 143(4):1917–1923. https://doi.org/10.1021/jacs.0c10792
Sun ZY, Wang XN, Cheng SQ, Su XX, Ou TM (2019) Developing novel G-quadruplex ligands: from interaction with nucleic acids to interfering with nucleic acid(-)protein interaction. Molecules 24(3):396. https://doi.org/10.3390/molecules24030396
Sun J, Wu G, Pastor F et al (2022) RNA helicase DDX5 enables STAT1 mRNA translation and interferon signalling in hepatitis B virus replicating hepatocytes. Gut 71(5):991–1005. https://doi.org/10.1136/gutjnl-2020-323126
Thakur S, Sasi S, Pillai SG et al (2022) SARS-CoV-2 mutations and their impact on diagnostics, therapeutics and vaccines. Front Med 9:815389–815389. https://doi.org/10.3389/fmed.2022.815389
Tse WC, Boger DL (2005) A fluorescent intercalator displacement assay for establishing DNA binding selectivity and affinity. Curr Protoc Nucleic Acid Chem. https://doi.org/10.1002/0471142700.nc0805s20
Tuesuwan B, Kern JT, Thomas PW et al (2008) Simian virus 40 large T-antigen G-quadruplex DNA helicase inhibition by G-quadruplex DNA-interactive agents. Biochemistry 47(7):1896–1909. https://doi.org/10.1021/bi701747d
V’Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V (2021) Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 19(3):155–170. https://doi.org/10.1038/s41579-020-00468-6
Wang M, Cao R, Zhang L et al (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30(3):269–271. https://doi.org/10.1038/s41422-020-0282-0
Wu J, Yuan X, Wang B et al (2020) Severe acute respiratory syndrome coronavirus 2: from gene structure to pathogenic mechanisms and potential therapy. Front Microbiol 11(1576):1576. https://doi.org/10.3389/fmicb.2020.01576
Xie X, Muruato A, Lokugamage KG et al (2020a) An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27(5):841-848.e3. https://doi.org/10.1016/j.chom.2020.04.004
Xie X, Muruato AE, Zhang X et al (2020b) A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat Commun 11(1):5214. https://doi.org/10.1038/s41467-020-19055-7
Xu S, Li Q, Xiang J et al (2016) Thioflavin T as an efficient fluorescence sensor for selective recognition of RNA G-quadruplexes. Sci Rep 6(1):24793. https://doi.org/10.1038/srep24793
Zhao C, Qin G, Niu J et al (2021) Targeting RNA G-quadruplex in SARS-CoV-2: a promising therapeutic target for COVID-19? Angew Chem Int Ed Engl 60(1):432–438. https://doi.org/10.1002/anie.202011419
Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F (2020) Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 6(1):14. https://doi.org/10.1038/s41421-020-0153-3
Acknowledgements
We thank Dr. Pei-Yong Shi (University of Texas Medical Branch, Galveston, TX) for providing the recombinant SARS-CoV-2-Nluc and -mNG viruses. This research was supported by the Bio & Medical Technology Development Program (2021M3A9I2080487 and 2021M3A9I2080488), and the Basic Research Laboratory Program (2020R1A4A1018019) of the National Research Foundation (NRF) funded by the Korean government (MSIT). It was also supported by NRF (2021R1A2C3011644 and 2020R1A2C2013827).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12272_2023_1458_MOESM1_ESM.docx
Supplementary file1 (DOCX 3375 kb)—Supplementary information accompanies the manuscript on the Archives of Pharmacal Research (https://www.springer.com/journal/12272)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Razzaq, M., Han, J.H., Ravichandran, S. et al. Stabilization of RNA G-quadruplexes in the SARS-CoV-2 genome inhibits viral infection via translational suppression. Arch. Pharm. Res. 46, 598–615 (2023). https://doi.org/10.1007/s12272-023-01458-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12272-023-01458-x