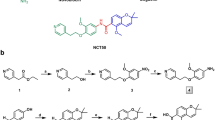
Abstract
The molecular chaperone Hsp90 has emerged as an attractive cancer therapeutic target due to its role in cellular homeostasis by modulating the stabilization and maturation of many oncogenic proteins. In this study, we designed and synthesized a series of Hsp90 inhibitors that hybridized NVP-AUY992 (2) and PU3 (3) in the chalcone scaffold using a structure-based approach. Our results indicate that compound 1g inhibited the proliferation of gefitinib-resistant non-small cell lung cancer (H1975) cells, downregulated the expression of client proteins of Hsp90 including EGFR, MET, Her2 and Akt, and up-regulated the expression of Hsp70. The compound 1g represents a new class of Hsp90 inhibitors with a chalcone structure. The design, synthesis, and evaluation of 1g are described herein.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the last two decades, several cancer drugs that target a single genetic abnormality have been developed as so called “targeted cancer drugs” and seem to be successful to cure tumors in more specific ways without harmful nonspecific side effects (Aggarwal 2010). Despite the robust efficacy of the targeted cancer drugs, the emergence of drug-resistance remains a major obstacle to battle against cancer. Most tumors are heterogeneous and have multiple active oncogenic pathways, which drive the uncontrolled proliferation of tumor cells. In this regard, it is being recognized that tumors might be invincible by single-target drugs and multi-target drugs have emerged as a new paradigm to eradicate tumors and overcome the resistance in drug discovery (Petrelli and Giordano 2008; Boran and Iyengar 2010). Alternatively, in the search of drugs that inhibit a single protein interfering with multiple signaling pathways, heat shock protein 90 (Hsp90) represents an attractive cancer therapeutic target. Hsp90 is a cancer nodal protein that is responsible for a variety of cellular processes, including conformational maturation, stability and function of its substrate proteins, referred to as “client” proteins. Many Hsp90 client proteins play significant roles in six essential hallmarks of a cancer cell, including EGFR, Her2, Met, Cdk4, Akt, HIF and MMP2 (Mahalingam et al. 2009; Hanahan and Weinberg 2000, 2011). Disruption of Hsp90 chaperone function induces client proteins degradation via the ubiquitin–proteasome pathway, which can ultimately lead to cell death.
Lung cancer is the leading cause of cancer deaths worldwide and the most common type of lung cancer is non-small cell lung cancer (NSCLC). Non-small cell lung cancer accounts for 85 % of lung cancer patients (Herbst et al. 2008). Despite advances in early detection and standard treatment, NSCLC is often diagnosed at an advanced stage and has a poor prognosis. Even with surgical resection at early diagnosis, approximately 50 % of NSCLC patients face recurring cancers (Kelsey et al. 2006), and 40–75 % of NSCLC patients are unfortunately projected to die within 5 years even with surgery (Shapiro 2012).
In the past decade, various molecular targeted therapies have been extensively developed for the treatment of NSCLC. The progress in the treatment of NSCLC has been made using small molecule inhibitors of gefitinib (Iressa, AstraZeneca) and erlotinib (Tarceva, Roche). Gefitinib and erlotinib are tyrosin kinase inhibitors of the epidermal growth factor receptor (EGFR) that interrupt signaling through EGFR. The activation of EGFR signaling promotes cancer cell proliferation and tumor progression in cancers, whereas the blockage of EGFR results in apoptotic cell death. However, the clinical efficacies of these EGFR kinase inhibitors in NSCLC are limited due to the development of drug resistance. The acquired drug resistance is reported to be associated with the ‘gatekeeper’ mutation of EGFR T790M, and the activation and amplification of Met tyrosine kinase receptor to maintain Akt signaling pathway (Sordella et al. 2004; Paez et al. 2004). Considering that EGFR, Met, and Akt are client proteins of Hsp90, the inhibitin of Hsp90 chaperoning function would induce the degradation of EGFR, Met, and Akt, which could eventually overcome the resistance by EGFR mutation and Met amplification. As a result, Hsp90 is ideally suited for a potential therapeutic target against drug resistance, and multiple Hsp90 inhibitors are currently under extensive investigation (Jeong et al. 2014; Lee and Seo 2013; Chiosis et al. 2001; Bhat et al. 2014; Seo 2015; Wang et al. 2010; Li et al. 2014; Chen et al. 2014). As part of our ongoing efforts to discover Hsp90 inhibitors, we have found that chalcone-based natural and synthetic products disrupt Hsp90 chaperoning function and impairs the growth of cancer cells (Seo and Park 2014; Jeong et al. 2014; Seo 2013; Lee and Seo 2013). Herein, we report the design, synthesis, and biological evaluation of a new class of Hsp90 inhibitors.
Materials and methods
Chemistry
Unless otherwise noted, all reactions were performed under an argon atmosphere in oven-dried glassware. All purchased materials were used without further purification. Thin layer chromatography (TLC) was carried out using Merck silica gel 60 F254 plates. TLC plates were visualized using a combination of UV, p-anisaldehyde, ceric ammonium molybdate, ninhydrin, and potassium permanganate staining. NMR spectra were obtained on a Bruker 400 (400 MHz for 1H; 100 MHz for 13C) spectrometer. 1H and 13C NMR chemical shifts are reported in parts per million (ppm) relative to TMS, with the residual solvent peak used as an internal reference. Signals are reported as m (multiplet), s (singlet), d (doublet), t (triplet), q (quartet), bs (broad singlet), bd (broad doublet), dd (doublet of doublets), dt (doublet of triplets), or dq (doublet of quartets); the coupling constants are reported in hertz (Hz). Final products were purified by MPLC (Biotage Isolera One instrument) on a silica column (Biotage SNAP HP-Sil). On the basis of NMR and analytical HPLC data (Shimadzu prominence, VP-ODS C18 column), purity for all the tested compounds was found to be >95 %.
1-(2,4-Dyhydroxy-5-isopropylphenyl)ethanone (6)
A mixture of 2,4-dihydroxyacetophenone (2.0 g, 13.1 mmol), 2-bromopropane (2.47 mL, 26.3 mmol), and aluminum chloride (3.5 g, 26.3 mmol) in CH2Cl2 was stirred at 50 °C for 12 h, equipped with a refluxing condenser under argon. 2-Bromopropane (2.47 mL, 26.3 mmol) was added to the reaction mixture three times every 6 h. The mixture was neutralized with 10 % NaOH to pH 5 and then extracted with ethyl acetate. The organic layer was washed with water three times, dried over Na2SO4, concentrated under reduced pressure, and purified by MPLC to afford compound 6 in 46 % yield. Rf = 0.33 (1:4 ethyl acetate: hexane). 1H NMR (400 MHz, CDCl3) δ 12.61 (s, 1H), 7.52 (s, 1H), 6.35 (s, 1H), 6.31 (s, 1H), 3.22–3.12 (m, 1H), 2.61 (s, 3H), 1.27 (d, J = 6.8 Hz, 6H). ESI MS (m/e) = 195 [M+1] +.
1-(2,4-Bis(allyloxy)-5-isopropylphenyl)ethanone (7)
A mixture of compound 6 (0.1 g, 0.52 mmol), allyl bromide (1.4 g, 1.3 mmol), and potassium carbonate (0.19 g, 1.3 mmol) in DMF was stirred at rt for 18 h. The solvent was removed under reduced pressure, and the remaining residue was dissolved in ethyl acetate. The organic layer was washed with saturated NaHCO3 solution, dried over Na2SO4, concentrated under reduced pressure, and purified by MPLC to afford compound 7 in 99 % yield. Rf = 0.45 (1:9 ethyl acetate: hexane). 1H NMR (400 MHz, CDCl3): δ 7.73 (s, 1H), 6.39 (s, 1H), 6.12–6.01 (m, 2H), 5.46 (s, 1H), 5.42 (s, 1H), 5.34–5.30 (m, 2H), 4.62–4.58 (m, 4H), 3.27–3.18 (m, 1H), 2.60 (s, 3H), 1.20 (d, J = 7.2 Hz, 6H). ESI MS (m/e) = 275 [M+1]+.
General procedure for preparing compounds (9a–g and 1a–g), as exemplified for compound 9g and 1g
A mixture of compound 7 (0.32 g, 1.15 mmol), 3,4,5-trimethoxybenzaldehyde (0.24 g, 1.26 mmol), KOH (4.0 g) in 2 mL of water and 10 mL of ethanol was stirred at rt for 1 h. The mixture was neutralized with 6 N HCl to pH 6 and then extracted with ethyl acetate. The organic layer was washed with saturated NaHCO3 solution three times, dried over Na2SO4, and concentrated under reduced pressure, and purified by MPLC (Biotage SNAP HP-Sil column) to afford compound 9g. Rf = 0.24 (1:4 ethyl acetate: hexane). 1H NMR (400 MHz, CDCl3) δ 7.75 (s, 1H), 7.62 (d, J = 15.6 Hz, 1H), 7.56 (d, J = 15.6 Hz, 1H), 6.82 (s, 2H), 6.44 (s, 1H), 6.09–6.02 (m, 2H), 5.49–5.42 (m, 2H), 5.34–5.24 (m, 2H), 4.62–4.61 (m, 4H), 3.88 (s, 9H), 3.32–3.20 (m, 1H), 1.24 (d, J = 5.6 Hz, 6H). 13C NMR (100 MHz, CDCl3) δ 190.3, 160.2, 157.6, 153.3, 141.5, 139.7, 132.8, 131.1, 130.3, 129.2, 126.9, 117.8, 117.5, 105.3, 97.3, 69.8, 68.9, 61.0, 56.1, 26.6, 22.6. ESI MS (m/e) = 453 [M+1] +. The resulting compound 9g was stirred under microwave irradiation (Biotage Initiator) for 30 min at 120 °C in the presence of PdCl2(PPh3)2 (30 mg) and ammonium formate (150 mg) in 4 mL of THF. The reaction mixture was diluted with ethyl acetate. The organic layer was washed with water, dried over Na2SO4, concentrated under reduced pressure, and purified by MPLC to afford compound 1g in 38 % yield in two steps. Rf = 0.21 (1:4 ethyl acetate: hexane). 1H NMR (400 MHz, CDCl3) δ 13.29 (bs, 1H), 7.80 (d, J = 15.2 Hz, 1H), 7.66 (s,1H), 7.44 (d, J = 15.2 Hz, 1H), 6.88 (s, 2H), 6.38(s, 2H), 3.94 (s, 6H), 3.91 (s, 3H), 3.21–3.14 (m, 1H), 1.29 (d, J = 6.8 Hz, 6H). 13C NMR (100 MHz, CDCl3) δ 191.8, 164.4, 160.7, 153.5, 144.5, 140.4, 130.4, 127.9, 126.8, 119.8, 114.1, 105.7, 103.6, 61.1, 56.3, 27.1, 22.6. ESI MS (m/e) = 373 [M+1] +.
(E)-1-(2,4-Bis(allyloxy)-5-isopropylphenyl)-3-phenylprop-2-en-1-one (9a)
Rf = 0.50 (1:4 ethyl acetate: hexane). 1H NMR (400 MHz, CDCl3) δ 7.71 (d, J = 15.8 Hz, 1H), 7.70 (s, 1H), 7.65 (d, J = 15.8 Hz, 1H), 7.60–7.58 (m, 2H), 7.39–7.36 (m, 3H), 6.45 (s, 1H), 6.12–6.02 (m, 2H), 5.48–5.42 (m, 2H), 5.34–5.26 (m, 2H), 4.63–4.60 (m, 4H), 3.12–3.25 (m, 1H), 1.24 (d, J = 7.3 Hz, 6H). 13C NMR (100 MHz, CDCl3) δ 190.7, 160.4, 157. 9, 141.5, 135.7, 132.9, 132.9, 130.3, 130.0, 129.3, 129.0, 128. 5, 127.7, 121.7, 118.1, 117.7, 97.5, 70.0, 69.1, 26.8, 22.7. ESI MS (m/e) = 363 [M+1] +.
(E)-1-(2,4-Dihydroxy-5-isopropylphenyl)-3-phenylprop-2-en-1-one (1a)
32 % yield in two steps. Rf = 0.25 (1:4 ethyl acetate: hexane). 1H NMR (400 MHz, CDCl3) δ 13.29 (bs, 1H), 7.89 (d, J = 15.4 Hz, 1H), 7.69–7.65 (m, 3H), 7.59 (d, J = 15.4 Hz, 1H), 7.46–7.43 (m, 3H), 6.39 (s, 1H), 6.20 (bs, 1H), 3.22-3.16 (m, 1H), 1.30 (d, J = 6.8 Hz, 6H). 13C NMR (100 MHz, CDCl3) δ 192.1, 164.3, 160.8, 144.4, 134.8, 130.7, 129.0, 128.6, 128.0, 127.0, 120.4, 114.2, 103.6, 26.9, 22.6. ESI MS (m/e) = 283 [M+1] +.
(E)-3-(4-(Allyloxy)phenyl)-1-(2,4-bis(allyloxy)-5-isopropylphenyl)prop-2-en-1-one (9b)
Rf = 0.24 (1:9 ethyl acetate: hexane). 1H NMR (400 MHz, CD3OD) δ 7.60–7.50 (m, 5H), 6.91 (d, J = 8.8 Hz, 2H), 6.57 (s, 1H), 6.10–6.00 (m, 3H), 5.47–5.36 (m, 3H), 5.28–5.22 (m, 3H), 4.64–4.52 (m, 6H), 3.28–3.21 (m, 1H), 1.20 (d, J = 6.8 Hz, 6H). 13C NMR (100 MHz, CD3OD) δ 192.5, 161.7, 159.2, 142.8, 134.2, 134.1, 132.5, 130.9, 130.6, 129.4, 129.0, 126.1, 122.0, 118.0, 117.5, 117.4, 115.9, 115.1, 98.5, 70.5, 69.8, 69.6, 27.5, 22.7. ESI MS (m/e) = 419 [M+1] +.
(E)-1-(2,4)-Dihydroxy-5-isopropylphenyl)-3-(4-hydroxyphenyl)prop-2-en-1-one (1b)
35 % yield in two steps. Rf = 0.29 (3:7 ethyl acetate: hexane). 1H NMR(400 MHz, CD3OD) δ (ppm) 7.76(d, J = 15.2 Hz, 1H), 7.12(s, 1H), 7.60(d, J = 2.0 Hz, 2H), 7.56(d, J = 15.2 Hz, 1H), 6.85(d, J = 8.8 Hz, 2H), 6.23(s, 1H), 3.29–3.23(m, 1H), 1.28(d, J = 7.2 Hz, 6H). 13C NMR (100 MHz, DMSO-d6) δ 1091.2, 163.5, 162.2, 159.9, 143.9, 131.4, 128.1, 126.9, 125.5, 117.2, 115.5, 115.4, 112.5, 102.1, 26.2, 22.2. ESI MS (m/e) = 299 [M+1] +.
(E)-1-(2,4-Bis(allyloxy)-5-isopropylphenyl)-3-(4-methoxyphenyl)prop-2-en-1-one (9c)
Rf = 0.35 (1:4 ethyl acetate: hexane). 1H NMR(400 MHz, CDCl3) δ (ppm) 7.70(s, 1H), 7.68(d, J = 15.6 Hz, 1H), 7.56–7.51 (m, 3H), 6.86 (d, J = 8.8 Hz, 2H), 6.47 (s, 1H), 6.12–6.04 (m, 2H), 5.50–5.44 (m, 2H), 5.36–5.28 (m, 2H), 4.64–4.62 (m, 4H), 3.76 (s, 3H), 3.30–3.24 (m, 1H), 1.26 (d, J = 7.2 Hz, 6H). 13C NMR (100 MHz, CDCl3) δ 190.5, 161.0, 159.9, 157.5, 141.2, 132.7, 132.6, 129.9, 129.8, 128.9, 128.1, 125.2, 121.7, 117.7, 117.3, 114.1, 97.3, 69.7, 68.7, 55.2, 26.5, 22.5. ESI MS (m/e) = 393 [M+1] +.
(E)-1-(2,4-Dihydroxy-5-isopropylphenyl)-3-(4-methoxyphenyl)prop-2-en-1-one (1c)
48 % yield in two steps. Rf = 0.33 (1:4 ethyl acetate: hexane). 1H NMR(400 MHz, CDCl3) δ 7.74 (d, J = 15.6 Hz, 1H), 7.60 (s, 1H), 7.54 (d, J = 8.8 Hz, 1H), 7.42 (d, J = 15.6 Hz, 1H), 6.87 (d, J = 8.8 Hz, 1H), 6.29 (s, 1H), 3.77 (s, 3H), 3.20–3.13 (m, 1H), 1.21 (d, J = 6.8 Hz, 6H). 13C NMR (100 MHz, CDCl3) δ 191.8, 163.6, 162.5, 161.5, 143.7, 130.1, 127.7, 127.4, 117.8, 114.2, 113.0, 102.4, 55.1, 26.5, 22.3. ESI MS (m/e) = 313 [M+1] +.
(E)-1-(2,4-Bis(allyloxy)-5-isopropylphenyl)-3-(2,4-dimethoxyphenyl)prop-2-en-1-one (9d)
Rf = 0.21 (1:4 ethyl acetate: hexane). 1H NMR (400 MHz, CDCl3) δ 7.98 (d, J = 16.0 Hz, 1H), 7.64 (s, 1H), 7.56 (d, J = 16.0 Hz, 1H), 7.51 (d, J = 8.4 Hz, 1H), 6.47 (dd, J = 8.4, 2.4 Hz, 1H), 6.43–6.42 (m, 2H), 6.12–6.00 (m, 2H), 5.45–5.40 (m, 2H), 5.30–5.22 (m, 2H), 4.60–4.57 (m, 4H), 3.81 (s, 3H), 3.80 (s, 3H), 3.30–3.21 (m, 1H), 1.22 (d, J = 6.8 Hz, 6H). 13C NMR (100 MHz, CDCl3) δ 191.2, 162.4, 159.9, 159.5, 157.2, 1376.0, 132.7, 129.9, 129.7, 128.8, 125.4, 122.1, 117.4, 117.3, 117.2, 105.1, 98.1, 97.4, 69.7, 68.7, 55.3, 26.4, 22.4. ESI MS (m/e) = 423 [M+1] +.
(E)-1-(2,4-Dihydroxy-5-isopropylphenyl)-3-(2,4-dimethoxyphenyl)prop-2-en-1-one (1d)
25 % yield in two steps. Rf = 0.31 (3:7 ethyl acetate: hexane). 1H NMR (400 MHz, CD3OD) δ 8.02 (d, J = 15.6 Hz, 1H), 7.68 (d, J = 16 Hz, 2H), 7.61 (d, J = 8.4 Hz, 1H), 7.56 (m, 2H), 6.32 (s, 1H), 3.93 (s, 3H), 3.84 (s, 3H), 3.25–3.18 (m, 1H), 1.28 (d, J = 7.2 Hz, 6H). 13C NMR (100 MHz, CD3OD/CDCl3) δ 192.6, 164.0, 163.4, 162.6, 160.5, 139.5, 131.0, 127.5, 127.3, 118.1, 116.6, 113.0, 105.6, 102.2, 98.0, 54.9, 54.7, 26.6, 21.8. ESI MS (m/e) = 343 [M+1] +.
(E)-1-(2,4-Bis(allyloxy)-5-isopropylphenyl)-3-(3,4-dimethoxyphenyl)prop-2-en-1-one (9e)
Rf = 0.31 (1:4 ethyl acetate: hexane). 1H NMR (400 MHz, CDCl3) δ 7.66 (s, 1H), 7.62 (d, J = 15.6 Hz, 1H), 7.51 (d, J = 15.6 Hz, 1H), 7.12–7.07 (m, 2H), 6.80 (d, J = 8.4 Hz, 1H), 6.40 (s, 1H), 6.05–5.96 (m, 2H), 5.44–5.38 (m, 2H), 5.28–5.19 (m, 2H), 4.58–4.54 (m, 4H), 3.85 (s, 3H), 3.84 (s, 3H), 3.27–3.19 (m, 1H), 1.18 (d, J = 6.8 Hz, 6H). 13C NMR (100 MHz, CDCl3) δ 190.1, 160.0, 157.4, 150.6, 148.9, 141.4, 132.6, 132.6, 130.0, 128.9, 128.3, 125.3, 122.9, 121.5, 117.5, 117.2, 110.8, 109.4, 97.1, 69.5, 68.7, 55.7, 55.6, 26.4, 22.4. ESI MS (m/e) = 423 [M+1] +.
(E)-1-(2,4-Dihydroxy-5-isopropylphenyl)-3-(3,4-dimethoxyphenyl)prop-2-en-1-one (1e)
52 % yield in two steps. Rf = 0.21 (1:4 ethyl acetate: hexane). 1H NMR (400 MHz, acetone-d6): δ 13.70 (s, 1H), 9.70 (s, 1H), 8.06 (s, 1H), 7.96 (s, 2H), 7.57–7.48 (m, 2H), 7.12 (d, J = 8.0 Hz, 1H), 6.55 (s, 1H), 4.02 (s, 3H), 4.00 (s, 3H), 3.39–3.31 (m, 1H), 1.39 (d, J = 6.8 Hz, 6H). 13C NMR (100 MHz, CDCl3) δ 191.9, 164.3, 163.9, 151.4, 149.2, 144.3, 144.2, 128.0, 127.7, 123.0, 118.5, 118.4, 113.4, 111.2, 110.8, 56.0, 56.0, 27.0, 21.1. ESI MS (m/e) = 343 [M+1] +.
(E)-3-(Benzo[d][1,3]dioxol-5-yl-1-(2,4-bis(allyloxy)-5-isopropylphenyl)prop-2-en-1-one (9f)
Rf = 0.30 (3:7 ethyl acetate: hexane). 1H NMR (400 MHz, CDCl3) δ 7.68(s, 1H), 7.63 (d, J = 15.6 Hz, 1H), 7.48 (d, J = 16.0 Hz, 1H), 7.09 (s, 1H), 7.06 (d, J = 8.4 Hz, 1H), 6.80 (d, J = 8.0 Hz, 1H), 6.43 (s, 1H), 6.11–6.02 (m, 2H), 5.99 (s, 2H), 5.45 (d, J = 17.6 Hz, 2H), 5.33–5.28 (m, 2H), 4.61 (t, J = 4.4 Hz, 4H), 3.30–3.18 (m, 1H), 1.22 (d, J = 6.8 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 190.5, 160.1, 157.6, 149.3, 148.3, 141.3, 132.8, 132.8, 130.1, 130.0, 129.1, 125.7, 124.8, 121.7, 118.0, 117.5, 108.6, 106.6, 101.5, 97.4, 69.9, 68.9, 26.6, 22.6. ESI MS (m/e) = 407 [M+1] +.
(E)-3-(Benzo[d][1,3]dioxol-5-yl-1-(2,4-dihydroxy-5-isopropylphenyl)prop-2-en-1-one (1f)
65 % yield in two steps. Rf = 0.22 (3:7 ethyl acetate: hexane). 1H NMR (400 MHz, CDCl3/CD3OD) δ 7.61 (d, J = 15.2 Hz, 1H), 7.50 (s, 1H), 7.30 (d, J = 15.2 Hz, 1H), 7.26 (s, 1H), 7.06 (d, J = 1.2 Hz, 1H), 7.01 (dd, J = 8.0 Hz, 1.2 Hz, 1H), 6.72 (d, J = 8.0 Hz, 1H), 6.20 (s, 1H), 5.90 (s, 2H), 3.22–3.04 (m, 1H), 1.11 (d, J = 6.8 Hz, 3H). 13C NMR (100 MHz, CDCl3/CD3OD) δ 191.8, 163.7, 162.8, 149.9, 148.3, 143.7, 129.2, 127.9, 125.3, 118.4, 113.0, 108.6, 106.6, 102.5, 101.6, 26.6, 22.4. MS (m/e) = 327 [M+1] +.
Docking studies
In silico docking of 1g with the 3D coordinates of the X-ray crystal structures of the N-terminal domain of human Hsp90 (PDB code: 1UYM) was accomplished using the AutoDock program downloaded from the Molecular Graphics Laboratory of the Scripps Research Institute. The AutoDock program was chosen because it uses a genetic algorithm to generate the poses of the ligand inside a known or predicted binding site utilizing the Lamarckian version of the genetic algorithm where the changes in conformations adopted by molecules after in situ optimization are used as subsequent poses for the offspring. In the docking experiments carried out, water was removed from the 3D X-ray coordinates while Gasteiger charges were placed on the X-ray structures of the N-terminal domain of Hsp90 along with 1g using tools from the AutoDock suite. A grid box centered on the N-terminal Hsp90 domain with definitions of 60_60_60 points and 0.375 Å spacing was chosen for ligand docking experiments. The docking parameters consisted of setting the population size to 150, the number of generations to 27,000, and the number of evaluations to 25,000,000, while the number of docking runs was set to 50 with a cutoff of 1 Å for the root-mean-square tolerance for the grouping of each docking run. The docking model of human Hsp90 with compound 1g was depicted in Fig. 5 and rendering of the picture was generated using PyMol (DeLano Scientific).
Biology
Antibodies specific for EGFR, Her2, Met, Akt, Hsp90, Hsp70, and β-actin were purchased from Cell Signaling Technology. Goat anti-rabit IgG horseradish peroxidase conjugate was purchased from Santa Cruz Biotechnology. Cell Titer 96 Aqueous One Solution cell proliferation assay kit was purchased from Promega.
Cell culture
H1975 cells were grown in RPMI 1640 with l-glutamine supplemented with streptomycin (500 mg/mL), penicillin (100 units/mL), and 10 % fetal bovine serum (FBS). Cells were grown to confluence in a humidified atmosphere (37 °C, 5 % CO2).
MTS assay
Cells were seeded at 3000 cells per well in a clear 96-well plate, the medium volume was brought to 100 μL, and the cells were allowed to attach overnight. The next day, the indicated concentration of compound or 1 % DMSO vehicle control was added to the wells. Cells were then incubated at 37 °C for 1, 2, and 3 days. Cell viability was determined using the Promega Cell Titer 96 Aqueous One Solution cell proliferation assay. After incubation with compounds, 20 μL of the assay substrate solution was added to the wells, and the plate was incubated at 37 °C for an additional 1 h. Absorbance at 490 nm was then read on Tecan Infinite F200 Pro plate reader, and values were expressed as percent of absorbance from cells incubated in DMSO alone.
Western blot
Cells were seeded in 60 mm culture dishes (5 × 105/dish), and allowed to attach overnight. Compound (1a–g) was added at the concentrations indicated in Fig. 3 or 4, and the cells were incubated for an additional 24 h. For comparison, cells were also incubated with DMSO (1 %) or geldanamycin (1 μM) for 24 h. Cells were harvested in ice-cold lysis buffer (23 mM Tris–HCl pH 7.6, 130 mM NaCl, 1 % NP40, 1 % sodium deoxycholate, 0.1 % SDS), and 20 μg of lysate per lane was separated by SDS-PAGE and followed by transferring to a PVDF membrane (Bio-Rad). The membrane was blocked with 5 % skim milk in TBST, and then incubated with the corresponding antibody (EGFR, Her2, Met, Akt, Hsp90, Hsp70, or β-Actin). After binding of an appropriate secondary antibody coupled to horseradish peroxidase, proteins were visualized by ECL chemiluminescence according to the instructions of the manufacturer (Thermo Scientific, USA).
Results and discussion
Design of Hsp90 inhibitors
Structural analysis of Hsp90 revealed that ATP binding pocket in the N-terminal domain of Hsp90 was composed of a hydrophilic region (P1) of Asp93, Asn51, and Thr184 residues and a hydrophobic region (P2) of Phe138, Tyr139, and Trp162 residues (Fig. 1). Many Hsp90 inhibitors bearing resorcinol ring or purine moiety were known to interact with Asp93, Asn51, and Thr184 residues of the hydrophilic region (P1), whereas the 3,4,5-trimethoxyphenyl group of PU3 (3) (Wright et al. 2004) formed a proximal contacts to Phe138, Tyr139, and Trp162 residues in the hydrophobic region (P2). To maximize ligand interactions in the ATP binding pocket of the Hsp90 N-terminus, we designed a small molecule that has the resorcinol ring of NVP-AUY922 (2) (Brough et al. 2008) and the 3,4,5-trimethoxyphenyl ring of PU3 (3) to target both P1 and P2. To connect the 3,4,5-trimethoxyphenyl ring to the resorcinol ring, we decided to use a chalcone scaffold as a core template (Fig. 2). Chalcones are natural products abundant in edible plants such as green tea and exhibit a wide spectrum of biological activities including anti-inflammatory, anti-viral, and anti-tumor activities (Dimmock et al. 1999; Batovska and Todorova 2010). Accordingly, chalcones are an important class of molecules and speculated as promising candidates as anti-cancer agents. With a hybrid inhibitor embedded in the chalcone scaffold, we envision that the resorcinol ring of the inhibitor would form a hydrogen bond with Asp93 and Asn51 in the hydrophobic region of ATP binding pocket and the 3,4,5-trimethoxyphenyl group of the inhibitor would project into the π-rich lipophilic region of the pocket.
Synthesis of Hsp90 inhibitors
To investigate our rationale for the hybrid design, we first pursued the synthesis of the compounds 1a–g. The synthetic route of the compounds was shown in Scheme 1. Initial attempts to install isopropyl group to benzene moiety were carried out with either TBS or allyl-protected acetophenone, which was synthesized by the treatment of 2,4-dihydroxyacetophenone (5) with TBSCl/DBU or allyl bromide/K2CO3. Then, we carried out Friedel–Crafts alkylation of the protected acetophenone with isopropyl bromide and aluminum chloride. Unexpectedly, the alkylation reactions provided complex mixtures and the protecting groups of TBS and allyl seemed to be cleaved during the reaction. Also, complex peaks at 0.8–1.2 ppm in 1H-NMR indicated that isopropyl bromide decomposed during the reaction. Therefore, we decided to introduce isopropyl group directly to 2,4-dihydroxyacetophenone (5) without any protection. Friedel–Crafts alkylation of acetophenone 5 with isopropyl bromide and aluminum chloride still was not satisfactory but only to provide a trace amount of the product 6. As we observed that isopropyl bromide decomposed during the reaction, we injected isopropyl bromide every 6 h during 24 h of the reaction, which provided the product 6 in 46 % yield. With the isopropyl compound in hand, we next protected compound 6 with allyl bromide in the presence of K2CO3 to furnish the allyl-protected ketone 7 in 99 % yield. Then, subsequent Claisen-Schmidt aldol condensation of compound 7 with the corresponding aromatic aldehydes 8a–g was performed in the presence of KOH in MeOH–H2O and provided enones 9a–g. Finally, the removal of allyl-protecting groups, using PdCl2(PPh3)2 and ammonium formate under microwave irradiation produced the resulting 3′-isopropyl-2′,4′-dihydroxychalcones (1a–g).
Biological evaluation of Hsp90 inhibitors
To investigate the effects of newly synthesized compounds (1a–g) on Hsp90 inhibition, we first screened the efficacy of these compounds by measuring the anti-proliferative activity against H1975 cell line, which is a gefitinib-resistant non-small cell lung cancer cell line. The drug resistance in this cell line is mediated by ‘gatekeeper’ the mutation T790 M-EGFR in combination with L858R (Kobayashi et al. 2005; Sordella et al. 2004). It is also reported that the resistance is related to Met amplification, compensating for the loss of EGFR signals (Bhat et al. 2014).
H1975 cells were incubated with each compound at various concentrations (0, 1, 5, 10, 30, 50, and 100 μM) for 72 h and cell viability was measured using MTS assay (Fig. 3a). The assay revealed that compound 1f and 1g more effectively inhibited cell proliferation in a concentration-dependent manner than others, and 1g impaired the growth of gefitinib-resistant cells by 30 and 85 % at the concentration of 30 μM and 100 μM, respectively. We next decided to screen the Hsp90 inhibition of compounds 1a–g by analyzing the cellular levels of EGFR, Met, Akt, Hsp70 and Hsp90 proteins, given that the proteasomal degradation of client proteins and the transcriptional induction of Hsp70 are molecular signatures of the Hsp90 inhibition (Wright et al. 2004). We treated H1975 cells with 40 μM of each compound (1a–g) for 24 h, and then we assessed the expression levels of Hsp90 client proteins and Hsp70 by western blot (Fig. 3b). Consistent with the result of anti-proliferative activity, 1g most effectively down-regulated Hsp90’s client proteins of EGFR, Met, and Akt, and up-regulated Hsp70, suggesting the observed toxicity of 1g was related to the Hsp90 inhibition.
a Effects of compounds 1a–g on cell proliferation of H1975 cells. Cell viability was determined with compound 1a–g. Cells were treated for 72 h at 0, 1, 5, 10, 30, 50, and 100 μM of compounds 1a–g and cell viability was measured by MTS assay. Data are presented as mean ±SD (n = 4). b Effects of compounds 1a–g on cellular biomarkers of Hsp90 inhibition. H1975 Cells were treated for 24 h with the indicated compound (40 μM) and the expression of the Hsp90 client proteins was analyzed by western blot. Geldanamycin (GA, 1 μM) and DMSO (D) were employed, respectively as positive and negative controls
To precisely determine the efficacy of compound 1g, we measured the dose and time dependent effect of 1g on the growth of H1975 non-small cell lung cancer. Cells were treated with various concentrations (0, 10, 30, 50, 70, and 100 μM) of 1g for 24, 48, or 72 h and cell proliferation was measured using MTS colorimetric assay (Fig. 4a). The data indicated that 50 μM of 1g decreased the cell proliferation by almost 50 %, compared with DMSO-treated cells. Treatment of cells with 100 μM of 1g completely inhibited the growth of cells.
a Anti-proliferative effect of 1g on H1975 cells. Cell proliferation was determined at 1, 2, and 3 days using MTS assay at the indicated concentrations of 1g. Data are presented as mean ±SD (n = 4). b Dose-dependent effect of compound 1g on cellular biomarkers of Hsp90 inhibition. H1975 cells were treated for 24 h with the indicated concentration of 1g and the expressions of the Hsp90’s client proteins and Hsp70 were analyzed by western blot. Geldanamycin (GA, 1 μM) and DMSO (D) were employed, respectively as positive and negative controls. c Analysis of Her2 and d Hsp70 expression in H1975 cells treated with 1g. Western blot analysis was performed on total cellular extracts after 24 h of treatment. Results of densitometry analysis were reported as normalized to β-actin ratio
To evaluate the dose-dependent potency of 1g for the down-regulation of Hsp90’s client proteins, cells was incubated with compound 1g (0, 1, 5, 10, 30, and 50 μM) for 24 h and the expression of Hsp90’s clients, EGFR, Her2, Met and Akt, along with Hsp70 and Hsp90 were measured by western blot (Fig. 4b). As expected, compound 1g demonstrated a robust degradation of EGFR, Her2, Met and Akt at a concentration of 30 μM, and up-regulated the cellular protein level of the cochaperone Hsp70, suggesting that compound 1g disrupted the Hsp90 protein folding machinery. The densitometry analysis of Her2 revealed that even 10 μM of 1g substantially depleted the expression level of Her2 by about 90 % (Fig. 4c), while the expression level of Hsp70 was increased in a dose-dependent manner and maximized at the concentration of 30 μM (Fig. 4d).
Docking studies of Hsp90 inhibitors
To determine the binding mode of 1g, in silico docking study was performed using the human-Hsp90 crystal structure (PDB code: 1UYM). Comparison of binding modes of 1g and PU3 revealed that the trimethoxy phenyl rings of two inhibitors shared common features (Fig. 5a–c). As the 3,4,5-trimethoxyphenyl ring of 1g was designed to bind in the hydrophobic pocket, the phenyl moiety of the 3,4,5-trimethoxyphenyl ring was stacked between the side chains of Phe138 and Leu107 through hydrophobic interactions. The binding mode of the 3,4,5-trimethoxyphenyl ring of 1g was literally equivalent to PU3. In addition to π–π interaction of the phenyl ring with Phe138, the methyls of the methoxy groups formed hydrophobic contacts with Trp162 and Trp139, and the oxygen atoms of the methoxy groups at C3 and C4 of the phenyl ring was positioned to interact with the nitrogen atom of Trp162 and the hydroxyl group of Tyr139 by hydrophilic contacts. The resorcinol ring of 1g was located in the hydrophilic pocket (P1) of Hsp90 as we designed. However, the binding conformation of the resorcinol ring was not exactly equivalent to NYP-AUY922. The para-hydroxy group at C4′ instead of the hydroxyl group at C2′ of 1g formed hydrogen-bonding with Asp93, while the carbonyl oxygen atom of 1g interacted with Asn51 through hydrogen bonding. Collectively, two hydrogen bonds in the hydrophilic region (P1) and hydrophobic interaction in the hydrophobic region (P2) contributed to the binding of compound 1g to ATP binding pocket of Hsp90, and the estimated binding energy (ΔGb) and inhibition constants (Ki) using the Lamarckian genetic algorithm result in −7.84 kcal/mol and 1.79 μM, respectively.
Comparative binding modes of 1g, ADP, PU3, and NYP-AUY922. a Molecular docking model of 1g with N-terminal Hsp90. b X-ray co-crystal structures of ADP (PDB code: 1BYQ), c PU3 (3, PDB code: 1UYM) and d NYP-AUY922 (2, PDB code: 2VCI) with Hsp90. The carbon atoms of 1g, ADP, PU3 and NYP-AUY922 are shown in yellow, sky blue, green, and pink, respectively. The oxygen, nitrogen, phosphine, and hydrogen atoms of 1g, ADP, PU3 and NYP-AUY922 are shown in red, blue, orange and gray, respectively. The side chains of binding site are colored by atom types (carbon, gray; nitrogen, blue; oxygen, red) and labeled with their residue name
Conclusions
A series of chalcone-templated Hsp90 inhibitors were rationally designed, synthesized, and evaluated against gefitinib resistant non-small cell lung cancer cells (H1975). Compound 1g appears to be most potent to impair H1975 cell proliferation. Consequently, 1g manifests considerable degradation of Hsp90’s client proteins of EGFR, Met, Her2 and Akt, and dose-dependent induction of Hsp70’s expression level, supporting that 1g targets the Hps90 chaperone machinery. Molecular docking analysis of compound 1g demonstrated that 1g occupied ATP binding pocket in the N-terminal domain of Hsp90. The resorcinol ring and the 3,4,5-trimethoxyphenyl ring of 1g bound to the hydrophilic region (P1) and the hydrophobic region (P2) of ATP binding pocket in the N-terminal domain of Hsp90, respectively. Further structure–activity relationship (SAR) studies are currently underway and will be reported in due course.
References
Aggarwal S (2010) Targeted cancer therapies. Nat Rev Drug Discov 9:427–428
Batovska DI, Todorova IT (2010) Trends in utilization of the pharmacological potential of chalcones. Curr Clin Pharmacol 5:1–29
Bhat R, Tummalapalli SR, Rotella DP (2014) Progress in the discovery and development of heat shock protein 90 (Hsp90) inhibitors. J Med Chem 57:8718–8728
Boran AD, Iyengar R (2010) Systems approaches to polypharmacology and drug discovery. Curr Opin Drug Discov Dev 13:297–309
Brough PA, Aherne W, Barril X, Borgognoni J, Boxall K, Cansfield JE, Cheung KM, Collins I, Davies NG, Drysdale MJ, Dymock B, Eccles SA, Finch H, Fink A, Hayes A, Howes R, Hubbard RE, James K, Jordan AM, Lockie A, Martins V, Massey A, Matthews TP, McDonald E, Northfield CJ, Pearl LH, Prodromou C, Ray S, Raynaud FI, Roughley SD, Sharp SY, Surgenor A, Walmsley DL, Webb P, Wood M, Workman P, Wright L (2008) 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer. J Med Chem 51:196–218
Chen D, Shen A, Li J, Shi F, Chen W, Ren J, Liu H, Xu Y, Wang X, Yang X, Sun Y, Yang M, He J, Wang Y, Zhang L, Huang M, Geng M, Xiong B, Shen J (2014) Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors. Eur J Med Chem 87:765–781
Chiosis G, Timaul MN, Lucas B, Munster PN, Zheng FF, Sepp-Lorenzino L, Rosen N (2001) A small molecule designed to bind to the adenine nucleotide pocket of Hsp90 causes Her2 degradation and the growth arrest and differentiation of breast cancer cells. Chem Biol 8:289–299
Dimmock JR, Elias DW, Beazely MA, Kandepu NM (1999) Bioactivities of chalcones. Curr Med Chem 6:1125–1149
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Herbst RS, Heymach JV, Lippman SM (2008) Lung cancer. N Engl J Med 359:1367–1380
Jeong CH, Park HB, Jang WJ, Jung SH, Seo YH (2014) Discovery of hybrid Hsp90 inhibitors and their anti-neoplastic effects against gefitinib-resistant non-small cell lung cancer (NSCLC). Bioorg Med Chem Lett 24:224–227
Kelsey CR, Clough RW, Marks LB (2006) Local recurrence following initial resection of NSCLC: salvage is possible with radiation therapy. Cancer J 12:283–288
Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B (2005) EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 352:786–792
Lee T, Seo YH (2013) Targeting the hydrophobic region of Hsp90’s ATP binding pocket with novel 1,3,5-triazines. Bioorg Med Chem Lett 23:6427–6431
Li Z, Jia L, Wang J, Wu X, Hao H, Wu Y, Xu H, Wang Z, Shi G, Lu C, Shen Y (2014) Discovery of diamine-linked 17-aroylamido-17-demethoxygeldanamycins as potent Hsp90 inhibitors. Eur J Med Chem 87:346–363
Mahalingam D, Swords R, Carew JS, Nawrocki ST, Bhalla K, Giles FJ (2009) Targeting HSP90 for cancer therapy. Br J Cancer 100:1523–1529
Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304:1497–1500
Petrelli A, Giordano S (2008) From single- to multi-target drugs in cancer therapy: when aspecificity becomes an advantage. Curr Med Chem 15:422–432
Seo YH (2013) Discovery of Licochalcone a as a natural product inhibitor of Hsp90 and its effect on gefitinib resistance in non-small cell lung cancer (NSCLC). Bull Korean Chem Soc 34:1917–1920
Seo YH (2015) Organelle-specific Hsp90 inhibitors. Arch Pharm Res 38:1582–1590
Seo YH, Park SY (2014) Synthesis of flavokawain analogues and their anti-neoplastic effects on drug-resistant cancer cells through Hsp90 inhibition. Bull Korean Chem Soc 35:1154–1158
Shapiro M (2012) The role of adjuvant chemotherapy in early-stage and locally advanced non-small cell lung cancer. Clevel Clin J Med 79(Electronic Suppl 1):eS42–eS45
Sordella R, Bell DW, Haber DA, Settleman J (2004) Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science 305:1163–1167
Wang Y, Trepel JB, Neckers LM, Giaccone G (2010) STA-9090, a small-molecule Hsp90 inhibitor for the potential treatment of cancer. Curr Opin Investig Drugs 11:1466–1476
Wright L, Barril X, Dymock B, Sheridan L, Surgenor A, Beswick M, Drysdale M, Collier A, Massey A, Davies N, Fink A, Fromont C, Aherne W, Boxall K, Sharp S, Workman P, Hubbard RE (2004) Structure-activity relationships in purine-based inhibitor binding to Hsp90 isoforms. Chem Biol 11:775–785
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2016R1A6A1A03011325 and 2016R1D1A1B01009559), and Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through High Value-added Food Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (316053-02).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jeong, J.H., Oh, Y.J., Kwon, T.K. et al. Chalcone-templated Hsp90 inhibitors and their effects on gefitinib resistance in non-small cell lung cancer (NSCLC). Arch. Pharm. Res. 40, 96–105 (2017). https://doi.org/10.1007/s12272-016-0848-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12272-016-0848-z