Abstract
C13-apocarotenoids are volatile organic compounds naturally derived from the oxidative cleavage of carotenoids. These small molecules form unique aromas in flowers, fruits, and plants. They are highly valued compounds in the flavor and fragrance industry. The microbial production of C13-apocarotenoids, such as β-ionone, α-ionone, and pseudoionone, is an emerging and promising approach with inherent advantageousness of scalable output to reach the goals as stated in the United Nations Sustainable Development Goals. Many engineering efforts have been implemented continuously but very few of them proved to be successful in achieving product titer at the grams-per-liter level with the least accumulated amount of precursor carotenoids and byproducts. The efficiency of oxidative cleavage of carotenoids conducted by carotenoid cleavage dioxygenases is suggested to be the critical metabolic node to reconstruct an economically viable C13-apocarotenoids biosynthetic pathway. In this regard, we review recent advances in improving microbial biosynthesis of C13-apocarotenoids by protein and metabolic engineering. The potential strategies that could be implemented further to achieve efficient C13-apocarotenoid production are also discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Plant-derived aromas and flavors are the sensory impressions formed by a mixture of secondary metabolites [1], including apocarotenoids, sesquiterpenes, phenolic derivatives, lipid derivatives, and amino acid derivatives. Apocarotenoids and their derivatives are molecules derived from the oxidative cleavage of carotenoids by carotenoid cleavage dioxygenases (CCDs) [2], and the cleaved products can be used as valuable nutrition [3], pigments [4,5,6], hormone precursors [7], and aroma volatiles [8, 9]. Among these CCD oxidative cleaved compounds, C13-apocarotenoids are significant contributors to the aroma found in the food, beverage, cosmetic, and tobacco industries due to their pleasant smell with extremely low odor thresholds (β-ionone: 0.007 μg/L in water; 0.09 μg/L in a model wine solution) [10]. The representative C13-apocarotenoids are α-ionone and β-ionone, which widely exist in various fruits and flowers.
C13-apocarotenoids are highly volatile molecules and are relatively less abundant in plant sources. The contents of natural α-ionone and β-ionone were reported to be ~ 10 μg/kg and ~ 1.7 mg/kg in ionone-rich raspberries [11, 12]. Extracting C13-apocarotenoids from natural sources is less economically stable and viable to meet the ever-increasing demand, considering that the supply of raw materials from plant cultivation is often affected by seasonal, climate, and other factors. Therefore, ionones are produced industrially from the aldol condensation of citral with acetone [8]. However, the chemical synthesis of C13-apocarotenoids would be processed under high temperature and pressure, which is non-environmentally friendly. In addition, it is challenging to produce the desired enantiomer (R)-α-ionone with a "more defined and purer" flavor via chemical synthesis [13]. More importantly, the products obtained from chemical synthesis cannot be marketed as natural ones obtained from plant extraction or biosynthesis. The value of natural flavor is often significantly higher than those produced by chemical synthesis due to consumers’ personal preferences, and the price of biosynthesized β-ionone could be ten times higher than the one produced using the chemical method [13].
de novo biosynthesis of natural C13-apocarotenoids from simple and renewable carbon sources (e.g., glucose, xylose, glycerol, ethanol, and acetate) using meticulously designed microbial cell factories (MCFs) offers a sustainable and alternative solution, and avoids the issues associated with chemical synthesis or plant-based extraction. The representative C13-apocarotenoids (e.g., β-ionone, α-ionone, and pseudoionone) have been successfully produced in engineered microbes. However, the reported studies have pointed out that the oxidative cleavage of carotenoids conducted by CCDs is the rate-limiting step that severely restricts the conversion of carotenoids to ionones in MCFs [11, 13,14,15]. In this review, we focus on the protein engineering of CCDs and the various metabolic engineering strategies to improve the production of ionones. In particular, we will review the efficient strategies to increase carotenoid conversion to ionones, such as optimizing the functional expression of CCDs, the improvement/alteration of CCDs’ catalytic activity/substrate specificity, the engineering of oleaginous yeasts to produce ionones, and strengthening substrate channeling and accessibility in MCFs. New potential engineering strategies for unlocking the potential of CCD-involved biosynthesis are also discussed briefly, which may offer insights into producing C13-apocarotenoids using MCFs.
2 Microbial biosynthesis of C13-apocarotenoids
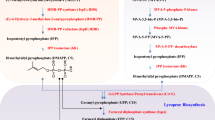
Heterologous microbial biosynthesis is a promising strategy to produce natural compounds economically and sustainably, such as artemisinin [16], taxol precursor [17], and amino acid [18]. In recent years, many efforts have been made to harness MCFs to overproduce C13-apocarotenoids with the well-characterized CCD catalytic module (Fig. 1). Carotenoids are the precursors of C13-apocarotenoids and could be biosynthesized from four natural or non-natural pathways. Starting from C2 to C6 substrates [19, 20], the biosynthesis of the key node metabolites of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) in isoprenoid biosynthetic pathways can be achieved through the native mevalonate (MVA) or methylerythritol phosphate (MEP) pathway [21]. The generated IPP and DMAPP would be further condensed iteratively to structurally diversified carotenoids. The MEP and MVA pathways are well-known natural pathways and have been extensively studied, reconstructed, and pathway-optimized in microbes to produce carotenoids, such as lycopene, β-carotene, and lutein [22,23,24]. The heterologous microbial biosynthesis of C13-apocarotenoid has also been employed to investigate the substrate specificities of CCDs in Escherichia coli by modifying the downstream carotenoid biosynthetic pathways to generate various substrates, such as lycopene, carotene, and lutein [25]. It is found that geranylacetone, pseudoionone, α-ionone, and β-ionone could be generated from cleaving phytoene, lycopene, α-carotene, and β-carotene, respectively, with Zea mays CCD1 (ZmCCD1) or Osmanthus fragrans CCD1 (OfCCD1) overexpressed in E. coli [13, 26, 27]. Besides, 3-OH-α-ionone and 3-OH-β-ionone could be produced by cleaving lutein using CCD2 (Fig. 1A) [28], providing a critical catalytic module to reconstruct the biosynthesis of hydroxylated C13-apocarotenoids in MCFs.
A The biosynthetic pathways of producing carotenoids-derived C13-apocarotenoids from possible C2-C6 carbon sources by engineered microbes. Schematic illustration of the biosynthesis of isoprenoids from central metabolites such as glyceraldehyde 3-phosphate, pyruvate, acetyl-CoA, or isopentenol. Conventional sugars, glycerol, or non-conventional substrates such as ethanol or acetate can be used by the engineered microbes to supply these important central metabolites. The isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), two essential building blocks of isoprenoids, can be biosynthesized via four biosynthetic pathways, including mevalonate (MVA) pathway, methylerythritol phosphate (MEP) pathway, and two alternative ones (isopentenol utilization pathway [IUP] and isoprenoid alcohol [IPA] pathway). They will be used to synthesize C40 carotenoids, which will be oxidatively cleaved by carotenoid cleavage dioxygenases (CCDs) to form the corresponding C13-apocarotenoids. B The CCD cleavage sites on both sides of several representative C40 carotenoids are highlighted in number, and the corresponding C13-apocarotenoids are illustrated on the right-hand side. The metabolite and enzymes abbreviations in this figure are as follows: DXP: 1-deoxy-d-xylulose-5-phosphate, CDP-ME: 4-diphosphocytidyl-2-C-methylerythritol, CDP-MEP: 4-diphosphocytidyl-2-C-methyl-d-erythritol 2-phosphate, MEC: 2-C-methyl-d-erythritol 2,4-cyclodiphosphate, HMBPP: (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate, HMG-CoA: 3-hydroxy-3-methylglutaryl-coenzyme A, MVAP: mevalonate-5-phosphate, MVAPP: mevalonate-5-pyrophosphate, IP: isopentenyl phosphate, DMAP: dimethylallyl phosphate, FPP: farnesyl pyrophosphate, GPP: geranyl pyrophosphate, GGPP: geranylgeranyl pyrophosphate, DXS: DOXP synthase, DXR: DXP reductoisomerase, CMS: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, CMK: 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase, MECS: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, HDS: HMBPP synthase, HDR: HMBPP reductase, HMGS: 3-hydroxy-3-methylglutaryl-CoA synthase, HMGR: 3-hydroxy-3-methylglutaryl-CoA reductase, MK: mevalonate-5-kinase, PMK: phosphomevalonate kinase, PMD: mevalonate pyrophosphate decarboxylase, IDI: isopentenyl diphosphate isomerase, IK: isopentenol kinase, IPK: isopentenyl phosphate kinase, GPPS: GPP synthase, FPPS: FPP synthase, GGPPS: GGPP synthase, CYP: cytochrome P450 monooxygenase, LCYeC: lycopene epsilon-cyclase, LCYbC: lycopene beta-cyclase, HYDB: β-carotene hydroxylase, TCA cycle: tricarboxylic acid cycle
To circumvent the inherent inefficiencies and complexities of native biosynthetic pathways, two synthetic pathways for isoprenoid production were developed. By harnessing isopentenyl phosphate kinases, low-cost C5 petrochemicals (isopentenol: isoprenol/prenol) could be converted into IPP or DMAPP by the isopentenol utilization pathway (IUP) via a two-step phosphorylation, which can avoid competing for intracellular metabolites such as acetyl-CoA during cell growth [29, 30]. The IUP could efficiently produce gram-per-liter isoprenoid titers in engineered microbes, such as E. coli, Saccharomyces cerevisiae, and Yarrowia lipolytica. The advantages and applications of the IUP have been well-summarized in a recently published review article [31]. Engineering microbes to produce isoprenoids can be limited by the competition between product formation and cell growth, and thus producing C13-apocarotenoids through the IUP is a promising way to bypass this limitation. It could be further developed using a whole-cell biotransformation strategy to replace the non-enantiomer-producing chemical processes. Since prenol could be synthesized from acetyl-CoA, Clomburg et al. [32] constructed a non-natural isoprenoid alcohol (IPA) pathway in E. coli by linking the formation of DMAPP from prenol with a cellular central metabolite (acetyl-CoA, Fig. 1A). The IPA pathway consumes 2 ATP and 2 NAD(P)H to convert acetyl-CoA to DMAPP and IPP with a carbon efficiency of 0.83 (one carbon loss from converting C6 to C5 intermediate). The MVA and MEP pathways have the same carbon efficiency but consume more ATP and NAD(P)H (MVA pathway: 3 ATP and 2 NAD(P)H; MEP pathways: 3 ATP and 3 NAD(P)H), and would require more carbon from the central metabolism to regenerate energy and redox molecules in the tricarboxylic acid cycle, thus lowering the product yield. The IUP or IPA pathway has been demonstrated successfully to efficiently produce lycopene [33], β-carotene [29], and pseudoionone [34]. Recently, Fan et al. [33] reconstructed both IUP and MVA pathway in E. coli to produce pseudoionone, with a titer of 20.61 mg/L reached in a shake flask, proving the feasibility of using IUP to boost the production of C13-apocarotenoids in MCFs.
3 Carotenoid cleavage dioxygenases
CCDs are categorized as non-heme iron-dependent enzymes (EC: 1.13.11.51), and can be classified into six CCD family members based on the phylogenetic analysis of their amino acid sequences (Fig. 2A). They include CCD1, CCD2, 9-cis-epoxy-carotenoid cleavage dioxygenase (NCED), CCD4, CCD7, and CCD8 [35]. CCD1 is the most studied CCD subfamily and could cleave a variety of carotenoid substrates symmetrically at varied sites [36]. For example, ZmCCD1 cleaves phytoene and ζ-carotene at 5,6/5′,6′ or 9,10/9′,10′; lycopene and δ-carotene at 5,6/5′,6′, 7,8/7′,8′, or 9,10/9′,10; β-carotene and zeaxanthin at 7,8/7′,8′ or 9,10/9′,10, which generates multiple volatile flavor compounds such as 6-methyl-5-hepten-2-one, geranylacetone, citral, pseudoionone, β-cyclocitral, β-Ionone, 3-hydroxy-β-cyclocitral, and 3-hydroxy-β-ionone [36].
A Phylogenetic analysis of 90 CCD protein sequences by maximum likelihood method in MEGA X. The CCD highlighted in blue was used as probe sequences to run a BLAST protein sequence similarity search in NCBI database. In each CCD family, 4 to 20 sequences with a sequence similarity score above 75% were selected and analyzed. The CCD highlighted in green has its protein structure publicly deposited (OfCCD1, Protein Data Bank ID: 7VUD). B Protein structure of CCDs visualized using PyMOL. Except for the protein structure of CsCCD2 obtained from modeling in SWISS-MODEL using OfCCD1 as a template, the others were modeled and retrieved from the AlphaFold Protein Structure Database. The presumed membrane-contacting/penetrating helix regions are highlighted in brown. CCD: carotenoid cleavage dioxygenase, NCED: 9-cis-epoxy-carotenoid cleavage dioxygenase, OfCCD1: Osmanthus fragrans CCD1, PhCCD1: Petunia hybrida CCD1, CsCCD2: Crocus sativus CCD2, AtCCD8: Arabidopsis thaliana CCD8, AtCCD7: Arabidopsis thaliana CCD7, AtCCD4: Arabidopsis thaliana CCD4, AtNCED: Arabidopsis thaliana NCED
CCD2 from the Crocus genus was identified to be from a novel CCD clade and cleaves zeaxanthin at C7 and C8 sites on both sides sequentially, generating two molecules of 3-hydroxy-β-cyclocitral and one molecule of crocetin dialdehyde, which could be converted to crocetin [26]. The NCED, a subfamily evolved with functional divergence from the CCD1 group (Fig. 2A) [37], is capable of cleaving 9'-cis-epoxycarotenoids [28, 38]. A NCED in Arabidopsis thaliana cleaves 9-cis-violaxanthin and 9-cis-neoxanthin at the C11 and C12 sites, generating 2-cis, 4-trans-xanthoxin. This compound is a precursor to an essential plant hormone, abscisic acid. The oxidative cleavage pattern of CCD4 is similar to CCD1. Characterization of several CCD4s from various plant sources has shown the ability to catalyze the oxidative cleavage of carotenoids at the sites of 5,6/5′,6′, 7,8/7′,8′, or 9,10/9′,10 [39, 40]. CCD7 and CCD8 are found to be related to cleaving the carotenoid isomer substrate and could catalyze 9-cis-β-carotene via a cascade reaction to form carlactone, which is a precursor of strigolactone [7], a novel plant hormone playing multiple physiological roles in plant development, hyphae branching regulation, and plant-fungal interaction promotion.
A. thaliana CCD1 (AtCCD1) was first purified by Schwartz et al. [41]. They validated its catalytic function on various carotenoids in vitro, and the amino acid sequence of AtCCD1 was then used as a probe to mine CCD1s from other plant sources, such as Petunia hybrid, Vitis vinifera, and Osmanthus fragrans [42,43,44]. Further enzyme characterizations subsequently demonstrated that most of the CCD1s exhibited a wide range of substrate preferences, which not only cleave carotenoids at the sites of 9,10/9′,10′ but also at the sites of 5,6/5′,6′ or 7,8/7′,8′ (Fig. 1B). The catalytic process of AtCCD1 was studied using an isotope labeling experiment [45]. Using 18O2-contained air, an abundant product of C17-dialdehyde with 18O labeled isotope was formed, suggesting that two oxygen molecules would react with the carbon double bond of carotenoid and generate an unstable dioxetane intermediate [46], which rapidly decays into two aldehydes.
Xue and Sharma [47] recently released the protein structure of OfCCD1 in the Protein Data Bank (PDB: 7VUD), offering valuable insights into the structural characteristics and catalytic function of CCD1. By using the SWISS-MODEL with OfCCD1 as the template, we structurally modeled Petunia hybrida CCD1 (PhCCD1), a commonly used catalytic module to biosynthesize β-ionone in MCFs (Table 1). By retrieving the protein structures of other CCD1s from the AlphaFold Protein Structure Database, a conserved seven-bladed β-sheet and two helix regions (in brown, Fig. 2B) as the basic motif can be observed throughout the protein structures of the CCD1 family [48]. In addition, four highly conserved histidine residues in the catalytic pocket of CCD1 were seen to coordinate with a Fe2+ ion in the catalytic pocket (Fig. 3), contributing to the cleavage of carbon double bond in the presence of oxygen molecules [49].
Structure illustration of the substrate-contacting and presumed membrane-contacting/penetrating regions in (A) OfCCD1 and (B) PhCCD1. The loop regions (highlighted in red) in the catalytic pocket substantially affect the enzyme catalytic activity and substrate preference, and the residues mutated in OfCCD1 with improved α-ionone production titer are shown in green. The membrane-contacting/penetrating helixes are highlighted in brown. The residue mutated in the α-helix of PhCCD1 with improved β-ionone production efficiency is shown in green. The oxtoxynol-10 ligand highlighted in yellow is a ligand-of-interest in OfCCD1 as presented in the PDB file (7VUD) designated by the Research Collaboratory for Structural Bioinformatics. For visualization of PhCCD1 with oxtoxynol-10, the PDB file of PhCCD1 from the AlphaFold Protein Structure Database was structurally aligned with OfCCD1 in PyMOL. The four histidine residues highlighted in orange reveal the enzyme catalytic center where they coordinate with a F.2+ ion. The amino acid symbols used in this figure are as follows: V: valine, D: aspartic acid, F: phenylalanine, K: lysine, N: asparagine, I: isoleucine, A: alanine, S: serine. OfCCD1: Osmanthus fragrans CCD1, PhCCD1: Petunia hybrida CCD1
4 Engineering strategies of C13-apocarotenoids biosynthesis
Given that CCD1 has been extensively characterized as the core catalytic module to reconstruct the microbial biosynthesis of C13-apocarotenoids, hereafter, as listed in Fig. 4, we will discuss the representative engineering strategies based on CCD1.
4.1 Functional expression of carotenoid cleavage dioxygenase 1
Previous studies validated that the functional, soluble expression of CCD1 in recombinant E. coli was insufficient [13]. CCD1 was often overexpressed with fusion protein tags to increase its expression level and solubility by simplifying or improving protein refolding. Fusing glutathione S-transferase (GST) at the N-terminal of CCD1 substantially improved its protein solubility and facilitated the in vitro functional characterization [50, 51]. Zhang et al. [13] fused three other protein tags (i.e., maltose binding protein, small ubiquitin-like modifier, thioredoxin) at the N-terminal of OfCCD1 to enhance the functional, soluble expression of CCD1 in an engineered E. coli. The results indicated that these fusions significantly increased the titer of α-ionone (from < 0.5 to > 10 mg/L). Among all the fusions, the TrxA-fused OfCCD1 exhibited the best solubility-enhancing effect, and a higher α-ionone titer was subsequently achieved (29.7 mg/L, a 50-fold increase compared with no tag fused) [13]. Although the underlying mechanism of protein tag to improve protein solubility needs to be further investigated, harnessing protein fusion partners to enhance the functional, soluble expression of CCD1 has been considered an essential step to improve the biosynthesis of C13-apocarotenoids in E. coli.
Modular pathway optimization of the combined genetic parts (e.g., ribosomal binding site [RBS], copy number, promoter, terminator, and operon) has been widely used to develop MCFs [52,53,54]. To improve the production of C13-apocarotenoids, Chen et al. [14] adopted an RBS library with a color-based colony screening strategy to fine-tune the expression of the TrxA-fused CCD1. They first identified that using an RBS with a moderate translation initiation rate produced a higher proportion of ε-carotene in total carotenoid products. Next, the authors used an RBS with a ~ twofold increased translation initiation rate to express the TrxA-fused OfCCD1, and the resulting strain led to a ~ 20% improvement of α-ionone titer. To increase the gene expression of PhCCD1, López et al. [55] integrated two additional gene copies of PhCCD1 into the genome of S. cerevisiae, and the titer of β-ionone produced increased ~ twofold with the enhanced expression level of PhCCD1.
A recent study showed that the expression level of CCD1 was critical in scaling up the production of α-ionone due to oxidative stress posed by CCD1 [12]. When the fermentation culture volume was increased from 1 mL in a tube to 10 mL in a shake flask, the α-ionone titer decreased from 96 to 19.3 mg/L. Subsequent investigations revealed that hydrogen peroxide accumulated when CCD1 was excessively overexpressed, which destroys an important precursor to α-ionone, lycopene. A hydroperoxide reductase (coding by ahpC/F) was overexpressed to react hydrogen peroxide to water and oxygen and the α-ionone titer was improved substantially. Finally, the fermentation process was scaled up in a 5-L bioreactor via fed-batch operation and the final α-ionone titer achieved was 700 mg/L [12].
4.2 Engineering carotenoid cleavage dioxygenase 1 with catalytic activity and substrate specificity
To a large extent, a lower catalytic activity of CCD1 would cause a decrease in carbon flux to product formations. Enzyme mining is still necessary to discover more CCD1s with higher catalytic activities. Zhang et al. [13] identified that the CCD1 from various plant sources significantly differed in catalytic efficiency and substrate specificity, and the β-carotene-producing E. coli strain overexpressing AtCCD1, Vitis vinifera CCD1(VvCCD1), and PhCCD1 achieved 0.1 mg/L, 13.0 mg/L, and 27.0 mg/L of β-ionone, respectively. This study first systematically compared the catalytic efficiencies of several plant-derived CCD1s and then identified that PhCCD1 had a higher catalytic efficiency for producing β-ionone. PhCCD1 was subsequently used for β-ionone biosynthesis in MCFs or via in vitro enzymatic reaction [15, 55, 56]. More sequence- or structure-based mining of CCD1 should be conducted in the protein database (e.g., UniProt, NCBI, and InterPro) to obtain more efficient C13-apocarotenoid biosynthesis modules and rationally guide the protein engineering process.
The loop regions formed in the substrate binding pocket were highly related to substrate selectivity and activity (Fig. 3). Based on the multiple protein sequence alignments of OfCCD1 with other identified CCD1s, Chen et al. [14] identified seven single mutation sites in OfCCD1 at the loop regions (K425). The OfCCD1 mutant library was introduced into E. coli, and the ε-carotene titer was evaluated for individual colonies. Two mutants on OfCCD1 (D424N and K425N, Fig. 3A) contributed to the improvement in α-ionone titer and the suppression of byproduct formation. The other mutants at the D424 site caused the change in substrate preference, lowering the formation of byproduct pseudoionone derived from the oxidative cleavage of lycopene. The results preliminarily validated that the residues at the loop region were highly related to OfCCD1’s substrate preference.
The α-helix of CCD1 is structurally conserved and plays an important role in interacting or penetrating regions with the cell membrane’s lipid layers where carotenoid substrates are often stored [57]. Enhancing the interaction or penetrating effect of CCD1 with the cell membrane may improve CCD1’s stability or activity, and further improve the production of C13-apocarotenoids. This hypothesis was partially supported by PhCCD1’s site-directed mutagenesis at the α-helix in S. cerevisiae [15]. Werner et al. [15] developed an in-house algorithm to guide the selection of the residues in the α1 helix of PhCCD1 and choosing structurally diversified amino acids for mutation. Next, the membrane-binding affinity and membrane penetration depth of each mutant were predicted using the positioning of proteins in membranes method [58]. The experimental validation of those mutants with the predicted, activity-improved parameters suggested that three mutations at the α1 helix K164 of PhCCD1 (Fig. 3B) were proved to improve enzyme activity, and the mutant K164L led to a ~ threefold increase in β-ionone titer. Modifying the α1 helix region of OfCCD1 also increased α-ionone production [14]. Chen et al. [14] swapped the first 25 amino acids of OfCCD1’s α-helix 1 with the first 28 amino acids of PhCCD1’s, and the hybrid mutant improved the production titer of the total C13-apocarotenoids by ~ 20%, possibly resulted from the reinforced membrane interaction or penetration. Thus, replacing CCD1’s α-helix with other non-CCD1-derived ones could enhance the enzyme subcellular localization efficiency (cell membrane in E. coli or membrane-based organelles in yeast) and improve CCD1’s stability and substrate accessibility.
Although protein engineering on CCD1 has improved C13-carotenoid biosynthesis in MCFs, a deeper understanding of the relationship between protein sequence and catalytic properties remains to be investigated. The reported studies on modifying the CCD1’s α-helix or loop regions still mostly rely on the in vivo characterization and using the product titer as the indicator. These mutations should be further titrated in vitro on their catalytic kinetics (i.e., Kcat, Km, Kcat/Km, and/or substrate/product inhibition) and enzyme stability.
4.3 Overexpression of CCDs in oleaginous microbial hosts for C13-carotenoids production
Oleaginous yeasts have been successfully developed for facilitating the production of hydrophobic carotenoids, such as carotene, lycopene, and squalene [59]. These molecules may be toxic to cells when accumulated to a large amount intracellularly, and storing them in lipid bodies or other subcellular structures in oleaginous yeasts could be an efficient natural solution when the produced titers reach a certain amount intracellularly [60]. Czajka et al. [11] integrated the genes coding the needed enzymes of the MVA and downstream β-carotene biosynthetic pathways into the genome of Y. lipolytica, and the resulting strain produced β-carotene at ~ 2.5 g/L in a 2-L bioreactor, which proved the potential of producing carotenoid using oleaginous yeasts. The authors further expressed the gene encoding OfCCD1 in this β-carotene-producing strain, which produced 380 mg/L of β-ionone.
Similarly, Lu et al. [56] performed a modular pathway engineering strategy on the MVA pathway to divert more acetyl-CoA flux to IPP/DMAPP in Y. lipolytica, and the titer of β-ionone reached 1 g/L in a 5-L bioreactor under the fed-batch operation. Next, the same group obtained 4 g/L of β-ionone titer in a shake flask by fermenting the engineered Y. lipolytica containing a PhCCD1 variant on food waste and sugarcane bagasse hydrolysates. This is the highest β-ionone titer reported thus far [61], but ~ 0.9 g/L of β-carotene was accumulated, suggesting further reinforcement of the PhCCD1-conducted step is required. Engineering oleaginous yeast as the microbial host enables the production of the carotenoid precursor at the gram-per-liter level. Oleaginous yeasts contain multiple membrane-based subcellular organelles (e.g., Endoplasmic Reticulum and Golgi), which provides more binding regions to facilitate the membrane interaction, subcellular localization, and substrate accessibility of CCD1, which suggests it is a more suitable host than E. coli to express CCD1 efficiently. Oleaginous yeast such as Rhodotorula toruloides naturally produces and accumulates a certain amount of γ-carotene and β-carotene in its lipid bodies [62,63,64]. In this regard, we believe that the applications of novel synthetic biology and metabolic engineering tools to modify the carotenogenic biosynthetic pathway of R. toruloides have enormous potential to overproduce C13-apocarotenoids.
4.4 Strengthening of substrate accessibility and channeling
Due to the intracellular self-aggregation, membrane-embedding, and liposoluble nature of carotenoids [57, 65], the substrate accessibility of CCD1 to the substrate is also highly essential. Ye et al. [66] constructed a fusion protein of PhCCD1 with an E. coli membrane-localizing protein (glycerol uptake facilitator [GlpF]), the engineered strain containing the GlpF-fused PhCCD1 produced β-ionone with an increase in ~ fourfold titer. In oleaginous yeasts, the produced hydrophobic carotenoids are often stored in lipid bodies [67], creating intracellular lipid droplet protein/peptide-fused CCD1 to enhance substrate accessibility could be feasible. Fusing CCD1 with an enzyme in the ionone biosynthetic pathway is an effective strategy to strengthen substrate channeling. Substrate channeling is defined here when a reaction intermediate generated from one enzyme is transferred to an adjacent enzyme that participates in an enzyme cascade reaction intracellularly without mixing enzymes in the bulk phase. Therefore, the overall reaction rate in the enzyme cascade may be increased [68]. Chen et al. [14] fused OfCCD1 at the C-terminal of lycopene ε-cyclase, a membrane-binding protein of converting lycopene to ε-carotene, but observed a decreased production of α-ionone, possibly caused by lower activity of lycopene ε-cyclase after fusion. Next, the authors systematically lengthened the glycine-serine linker. They found that the titer of α-ionone outperformed the control when the linker length increased to twenty amino acids (GGGGSGGGGSGGGGSGGGGS), and a ~ 10% improvement of α-ionone titer was obtained.
5 Perspective
The advancements in synthetic biology, metabolic engineering, protein engineering, and strain engineering have extensively promoted the microbial production of C13-apocarotenoids. Recently, the microbial production of β-carotene is reaching an industrial scale-up level (39.5 g/L) using an engineered Y. lipolytica [60], generating ample exploration space in MCFs for producing C13-apocarotenoids. In this study, Ma et al. [60] developed a structure-guided protein engineering strategy that yielded a variant that completely removed substrate inhibition while maintaining the enzymatic activity, and unlocked the constraint of converting lycopene to β-carotene. The substrate inhibition of CCD could also be a limiting factor in C13-apocarotenoid biosynthesis when a large amount of carotenoid substrates is provided for CCD, and in vitro experimental validation should have proceeded ahead of using this protein engineering strategy.
C13-apocarotenoids can be further derivatized enzymatically to other high-end aroma molecules. Chen et al. [69] developed a new-to-nature biosynthetic pathway by coupling OfCCD1 and bifunctional methyltransferase to convert lycopene to cis-α-irone, a valuable perfume-use molecule. Expanding the product spectrum of C13-apocarotenoids in MCFs is an appealing direction to be explored. For instance, converting β-ionone or α-ionone to hydroxylated products can be achieved using the targeted, protein-engineered cytochrome P450 monooxygenase [70]. In this way, the hydroxylated C13-apocarotenoids will be further glycosylated [71] or esterified [72] enzymatically to change the physicochemical properties or reduce their cytotoxicity [8], subsequently enhancing the production efficiency in MCFs.
6 Conclusion
In this review, we systematically discuss the natural and artifact-designed biosynthetic pathways to produce C13-apocarotenoids and the key catalytic module CCDs, which have been widely demonstrated to be the rate-limiting enzymes that directly control the conversion rate of carotenoids to the corresponding C13-apocarotenoid. The protein and metabolic engineering strategies developed centering on CCDs to enhance the oxidative cleavage of structurally diversified carotenoids are included in detail, and providing the outlook of the promising approaches to be further explored to unlock the potential of harnessing CCDs. Collectively, with the aid of the engineering strategies mentioned, the microbial biosynthesis of C13-apocarotenoids would be continuously reinforced to provide a sustainable solution for producing natural, customized C13-apocarotenoids molecules.
References
Walter MH, Floss DS, Strack D (2010) Apocarotenoids: hormones, mycorrhizal metabolites and aroma volatiles. Planta 232:1–17. https://doi.org/10.1007/s00425-010-1156-3
Schmidt K, Kunkel K, Szweda R et al (2013) Biotechnological production of norisoprenoid aroma compounds. In: Winterhalter P, Ebeler SE (eds) Carotenoid cleavage products. American Chemical Society, Washington
Belyaeva OV, Adams MK, Popov KM et al (2019) Generation of retinaldehyde for retinoic acid biosynthesis. Biomolecules 10:5. https://doi.org/10.3390/biom10010005
da Costa DP, Miranda-Filho KC (2020) The use of carotenoid pigments as food additives for aquatic organisms and their functional roles. Rev Aquacult 12:1567–1578. https://doi.org/10.1111/raq.12398
Sánchez AM, Winterhalter P (2013) Carotenoid cleavage products in saffron (Crocus sativus L). In: Winterhalter P, Ebeler SE (eds) Carotenoid cleavage products. American Chemical Society, Washington
Rodrigo MJ, Alquézar B, Alós E et al (2013) A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. J Exp Bot 64:4461–4478. https://doi.org/10.1093/jxb/ert260
Wu S, Ma X, Zhou A et al (2021) Establishment of strigolactone-producing bacterium-yeast consortium. Sci Adv 7:eabh4048. https://doi.org/10.1126/sciadv.abh4048
Cataldo VF, López J, Cárcamo M et al (2016) Chemical vs. biotechnological synthesis of C13-apocarotenoids: current methods, applications and perspectives. Appl Microbiol Biotechnol 100:5703–5718. https://doi.org/10.1007/s00253-016-7583-8
Dickinson AJ, Lehner K, Mi J et al (2019) β-Cyclocitral is a conserved root growth regulator. Proc Natl Acad Sci U S A 116:10563–10567. https://doi.org/10.1073/pnas.1821445116
Serra S (2015) Recent advances in the synthesis of carotenoid-derived flavours and fragrances. Molecules 20:12817–12840. https://doi.org/10.3390/molecules200712817
Czajka JJ, Nathenson JA, Benites VT et al (2018) Engineering the oleaginous yeast Yarrowia lipolytica to produce the aroma compound β-ionone. Microb Cell Fact 17:136. https://doi.org/10.1186/s12934-018-0984-x
Huang CN, Lim X, Ong L et al (2022) Mediating oxidative stress enhances α-ionone biosynthesis and strain robustness during process scaling up. Microb Cell Fact 21:246. https://doi.org/10.1186/s12934-022-01968-1
Zhang C, Chen X, Lindley ND et al (2018) A “plug-n-play” modular metabolic system for the production of apocarotenoids. Biotechnol Bioeng 115:174–183. https://doi.org/10.1002/bit.26462
Chen X, Shukal S, Zhang C (2019) Integrating enzyme and metabolic engineering tools for enhanced α-ionone production. J Agric Food Chem 67:13451–13459. https://doi.org/10.1021/acs.jafc.9b00860
Werner N, Ramirez-Sarmiento CA, Agosin E (2019) Protein engineering of carotenoid cleavage dioxygenases to optimize β-ionone biosynthesis in yeast cell factories. Food Chem 299:125089. https://doi.org/10.1016/j.foodchem.2019.125089
Paddon CJ, Westfall PJ, Pitera DJ et al (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496:528–532. https://doi.org/10.1038/nature12051
Ajikumar PK, Xiao WH, Tyo KE et al (2010) Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330:70–74. https://doi.org/10.1126/science.1191652
Ma X, Gözaydın G, Yang H et al (2020) Upcycling chitin-containing waste into organonitrogen chemicals via an integrated process. Proc Natl Acad Sci U S A 117:7719–7728. https://doi.org/10.1073/pnas.1919862117
Ma X, Liang H, Panda S et al (2022) C2 feedstock-based biomanufacturing of value-added chemicals. Curr Opin Biotechnol 73:240–245. https://doi.org/10.1016/j.copbio.2021.08.017
Tan N, Ong L, Shukal S et al (2023) High-yield biosynthesis of trans-nerolidol from sugar and glycerol. J Agric Food Chem 71:8479–8487. https://doi.org/10.1021/acs.jafc.3c01161
Wang C, Zhao S, Shao X et al (2019) Challenges and tackles in metabolic engineering for microbial production of carotenoids. Microb Cell Fact 18:55. https://doi.org/10.1186/s12934-019-1105-1
Fordjour E, Mensah EO, Hao Y et al (2022) Toward improved terpenoids biosynthesis: strategies to enhance the capabilities of cell factories. Bioresour Bioprocess 9:6. https://doi.org/10.1186/s40643-022-00493-8
Daletos G, Katsimpouras C, Stephanopoulos G (2020) Novel strategies and platforms for industrial isoprenoid engineering. Trends Biotechnol 38:811–822. https://doi.org/10.1016/j.tibtech.2020.03.009
Wang L, Liu Z, Jiang H et al (2021) Biotechnology advances in β-carotene production by microorganisms. Trends Food Sci Technol 111:322–332. https://doi.org/10.1016/j.tifs.2021.02.077
Gómez-Gómez L, Diretto G, Ahrazem O et al (2020) Determination of in vitro and in vivo activities of plant carotenoid cleavage oxygenases. Methods Mol Biol 2083:63–74. https://doi.org/10.1007/978-1-4939-9952-1_5
Vogel JT, Tan BC, McCarty DR et al (2008) The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J Biol Chem 283:11364–11373. https://doi.org/10.1074/jbc.M710106200
Jiang R, Chen X, Lian J et al (2019) Efficient production of Pseudoionone with multipathway engineering in Escherichia coli. J Appl Microbiol 126:1751–1760. https://doi.org/10.1111/jam.14245
Frusciante S, Diretto G, Bruno M et al (2014) Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc Natl Acad Sci U S A 111:12246–12251
Ma X, Liang H, Pan Q et al (2022) Optimization of the isopentenol utilization pathway for isoprenoid synthesis in Escherichia coli. J Agric Food Chem 70:3512–3520. https://doi.org/10.1021/acs.jafc.2c00014
Chatzivasileiou AO, Ward V, Edgar SM et al (2019) Two-step pathway for isoprenoid synthesis. Proc Natl Acad Sci U S A 116:506–511. https://doi.org/10.1073/pnas.1812935116
Zhang X, Wang X, Zhang Y et al (2023) Development of isopentenyl phosphate kinases and their application in terpenoid biosynthesis. Biotechnol Adv 64:108124. https://doi.org/10.1016/j.biotechadv.2023.108124
Clomburg JM, Qian S, Tan Z et al (2019) The isoprenoid alcohol pathway, a synthetic route for isoprenoid biosynthesis. Proc Natl Acad Sci U S A 116:12810–12815. https://doi.org/10.1073/pnas.1821004116
Luo Z, Liu N, Lazar Z et al (2020) Enhancing isoprenoid synthesis in Yarrowia lipolytica by expressing the isopentenol utilization pathway and modulating intracellular hydrophobicity. Metab Eng 61:344–351. https://doi.org/10.1016/j.ymben.2020.07.010
Fan X, Qi Z, Zhang X et al (2022) Combinatorial engineering of upper pathways and carotenoid cleavage dioxygenase in Escherichia coli for pseudoionone production. Appl Biochem Biotechnol 194:5977–5991. https://doi.org/10.1007/s12010-022-04078-1
Harrison PJ, Bugg TD (2014) Enzymology of the carotenoid cleavage dioxygenases: reaction mechanisms, inhibition and biochemical roles. Arch Biochem Biophys 544:105–111. https://doi.org/10.1016/j.abb.2013.10.005
Sun Z, Hans J, Walter MH et al (2008) Cloning and characterisation of a maize carotenoid cleavage dioxygenase (ZmCCD1) and its involvement in the biosynthesis of apocarotenoids with various roles in mutualistic and parasitic interactions. Planta 228:789–801. https://doi.org/10.1007/s00425-008-0781-6
Zhou Q, Li Q, Li P et al (2019) Carotenoid cleavage dioxygenases: identification, expression, and evolutionary analysis of this gene family in tobacco. Int J Mol Sci 20:5796. https://doi.org/10.3390/ijms20225796
Ohmiya A (2009) Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnol 26:351–358. https://doi.org/10.5511/plantbiotechnology.26.351
Mathieu S, Terrier N, Procureur J et al (2005) A carotenoid cleavage dioxygenase from Vitis vinifera L.: functional characterization and expression during grape berry development in relation to C13-norisoprenoid accumulation. J Exp Bot 56:2721–2731. https://doi.org/10.1093/jxb/eri265
Zhang B, Liu C, Wang Y et al (2015) Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol 206:1513–1526. https://doi.org/10.1111/nph.13335
Schwartz SH, Qin X, Zeevaart JA (2001) Characterization of a novel carotenoid cleavage dioxygenase from plants. J Biol Chem 276:25208–25211. https://doi.org/10.1074/jbc.M102146200
Simkin AJ, Underwood BA, Auldridge M et al (2004) Circadian regulation of the PhCCD1 carotenoid cleavage dioxygenase controls emission of beta-ionone, a fragrance volatile of petunia flowers. Plant Physiol 136:3504–3514. https://doi.org/10.1104/pp.104.049718
Huang FC, Horváth G, Molnár P et al (2009) Substrate promiscuity of RdCCD1, a carotenoid cleavage oxygenase from Rosa damascena. Phytochemistry 70:457–464. https://doi.org/10.1016/j.phytochem.2009.01.020
Baldermann S, Kato M, Kurosawa M et al (2010) Functional characterization of a carotenoid cleavage dioxygenase 1 and its relation to the carotenoid accumulation and volatile emission during the floral development of Osmanthus fragrans Lour. J Exp Bot 61:2967–2977. https://doi.org/10.1093/jxb/erq123
Schmidt H, Kurtzer R, Eisenreich W et al (2006) The carotenase AtCCD1 from arabidopsis thaliana is a dioxygenase. J Biol Chem 281:9845–9851. https://doi.org/10.1074/jbc.M511668200
Sui X, Kiser PD, Jv L et al (2013) Structural basis of carotenoid cleavage: from bacteria to mammals. Arch Biochem Biophys 539:203–213. https://doi.org/10.1016/j.abb.2013.06.012
Sharma D, Xue B (2021) Carotenoid cleavage dioxygenase 1 from Osmanthus fragrans. https://doi.org/10.2210/pdb7vud/pdb
Cui H, Wang Y, Qin S (2012) Genomewide analysis of carotenoid cleavage dioxygenases in unicellular and filamentous cyanobacteria. Comp Funct Genomics 2012:164690. https://doi.org/10.1155/2012/164690
Daruwalla A, Kiser PD (2020) Structural and mechanistic aspects of carotenoid cleavage dioxygenases (CCDs). Biochim Biophys Acta Mol Cell Biol Lipids 1865:158590. https://doi.org/10.1016/j.bbalip.2019.158590
Mathieu S, Bigey F, Procureur J et al (2007) Production of a recombinant carotenoid cleavage dioxygenase from grape and enzyme assay in water-miscible organic solvents. Biotechnol Lett 29:837–841. https://doi.org/10.1007/s10529-007-9315-8
Schilling M, Patett F, Schwab W et al (2007) Influence of solubility-enhancing fusion proteins and organic solvents on the in vitro biocatalytic performance of the carotenoid cleavage dioxygenase AtCCD1 in a micellar reaction system. Appl Microbiol Biotechnol 75:829–836. https://doi.org/10.1007/s00253-007-0878-z
Jeschek M, Gerngross D, Panke S (2017) Combinatorial pathway optimization for streamlined metabolic engineering. Curr Opin Biotechnol 47:142–151. https://doi.org/10.1016/j.copbio.2017.06.014
Ma X, Liang H, Cui X et al (2019) A standard for near-scarless plasmid construction using reusable DNA parts. Nat Commun 10:3294. https://doi.org/10.1038/s41467-019-11263-0
Zhang C, Seow VY, Chen X et al (2018) Multidimensional heuristic process for high-yield production of astaxanthin and fragrance molecules in Escherichia coli. Nat Commun 9:1858. https://doi.org/10.1038/s41467-018-04211-x
López J, Bustos D, Camilo C et al (2020) Engineering Saccharomyces cerevisiae for the overproduction of β-ionone and its precursor β-carotene. Front Bioeng Biotechnol 8:578793. https://doi.org/10.3389/fbioe.2020.578793
Lu Y, Yang Q, Lin Z et al (2020) A modular pathway engineering strategy for the high-level production of β-ionone in Yarrowia lipolytica. Microb Cell Fact 19:49. https://doi.org/10.1186/s12934-020-01309-0
Cerezo J, Zúñiga J, Bastida A et al (2013) Conformational changes of β-carotene and zeaxanthin immersed in a model membrane through atomistic molecular dynamics simulations. Phys Chem Chem Phys 15:6527–6538. https://doi.org/10.1039/c3cp43947j
Lomize MA, Pogozheva ID, Joo H et al (2012) OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 40:D370–D376. https://doi.org/10.1093/nar/gkr703
Liu HH, Ji XJ, Huang H (2015) Biotechnological applications of Yarrowia lipolytica: past, present and future. Biotechnol Adv 33:1522–1546. https://doi.org/10.1016/j.biotechadv.2015.07.010
Ma Y, Liu N, Greisen P et al (2022) Removal of lycopene substrate inhibition enables high carotenoid productivity in Yarrowia lipolytica. Nat Commun 13:572. https://doi.org/10.1038/s41467-022-28277-w
Chen S, Lu Y, Wang W et al (2022) Efficient production of the β-ionone aroma compound from organic waste hydrolysates using an engineered Yarrowia lipolytica strain. Front Microbiol 13:960558. https://doi.org/10.3389/fmicb.2022.960558
Park YK, Nicaud JM, Ledesma-Amaro R (2018) The engineering potential of Rhodosporidium toruloides as a workhorse for biotechnological applications. Trends Biotechnol 36:304–317. https://doi.org/10.1016/j.tibtech.2017.10.013
Yaegashi J, Kirby J, Ito M et al (2017) Rhodosporidium toruloides: a new platform organism for conversion of lignocellulose into terpene biofuels and bioproducts. Biotechnol Biofuels 10:241. https://doi.org/10.1186/s13068-017-0927-5
Liu F, Lu Z, Lu T et al (2023) Metabolic engineering of oleaginous yeast in the lipogenic phase enhances production of nervonic acid. Metab Eng 80:193–206. https://doi.org/10.1016/j.ymben.2023.10.001
Liu Y, Low ZJ, Ma X et al (2020) Using biopolymer bodies for encapsulation of hydrophobic products in bacterium. Metab Eng 61:206–214. https://doi.org/10.1016/j.ymben.2020.04.006
Ye L, Zhu X, Wu T et al (2018) Optimizing the localization of astaxanthin enzymes for improved productivity. Biotechnol Biofuels 11:278. https://doi.org/10.1186/s13068-018-1270-1
Ma T, Shi B, Ye Z et al (2019) Lipid engineering combined with systematic metabolic engineering of Saccharomyces cerevisiae for high-yield production of lycopene. Metab Eng 52:134–142. https://doi.org/10.1016/j.ymben.2018.11.009
Zhang YH (2011) Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol Adv 29:715–725. https://doi.org/10.1016/j.biotechadv.2011.05.020
Chen X, Esque J et al (2022) Total enzymatic synthesis of cis-α-irone from a simple carbon source. Nat Commun 13:7421. https://doi.org/10.1038/s41467-022-35232-2
Urlacher VB, Makhsumkhanov A, Schmid RD (2006) Biotransformation of beta-ionone by engineered cytochrome P450 BM-3. Appl Microbiol Biotechnol 70:53–59. https://doi.org/10.1007/s00253-005-0028-4
Sun G, Liao J, Kurze E et al (2023) Apocarotenoids are allosteric effectors of a dimeric plant glycosyltransferase involved in defense and lignin formation. New Phytol 238:2080–2098. https://doi.org/10.1111/nph.18875
Ziarani GM, Gholamzadeh P, Asadiatouei P et al (2015) The role of Pseudomonas cepacia lipase in the asymmetric synthesis of heterocyclic based compounds. J Mol Catal B Enzym 122:93–116. https://doi.org/10.1016/j.molcatb.2015.08.022
Acknowledgements
This work was supported by the Key R&D Project of China National Tobacco Corporation (110202202037), the Start-up fund of Shanghai Jiao Tong University (WH220415004), the Shanghai Collaborative Innovation Center of Agri-Seeds (ZXWH3150101/002). The partial materials of the schematic diagram in Figs. 1 and 4 are provided by Biorender. We also thank Dr. Xiaoqiang Ma, Dr. Vincent Fung Kin Yuen, and Dr. Dianqi Yang for their help in revising the manuscript.
Author information
Authors and Affiliations
Contributions
Jiawei Huang, Jiaying Lou, Jing Cao, Da Wu, and Jiale Wang analyzed the literature and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Neither ethical approval nor informed consent was required for this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, J., Lou, J., Cao, J. et al. C13-apocarotenoids biosynthesis with engineered microbes. Biotechnol Bioproc E 29, 601–612 (2024). https://doi.org/10.1007/s12257-024-00030-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-024-00030-8