Abstract
Over the last decade, the rho-associated kinases and several metastasis-associated microRNAs have emerged as important contributors of tumor invasion. However, despite prominence, our understanding of their involvement in the metastatic potential of Ewing Sarcoma (EWS) is incomplete. The expression profiles of ROCK1 or ROCK2 and miR-124-3p, miR-138-5p, miR-139-5p, miR-335-5p and miR-584-5p (all of which were previously predicted or validated to regulate these kinases) were evaluated through qRT-PCR and associated with clinical parameters. In vitro assays to evaluate colony formation and invasion/migration capacieties were performed on SK-ES-1 cells transfected with pre-miR mimics. ROCK1 expression was significantly reduced in EWS tissues, though there was no association with pathological parameters. miR-124-3p, miR-139-5p and miR-335-3p were also found significantly downregulated and positively correlated with ROCK1. Stratification indicated an association between lower levels of miR-139-5p and miR-584-5p with disease progression (p < 0.05), while reduced expression of the former and miR-124-3p were associated with reduced survival. In vitro miR-139-5p overexpression yielded inconsistent results: while mir-139-5p restoration significantly reduced invasion, the clonogenic capacity of cells was increased. Our study demonstrated that down-regulation of miR-124-3p, miR-139-5p and miR-584-5p are associated with disease progression in EWS and may serve as a risk assessment biomarkers though, as seen for mir-139-5p, their specific role remain to be elucidated for considering tailoring treatment options.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ewing’s sarcoma (EWS) is the second most common bone cancer in children and adolescents. This tumor is characterized by undifferentiated, small and round cells of neuroepithelial origin, that in 90% of patients, harbor a reciprocal chromosomal translocation that results in the expression of the oncogenic EWS/FLI1chimeric protein [1].
Standard treatment consists of pre- and postoperative chemotherapy, followed by surgery and/or radiotherapy. Despite this, in patients with metastatic disease, the 5-year survival diminishes from 60 to 70% to 10–40%. Moreover, the dissemination of tumor cells is such that even in patients without metastasis at diagnosis there may be circulating tumor cells [2].
For metastasis occurring, tumor cells must first detach from their tissue of origin, migrate and invade blood vessels, be able to adhere, proliferate and colonize a secondary site [3]. Among the regulators of these processes are ROCK1 and ROCK2, effectors of the Rho family of GTPases that are responsible for cytoskeleton control and whose dysregulation aggravates phenotypes associated with cancer and other diseases [4].
Thus, in view of the intrinsic metastatic nature of EWS, the present study aimed to verify the expression profiles of ROCK1 or ROCK2 in tumor samples and to evaluate metastasis-associated microRNAs predicted or validated to regulate these kinases (Online Resource 1).
Materials and Methods
Clinical Samples
EWS samples (n = 19) were obtained from surgeries at the Clinics University Hospital (Ribeirao Preto School of Medicine – University of Sao Paulo), performed from 2006 to 2015 (survival analysis was followed until June 2016). None of the patient received chemotherapy or radiotherapy prior to biopsy. The patients corresponded to 7 women and 12 men with a mean age at diagnosis of 14 years (range: 4–28 years old). Seven patients presented metastasis, and ten had relapse. Non-tumor bone tissues were obtained from malformation bone corrective surgery. All specimens were snap frozen in liquid nitrogen and stored at −80 °C for further analysis. Signed statement of informed consent was obtained from each participant or their parents/guardians. This study was approved by the local Ethics Committee that follows the Helsinki convention criteria (Comitê de Ética em Pesquisa da FFCLRP n° 43,619,215.9.0000.5407).
Cell Lines and Reagents
The EWS cell lines SK-ES-1 and RD-ES were purchased from Rio de Janeiro Cell Bank (BCRJ, Federal University of Rio de Janeiro, Brazil). MRC-5 (normal fibroblast) was obtained from the ATCC (American Type Culture Collection, Rockville, MD). Cells were cultivated with recommended media supplemented with 10% of fetal bovine serum (FBS), penicillin (100 U/mL) and streptomycin (100μg/mL) and kept at 37 °C in a humidified 5% CO2 incubator.
RNA Isolation, Reverse Transcription and Quantitative Real-Time PCR of mRNA and microRNA
Total RNA of tumor and non-tumor samples was extracted using TRIzol® Reagent (Invitrogen, Karlsruche, Germany) following the manufacturer’s protocol. The quality and quantity of the RNA was evaluated by a ND-1000 NanoDrop spectrophotometer (NanoDrop Technologies). cDNA was synthetized using the High Capacity kit (Applied Biosystems, MA, USA) according to the manufacturer’s instructions. For microRNA quantification, the complementary strands were obtained using microRNA-specific primers. qRT-PCRwas performed using Taqman® microRNA and gene assays [ROCK1 (Hs01127699-m1), ROCK2 (Hs00178154-m1), miR-124-3p (001182), miR-138-5p (002284), miR-139-5p (002289), miR-335-5p (000546) and miR-584-5p (001624)]according to the manufacturer’s protocol on a 7500 Real Time PCR System (Applied Biosystems, Waltham, MA, USA). As internal controls, small nuclear RNU6B and RNU48 were used to normalize the microRNAs level. GAPDH and GUSβ were used to normalize the gene levels. The MRC5 cell line was used as calibrator. Relative expression was calculated by 2-ΔΔCT analysis method.
Transfection of microRNA
TheSK-ES-1 cell line was transfected with pre-microRNA miR-139-5p (mirVanaTMmicroRNA mimic ID: MC11749, Ambion®) and control (mirVanaTMmicroRNA mimic negative control, Ambion®) using Lipofectamine® RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. microRNA transfection efficiency was monitored by qRT-PCR after 48 h.
Colony Formation Assay
Single cell suspensions of 500 cells were seeded in 6-well plates 24 h after the transfection and incubated at 37 °C for 10 days. Colonies were then washed, fixed with methanol and stained with Giemsa 3%. Only colonies with >50 cells were counted by direct visual inspection with a stereo microscope at 20× magnification. Assays were performed in triplicate.
Invasion Assay
Cell invasion was measured by invasion of the cells through Matrigel-coated Transwell inserts (Becton Dickinson & Co., NJ, USA). Cells were harvested after 24 h of transfection, re-suspended in serum-free medium and seeded on the top of coated invasion 8um pore size chambers (5 × 105 cell/insert).The bottom chamber was filled with media containing 10% of FBS. After 24 h of incubation, noninvasive cells were removed from the membrane upper surface with swabs. The invaded cells attached to the lower side of the membrane were fixed with 100% methanol and stained with Giemsa 3%. The membranes were removed from the inserts and placed on microscope slides with Entelan (Merk, NY, USA) and photographed at 20× magnification (ten random fields of each membrane). The invaded cells were counted with the ImageJ software(Schneider et al., 2012). Assays were performed in triplicate.
Boyden Migration Assay
Migration was assessed using a 48-wells Boyden Chamber containing a polycarbonate membrane with 8 μm pores (Neuroprobe Inc., MD, USA). Each membrane was coated with 5 mg/mL of fibronectin (Sigma-Aldrich, St. Louis, MO, USA) overnight before the assay. The bottom chambers were filled with FBS-free medium or medium containing FBS. Cell lines were harvested after 24 h of transfection, re-suspended in serum-free medium and seeded on the upper chambers (2.5 × 104 cells/well). After 24 h of incubation, non-migrating cells were scraped from the upper surface of the membrane. Cells on the lower surface were fixed with 100% methanol and stained with hematoxylin. The membrane was placed on microscope slides and photographed at 20× magnification (four random fields of each well). The invaded cells were counted with the ImageJ® software. Assays were performed in triplicate.
Western Blot
Total protein samples were extracted from transfected (control and pre-miR-139-5p) SK-ES-1 cells after 48 h with RIPA buffer (Thermo Scientific, CA, USA) according to manufacturer instructions. Heat-denatured protein samples (50 mg per lane) were resolved by 10% SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Amersham Pharmacia Biotech, Piscataway, NJ, EUA). The membranes were incubated for 1 h in TBS containing 0.1% of Tween 20 and 5% of blocker solution followed by incubation for 12 h at 4 °C with primary rabbit monoclonal anti-ROCK1 (Ab45171 at 1:500 dilution, Abcam, Cambridge, United Kingdom) and primary rabbit monoclonal anti-ROCK2 (Ab125025 at 1:10,000 dilution, Abcam) antibodies. The GAPDH expression level was measured as a loading control, using rabbit monoclonal anti-GAPDH antibody (Ab128915 at 1:10,000 dilution, Abcam) incubated for 1 h. The membrane was then incubated with goat anti-rabbit secondary antibody (Ab6721 at 1:5000 dilution, Abcam) for 1 h, and detected with the ECL Western Blotting Substrate Kit (Abcam, Cambridge, United Kingdom) and visualized in a ChemiDocBioimaging System (Bio-Rad, Hercules, California, EUA). Expression levels were quantified using ImageJ® software and normalized to loading control. Assays were performed in triplicate.
Statistical Analysis
The association between the following variables: age (< 14 years versus 14 years old); sex; EWS/FLI1 status; necrosis stage after chemotherapy – HUVOS levels 1 and 2 (less than 90% of necrotic areas) versus Huvos levels 3 and 4 (more than 90% necrotic areas); metastasis (presence versus absence); relapse (presence versus absence); disease progression (relapse/metastasis versus complete remission), death (alive versus deceased); and expression levels of each microRNA and genes was determined by Mann-Whitney tests. Survival analysis was carried out based on Log-Rank test represented on Kaplan-Meier curves using the median of the microRNA/gene expression observed as cutoff. Correlations between gene and miRNA expression were evaluated by Spearman’s test and presented as a heatmap using the online platform Plotly 2.0. The functional assays data were statistically analyzed by Student’s two-tailed t-test or One-Way Repeated Measures Analysis of Variance (ANOVA) followed by the Bonferroni Pairwise Multiple Comparison. All tests were carried out for 훼=0.05. All analyses were performed using the SPSS 21.0 software (SPSS Inc., IL, USA) and expressed as the mean ± standard deviation.
Results
ROCKs and ROCK-Associated microRNAs are Dysregulated in Ewing Sarcoma
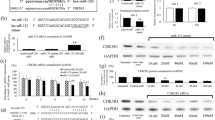
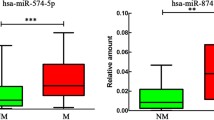
qRT-PCRwas performed on nineteen EWS samples and twelve non-tumor bone samples from paired-age individuals to evaluate the expression levels of ROCKs and ROCK-associated microRNAs. As seen in Fig. 1a, ROCK1 expression was significantly reduced in EWS tissues compared with the control.Among the selected set of microRNAs, three of them (miR-124-3p, miR-139-5p and miR-335-3p) were found significantly downregulated compared to the control.
Analysis of ROCK-1 and -2 and microRNAs related expression in Ewing Sarcoma. a Dysregulation of ROCK1, miR-124a-3p, miR-139-5p and miR-335-5p expression in EWS tissue and cell lines compared to non-tumor tissue. b Spearman Correlation showed positive correlations between ROCK1 and the following miRNAs: miR-124a-3p, miR-139-5p and miR-355-5pc. Downregulation of miR-139-5p associated with tumor progression, relapse and shorter event-free survival. d Downregulation of miR-584-5p associated with tumor progression and relapse. e Lower expression of miR-124-3p associated with shorter event-free survival
Similar expression patterns were observed for both EWS cell lines with decreased expression of ROCK1, miR124a-3p and miR139-5p. Conversely, no significant alterations were found to ROCK2, miR138-5p and miR584-5p (Fig. 1a).
miR-124a-3p, miR139-5p and miR-584-5p Expression Levels are Associated with EWS Progression
Associations between ROCKs and microRNA expression in EWS samples and clinical features were also investigated (Table 1). Contrary to the expected, our results showed a significant positive correlation between ROCK1 and miR-124a-3p, miR139-5p and miR-584-5p expression (Fig. 1b). Moreover, lower miR-139-5p and miR-584-5p expression was found in tumors from patients who presented relapse after treatment or any signals of tumor progression such as metastasis or death (Fig. 1c, d). Event-free survival analysis also showed that lower expression levels of miR-139-5p and miR-124a-3pare also correlated with worse prognosis (Fig. 1c, e).
Restoration of miR-139-5p Stimulates EWS Clonogenic Capacity but Inhibits Invasion and Migration
Once miR-139-5p was differentially expressed and related with tumor progression, we selected it for further functional analysis. In order to elucidate its contribution on cell growth and metastasis, we induced miR-139-5p expression in SK-ES-1 cell line. Our results suggested that this microRNA affects cell growth and invasion in a different manner. Overexpression of miR-139-5p leads to an increase on colony formation while reduces the cell line invasion capacity, and such results were obtained from 3 independent experiments (Fig. 2a, b). Moreover, miR-139-5p seems to suppress chemoattractive-independent migration but does not affect this process when stimulated by FBS (Fig. 2c).Also, despite the predicted positive correlation, after miR-139-5p overexpression, ROCK1 and ROCK2 levels were not significantly altered (Fig. 2d).
Effects of miR-139-5p overexpression in SK-ES-1 cell line. a Overexpression of miR-139-5p stimulates the clonogenic capacity in vitro. b Restoration of miR-139-5p expression suppresses SK-ES-1 cells invasion through Matrigel towards chemoattractant. c In the absence of chemoattractant (FBS), miR-139-5p overexpression decreases SK-ES-1 cells migration ability. d Forced miR-139-5p expression does not alter ROCK1 and ROCK2 protein levels
Discussion
The high metastatic potential of EWS remains a major challenge in clinics and, despite aggressive treatment, 80% of patients with metastatic disease die due to cancer progression [3].
The Rho-associated kinases, ROCK1 and ROCK2, frequently associated with the regulation of invasion through their ability to control cytoskeleton, cell adhesion and motility, have frequently been described as dysregulated in cancer over recent years [5]. Both kinases were found significantly increased and associated with poor differentiation, metastasis and short survival in primary bladder tumors [6]. Similarly, ROCK1 has been described as overexpressed in metastatic breast cancer and related with overall patient survival [7]. ROCK2, was also found hyperexpressed in intrahepatic metastasis [8]. Nonetheless, ROCK involvement in EWS remains elusive.
Our results showed downregulation of ROCK1 in EWS samples. Similar results were previously observed in vulvar cancer, in which ROCK1 levels were also related with low survival rates [9]. Comparatively, ROCK1 was described as poorly expressed in invasive human melanomas and cutaneous metastases [10], suggesting a tumor dependent participation of Rho-associated kinases in cancer development. More importantly, the absence of associations between either ROCK1 or ROCK2 expression with clinical features or their expression pattern itself do not obligatorily reflect their activity, since they are controlled by several positive and negative regulators and that mainly occur on the posttranslational level [11].
Thus, in our study, we also investigated the expression of five microRNAs (miR-124a-3p, miR-138-5p, miR-139-5p, miR-335-5p and miR-584-5p) already validated to target ROCK1 or ROCK2. Our results showed that among them, miR-124a-3p, miR-139-5p and miR584-5p were also found downregulated EWS tumor samples.
Lower levels of miR-124a-3pwere also related with shorter event-free survival in our cohort. In glioma samples, this microRNA was also underexpressed and negatively correlated with malignancy grade [12]. Dysregulation of miR-124a-3p has been reported in liver and cervical cancer, as well as medulloblastoma and acute leukemia [13,14,15,16]. Several studies showed that miR-124a-3prestoration inhibits cell proliferation and invasion in vitro [17, 18]. Mostly, the tumor suppressor effects of this microRNA were explained by its capacity to target ROCK1 [18, 19]. Nonetheless, our results showed a positive correlation between these genes, discouraging further analysis.
Even though there are few articles in the literature, miR-584-5p anti-invasive properties have also been associated with ROCK1 [20,21,22]. The expression of this microRNA was found downregulated in renal cell and papillary thyroid carcinoma cell lines [20, 21]. Additionally, Xue et al. [22] reported higher postoperative survival in glioma patients with high miR-584-5p expression. Those results are in agreement with our findings in which low miR-584-5pexpression levels were associated with tumor progression (metastasis, relapse or deceased). Nonetheless, we were unable to find any correlations between this microRNA and its predicted target in EWS samples.
In parallel, miR-139-5ppresented the most remarkable results. Low levels of this microRNA were not only evinced in EWS tumor compared with non-neoplastic tissue, but also associated with tumor progression and reduced event-free survival. miR-139-5p displays multifunction in human tumorigenesis, being described as relevant in several tumor types and significantly downregulated in breast, and laryngeal carcinomas [23,24,25,26]. Moreover, this microRNA was positively related with overall survival in colorectal cancer [25] and described as a potent tumor suppressor in acute myeloid leukemia [27]. In vitro, we showed that restoration of miR-139-5pwas able to impair EWS migration and invasion. Several reports have indicated cancer metastasis as the most affected process by miR-139-5p. Its forced expression leads to reduced rates of invasion and migration in colorectal cancer [25], in laryngeal and hepatocellular carcinomas [24]. However, its involvement in ROCKs regulation might be variable. On hepatocellular carcinoma, miR-139-5p suppresses metastasis by downregulating ROCK2 [8]. Our data showed a positive correlation between miR-139-5p and ROCK1 but not with ROCK2, nonetheless these protein levels were not altered after miR-139-5p overexpression, suggesting that other targets must be implicated on miR-139-stimulated invasion reduction. In gastric cancer, for instance, miR-139-5p downregulation is responsible for the increased expression of its target CXCR4, a transmembrane G-protein-coupled receptor also associated with cell invasion and metastasis [28]. On colorectal cancer, AMFR and NOTCH1 were suppressed by miR-139-5p reducing cell invasion [25]. Moreover, in non-small cell lung cancer, ectopic expression of miR-139-5p inhibited matrix metalloproteinases (MMP)-7 and −9, but also cyclin D1 and BCL2, which resulted on tumor growth inhibition and apoptosis [26]. Conflictingly, our results demonstrated that miR-139-5p expression, instead of inhibiting, stimulated the clonogenic capacity of cells.
One of the most challenging attributes of cancer is the extreme intratumoral heterogeneity at genomic, epigenomic and proteomic levels, which is also a common feature of EWS [29]. This heterogeneity might in turn, be reflected in derivative cell lines. In fact, side populations (SP) with stem cell characteristics cells have been identified in several tumor cell lines including EWS. Komuro et al., [30] demonstrated that SP cells constituted up to 5.5% of the cells in 5 EWS cell lines. The SK-ES-1 cell lines is believed to possess 1.2% of such cells and functional analysis characterized them as the more clonogenic and invasive within the culture [31].
Tumor plasticity also implies the coexistence of several distinct cell types, including highly proliferative cells and highly invasive cells. Nonetheless, proliferation and invasion seem to be mutually incompatible processes, and were shown to involve different signaling pathways [32]. Experimental data shows that different cell clones behave differently under different environmental conditions: while the proliferative phenotype exhibits the highest performance rate under a normal or high resource level, the invasive phenotype shows the highest success under low levels of nutrients and oxygen [33].
In the absence of FBS cells with restored expression of mir-139-5p lost motility and invaded through matrigel. No alteration on cell movement was observed in the presence of the supplement. Nonetheless, whether our contrasting results are a consequence of different subclones within the culture or whether mir-139-5p is part of the genetic makeup that influences through its innumerous targets a phenotypic proliferation-invasion dichotomy remains to be unveiled in detail.
References
Gonzales EHH, Betancourt GM, Martinez OQ, Cabezaz IH (2013) Sarcoma de Ewing. AMC 17:n5:623–640
Rodriguez-Galindo C, Spunt SL, Pappo AS (2003) Treatment of Ewing sarcoma family of tumors: current status and outlook for the future. Med Pediatr Oncol 40:276–287
Kim JY, Nam JK, Lee SA, Lee MS, Cho SK, Park ZY et al (2011) Proteasome inhibition causes epithelial-mesenchymal transition upon TM4SF5 expression. J Cell Biochem 112:782–792
Tseliou M, Al-Qahtani A, Alarifi S, Alkahtani SH, Stournaras C, Sourvinos G (2016) The role of RhoA, RhoB and RhoCGTPases in cell morphology, proliferation and migration in human cytomegalovirus (HCMV) infected glioblastoma cells. Cell Physiol Biochem 38:94–109
Amano M, Nakayama M, Kaibuchi K (2010) Rho-kinase/ROCK. A key regulator of the cytoskeleton and cell polarity. Cytoskeleton (Hoboken) 67:545–554
Kamai T, Tsujii T, Arai K, Takagi K, Asami H, Ito Y et al (2003) Significant association of rho/ROCK pathway with invasion and metastasis of bladder cancer. Clin Cancer Res 9:2632–2641
Bottino J, Gelaleti GB, Maschio LB, Jardim-Perassi BV, de Campos Zuccari DA (2014) Immunoexpression of ROCK-1 and MMP-9 as prognostic markers in breast cancer. Acta Histochem 116:1367–1373
Wong CC, Wong CM, Tung EK, SL A, Lee JM, Poon RT et al (2011) The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating rho-kinase 2. Gastroenterology 140:322–331
Akagi EM, Lavorato-Rocha AM (2014) Maia BeM, Rodrigues IS, Carvalho KC, Stiepcich MM et al. ROCK1 as a novel prognostic marker in vulvar cancer. BMC Cancer 14:822
Raimo M, Orso F, Grassi E, Cimino D, Penna E, De Pittà C et al (2016) miR-146a exerts differential effects on melanoma growth and Metastatization. Mol Cancer Res 14:548–562
Loirand G (2015) Rho kinases in health and disease: from basic science to translational research. Pharmacol Rev 67:1074–1095
SH L, Jiang XJ, Xiao GL, Liu DY, Yuan XR (2014) miR-124a restoration inhibits glioma cell proliferation and invasion by suppressing IQGAP1 and β-catenin. Oncol Rep 32:2104–2110
Agirre X, Vilas-Zornoza A, Jiménez-Velasco A, Martin-Subero JI, Cordeu L, Gárate L et al (2009) Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res 69:4443–4453
Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J (2010) miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 31:766–776
Pierson J, Hostager B, Fan R, Vibhakar R (2008) Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neuro-Oncol 90:1–7
Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA et al (2010) Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer 9:167
Chen X, He D, Dong XD, Dong F, Wang J, Wang L et al (2013) MicroRNA-124a is epigenetically regulated and acts as a tumor suppressor by controlling multiple targets in uveal melanoma. Invest Ophthalmol Vis Sci 54:2248–2256
CB H, Li QL, JF H, Zhang Q, Xie JP (2014) Deng.miR-124 inhibits growth and invasion of gastric cancer by targeting ROCK1. Asian Pac J Cancer Prev 15:6543–6546
An L, Liu Y, Wu A, Guan Y (2013) microRNA-124 inhibits migration and invasion by down-regulating ROCK1 in glioma. PLoS One 8:e69478
Ueno K, Hirata H, Shahryari V, Chen Y, Zaman MS, Singh K et al (2011) Tumour suppressor microRNA-584 directly targets oncogene Rock-1 and decreases invasion ability in human clear cell renal cell carcinoma. Br J Cancer 104:308–315
Xiang J, Wu Y, Li DS, Wang ZY, Shen Q, Sun TQ et al (2015) miR-584 suppresses invasion and cell migration of thyroid carcinoma by regulating the target oncogene ROCK1. Oncol Res Treat 38:436–440
Xue H, Guo X, Han X, Yan S, Zhang J, Xu S et al (2016) MicroRNA-584-3p, a novel tumor suppressor and prognostic marker, reduces the migration and invasion of human glioma cells by targeting hypoxia-induced ROCK1. Oncotarget 7:4785–4805
Hua W, Sa KD, Zhang X, Jia LT, Zhao J, Yang AG et al (2015) MicroRNA-139 suppresses proliferation in luminal type breast cancer cells by targeting topoisomerase II alpha. Biochem Biophys Res Commun 463:1077–1083
Luo HN, Wang ZH, Sheng Y, Zhang Q, Yan J, Hou J et al (2014) MiR-139 targets CXCR4 and inhibits the proliferation and metastasis of laryngeal squamous carcinoma cells. Med Oncol 31:789
Song M, Yin Y, Zhang J, Zhang B, Bian Z, Quan C et al (2014) MiR-139-5p inhibits migration and invasion of colorectal cancer by downregulating AMFR and NOTCH1. Protein Cell 5:851–861
Sun C, Sang M, Li S, Sun X, Yang C, Xi Y et al (2015) Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated with down-regulation of c-met. Oncotarget 6:39756–39792
Krowiorz K, Ruschmann J, Lai C, Ngom M, Maetzig T, Martins V et al (2016) MiR-139-5p is a potent tumor suppressor in adult acute myeloid leukemia. Blood Cancer J 6:e508
Bao W, Fu HJ, Xie QS, Wang L, Zhang R, Guo ZY et al. HER2 interacts with CD44 to up-regulate CXCR4 via epigenetic silencing of microRNA-139 in gastric cancer cells. Gastroenterology. 2011; 141:2076–2087.e2076
Bühnemann C, Li S, Yu H, Branford WH, Schäfer KL, Llombart-Bosch A et al (2014) Quantification of the heterogeneity of prognostic cellular biomarkers in ewing sarcoma using automated image and random survival forest analysis. PLoS One 9:e107105
Komuro H, Saihara R, Shinya M, Takita J, Kaneko S, Kaneko M et al (2007) Identification of side population cells (stem-like cell population) in pediatric solid tumor cell lines. J Pediatr Surg 42:2040–2045
Yang M, Zhang R, Yan M, Ye Z, Liang W, Luo Z (2010) Detection and characterization of side population in Ewing's sarcoma SK-ES-1 cells in vitro. Biochem Biophys Res Commun 391:1062–1066
Gao CF, Xie Q, Su YL, Koeman J, Khoo SK, Gustafson M et al (2005) Proliferation and invasion: plasticity in tumor cells. Proc Natl Acad Sci USA 102:10528–10533
Hecht I, Natan S, Zaritsky A, Levine H, Tsarfaty I, Ben-Jacob E (2015) The motility-proliferation-metabolism interplay during metastatic invasion. Sci Rep 5:13538
Acknowledgments
FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo): Grant 2014/03877-3 and LEAD and GMV fellowships, 2014/07117-3 and 2014/07118-0.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
None declared.
Electronic supplementary material
Online Resource 1
(DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Roberto, G.M., Delsin, L.E.A., Vieira, G.M. et al. ROCK1-PredictedmicroRNAs Dysregulation Contributes to Tumor Progression in Ewing Sarcoma. Pathol. Oncol. Res. 26, 133–139 (2020). https://doi.org/10.1007/s12253-017-0374-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-017-0374-4