Abstract
Colorectal sessile serrated adenomas (SSA) are hypothesized to be precursor lesions of an alternative, serrated pathway of colorectal cancer, abundant in genes with aberrant promoter DNA hypermethylation. In our present pilot study, we explored DNA methylation profiles and examined selected gene mutations in SSA. Biopsy samples from patients undergoing screening colonoscopy were obtained during endoscopic examination. After DNA isolation and quality analysis, SSAs (n = 4) and healthy controls (n = 5) were chosen for further analysis. DNA methylation status of 96 candidate genes was screened by q(RT)PCR using Methyl-Profiler PCR array system. Amplicons for 12 gene mutations were sequenced by GS Junior Instrument using ligated and barcoded adaptors. Analysis of DNA methylation revealed 9 hypermethylated genes in both normal and SSA samples. 12 genes (CALCA, DKK2, GALR2, OPCML, PCDH10, SFRP1, SFRP2, SLIT3, SST, TAC1, VIM, WIF1) were hypermethylated in all SSAs and 2 additional genes (BNC1 and PDLIM4) were hypermethylated in 3 out of 4 SSAs, but in none of the normal samples. 2 SSAs exhibited BRAF mutation and synchronous MLH1 hypermethylation and were microsatellite instable by immunohistochemical analysis. Our combined mutation and DNA methylation analysis revealed that there is a common DNA methylation signature present in pre-neoplastic SSAs. This study advocates for the use of DNA methylation as a potential biomarker for the detection of SSA; however, further investigation is needed to better characterize the molecular background of these newly recognized colorectal lesions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal serrated polyps have become an area of intense focus for gastroenterologists and pathologists over the past several years [1]. Serrated polyps are subdivided into three histological subtypes: sessile serrated adenomas (SSA), traditional serrated adenomas (TSA), and hyperplastic polyps (HP). Whereas the most frequent subtype, HP is regarded as self-limiting and non-neoplastic; SSA and TSA are considered to be pre-neoplastic and hypothesized to be precursor lesions of an alternative pathway leading to colorectal cancer (CRC) [2]. As compared to the classical adenoma-carcinoma pathway driven and characterized by several genetic mutations [3], serrated pathway arises from distinct pre-neoplastic lesions, and it is abundant in genes with aberrant promoter hypermethylation, and termed CpG island methylator phenotype (CIMP) [2]. DNA hypermethylation in the promoter region can lead through several mechanisms to down-regulation of the affected genes and contribute to colorectal carcinogenesis [4]. Its prominent role in colorectal carcinogenesis has also been utilized as a non-invasive prescreening method for CRC in clinical practice, as it can be detected both in colonic tissue and plasma [5, 6]. SSAs are histologically characterized by a sawtoothed growth pattern, horizontal crypt extensions (inverted T- or L-shaped) at the crypt bases and inverted crypts lying on the mucosal muscle layer (Supplementary Fig. 1) [2]. Patients with SSAs have an increased risk for developing interval CRCs [7, 8] as they are frequently missed by screening colonoscopy due to their flat or sessile morphology and pale color, making them similar to normal colonic mucosa [2]. As colonoscopy is the gold standard method for CRC screening [9], the above observations explain the relatively lower effectiveness of colonoscopy against right-sided CRC (where SSAs tend to be the precursor lesion more commonly) as compared to left-sided CRC [10]. Serrated polyps have also been shown to be poorly detected by fecal occult blood test [11], another important CRC screening method. These observations underline the importance to find alternative methods to improve the detection of serrated polyps, and consequently to optimize CRC screening. Given the key regulatory role of DNA methylation in serrated pathway [2], one can speculate that DNA methylation might be a good biomarker for the early detection of serrated lesions. So far many genes have been examined for DNA hypermethylation in serrated polyps [12–21], but only few studies have focused on multiple genes [22, 23].
The role of genetic and epigenetic alterations in the pathogenesis of serrated adenomas is not fully understood. To further explore this issue, we aimed to analyze promoter DNA methylation status of 96 genes in parallel with 12 frequently described gene mutations (APC, BRAF, CTNNB1, EGFR, FBXW7, KRAS, MSH6, NRAS, PIK3CA, SMAD2, SMAD4, TP53) in serrated polyps.
Materials and Methods
Patient Selection and Ethical Considerations
The study was conducted according to the declaration of Helsinki and approved by local ethics committee and government authorities (Regional and Institutional Committee of Science and Research Ethics (TUKEB) Nr.: 69/2008). All subjects provided written informed consent before entering the study, and anonymity was maintained by tracing patients through their clinical history number. Detailed medical history was taken and thorough physical examinations were performed for all participants. The study population encompassed all patients who underwent colonoscopy between January 1, 2011 and December 31, 2012 at the Endoscopy Unit of our Department. Pathological records were searched for “serrated polyps” in the archive of the 1st Department of Pathology and Experimental Cancer Research, Semmelweis University.
DNA Isolation and Sample Selection
DNA was isolated using High Pure PCR Template Preparation Kit (Roche Diagnostics, Penzberg, Germany) according to the manufacturer’s instructions as previously described [24]. As further experiments required high-purity DNA, quality check followed (A260/280 above 1.8 and A260/230 above 1.7) with NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Ultimately 4 SSAs, 1 TSA and 5 samples from normal colonic mucosa were chosen for further analysis. Clinical data is shown in Table 1.
DNA Methylation Analysis
DNA methylation of 96 genes were evaluated (Supplementary Table 1) using Methyl-Profiler (EpiTect Methyl qPCR Array System, Qiagen, Hilden, Germany) as described previously [24]. This bisulfite-free DNA methylation analysis is based on the detection of remaining DNA input after cleavage with restriction enzyme digestion followed by quantitative PCR (Supplementary Fig. 2) [25]. Methylation-sensitive restriction enzyme selectively digests the unmethylated DNA, whereas methylation-dependent restriction enzyme cleaves the methylated DNA (Supplementary Fig. 3). Treatment with both enzymes serves as a control for how much DNA is amplified in the assay (Supplementary Fig. 3) [24, 25]. Following enzyme digestion, samples were analyzed by fluorescence-based, quantitative PCR (Supplementary Fig. 2) using LightCycler 480 (Roche) instrument [24]. After the cycling program completed, CT values were copied into an Microsoft Office Excel (Microsoft, Redmond, WA, USA) file provided by the manufacturer, and ΔCT analysis was performed. The threshold for DNA hypermethylation was set at 15% [24].
Mutation Analysis
Mutation hotspots of 12 frequently described gene mutations in CRC (APC, BRAF, CTNNB1, EGFR, FBXW7, KRAS, MSH6, NRAS, PIK3CA, SMAD2, SMAD4, TP53) were amplified by own designed PCR primers [26]. Amplicons were sequenced with a GS Junior Instrument (Roche). In the course of library preparation, rapid Library Molecular Identifier (RL_MID) adaptors were ligated to the PCR products. After ligation, quality of PCR libraries was assessed with the High Sensitivity DNA Chip on Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). Emulsion PCR amplification of the amplicon libraries was performed using the Lib-L. emPCR Kit (Roche), with 2 DNA molecules per bead ratio following the manufacturer’s instructions. GS Junior Titanium Sequencing Kit (Roche) was used for bead enrichment and sequencing steps with the method described in the Sequencing Method Manual, GS FLX Titanium Series. Variants were identified with the Amplicon Variant Analyzer software.
Immunohistochemical Analysis
Immunohistochemical analysis was performed as described previously [24, 27]. Briefly, following dewaxing and rehydration, endogenous peroxidase activity was inhibited with 2.5% hydrogen peroxide in methanol for 30 min, then antigen retrieval was performed in an electric pressure cooker for 40 min in TRIS-EDTA buffer (pH 9). After blocking in 1% bovine serum albumin, slides were incubated overnight at 58 °C with monoclonal antibodies for MLH1 (1:80) (Santa Cruz Biotechnology, Dallas, TX, USA) and SFRP1 (1:150, Abcam, Cambridge, UK). After antibody detection (HISTOLS-MR-T kit, Hisztopatológia Kft, Pécs, Hungary) and visualization (Dako Cytomation, Glostrup, Denmark), slides were counterstained with haematoxylin, dehydrated with xylol, and mounted. After digital archiving (Pannoramic Flash 250 instrument, 3DHISTECH Ltd., Budapest, Hungary), stainings were evaluated with a Pannoramic Viewer digital microscope (software version 1.15; 3DHISTECH) by an experienced researcher (GV).
Results
DNA Methylation Analysis of Serrated Polyps
Analysis of DNA methylation revealed 9 hypermethylated genes (BAGE, CCNA1, H19, MAGEA1, MGX1, PTGIS, RUNX3, SPARC, UGT1A1) in both normal and SSA samples (data not shown). 12 genes (CALCA, DKK2, GALR2, OPCML, PCDH10, SFRP1, SFRP2, SLIT3, SST, TAC1, VIM, WIF1) were hypermethylated in all SSAs and 2 additional genes (BNC1 and PDLIM4) were hypermethylated in 3 out of 4 SSAs (Table 2), but in none of the normal samples. Two SSAs (SSA 1 and SSA 2) had hypermethylation in MLH1, these samples were also BRAF mutant and microsatellite instable (see later). Two additional genes (APC and MGMT) were hypermethylated in one SSA (SSA 4). TSA showed hypermethylation for only two genes CALCA and SST, which the only two genes that were hypermethylated in all serrated polyps (Table 2).
Mutation Analysis of Serrated Polyps
Mutation analysis of our samples showed that those two SSAs (SSA 1 and SSA 2), which had also MLH1 hypermethylation (Table 2) and were microsatellite instable (see later), exhibited only BRAF mutation (Table 3). SSA 4, which also had the most hypermethylated genes (Table 2), had three mutations (APC, FBXW7, TP53) and SSA 4, similarly to the TSA sample, did not exhibit any mutations (Table 3). Of note, no KRAS mutation was detected (Table 3).
Immunohistochemical Analyses of MLH1 and SFRP1
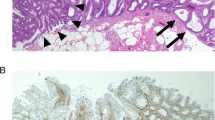
Protein expression for MLH1 was analyzed by immunohistochemistry to assess microsatellite status. Retained MLH1 protein expression (Fig. 1a) translates to proficient mismatch repair system and microsatellite stable status, whereas loss of MLH1 protein expression (Fig. 1b) corresponds to deficient mismatch repair system and microsatellite instable status. In SSA 1 and SSA 2, that also exhibited MLH1 methylation (Table 2) and BRAF mutation (Table 3), MLH1 protein was absent (Fig. 1b), so these samples were declared microsatellite instable. The remaining samples (SSA3, SSA4, TSA and the normal samples) retained MLH1 staining (Fig. 1a), and were considered to be microsatellite stable.
SFRP1, one of the genes that were hypermethylated in all SSA samples, was chosen to be analyzed at protein level by immunohistochemistry. Accordingly, SFRP1 protein expression seen in normal colonic mucosa samples (Fig. 1c) was reduced in SSA samples (Fig. 1d).
Discussion
Our combined mutation and methylation analysis showed that there is a common DNA methylation signature present in pre-neoplastic SSAs as opposed to TSA and normal controls. These findings are in line with our previous study [24], where we showed that pre-neoplastic lesions and CRCs arising in the classical adenoma-carcinoma pathway have a characteristic DNA methylation profile, as well. Interestingly, 6 genes (BNC1, SFRP1, SFRP2, SLIT3, SST, WIF1) were found to be frequently hypermethylated not only in CRC, but in both pre-neoplastic traditional adenomas and sessile serrated lesions, that warrants further investigation with larger sample size. As serrated polyps have recently been shown to be poorly detected by fecal occult blood test [11], these hypermethylated genes could be exploited as novel stool or blood biomarkers for the non-invasive detection of these polyps.
Previous studies examining DNA methylation in serrated polyps have either analyzed a single gene [16, 18, 20, 21] or a few well-described (usually CIMP) genes [12–15, 17, 19, 22, 23]. Compared to these previous works, we managed to identify some additional candidate hypermethylated genes.
Majority of earlier studies evaluated BRAF and KRAS mutation status, as BRAF mutation was found to be a characteristic for SSA and KRAS mutation for TSA [14], although the specificity and the sensitivity is very low. In addition to KRAS and BRAF, we analyzed 10 other mutations that had been frequently described in the classical adenoma-carcinoma pathway. Although due to the small sample size we cannot draw firm conclusions from our results, it seems that these examined additional genetic mutations are scarce in serrated carcinogenesis, but this should be further examined in subsequent studies.
Similarly to earlier observations, we found that MLH1 hypermethylation leads to loss of MLH1 expression at protein level. It was recently shown that promoter methylation and loss of protein expression are not parallel processes, and MLH1 hypermethylation is an earlier molecular event that occurs also in HP, whereas loss of protein expression is a much later step [28].
There are several limitations to our study. Samples were collected in a single center, and only pre-neoplastic serrated polyps were included. The major drawback of our study is its relatively small sample size. However, in contrast to earlier molecular studies on serrated polyps where mostly only a single or a few genes were examined, we followed a different approach and analyzed the genetic and epigenetic alterations of more than 100 genes. It is also highly debatable, that we included only a single TSA into our study, however as it was shown in a recent, large cohort study from the Netherlands [29], TSA is an extremely rare type of serrated polyps, even in internationally acknowledged, expert centers. In this well-described cohort, among 744 serrated polyps, only one lesion was diagnosed as a TSA, comprising 0.1% of all colorectal polyps. During our study period 2 TSAs were identified; however, the other TSA was excluded from the study during the preanalytical phase due to inappropriate quality of the isolated DNA. We still decided to include a single TSA because of its vastly different mutation and methylation profile, and to stimulate further molecular studies to support or disprove our observations on this enigmatic polyp.
In conclusion, we demonstrated that in line with previous results, DNA methylation is a frequent event in SSAs and several novel genes have been described for the first time. We hope that our results can provide data for subsequent studies and inspire further research on this topic, including genome-wide screening of DNA methylation [30] in serrated polyps. This would provide important insights into the pathogenesis of serrated polyps and subsequent cancers, and also yield potential new biomarkers that could be used for the better detection of these lesions, which could ultimately lead to improved CRC screening and better management of this common, but preventable disease.
References
East JE, Vieth M, Rex DK (2015) Serrated lesions in colorectal cancer screening: detection, resection, pathology and surveillance. Gut 64:991–1000
Patai AV, Molnár B, Tulassay Z, Sipos F (2013) The serrated pathway: the alternative route to colorectal cancer. World J Gastroenterol 19:607–615
Fearon ER, Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell 61:759–767
Patai AV, Molnár B, Kalmár A, Schöller A, Tóth K, Tulassay Z (2012) Role of DNA methylation in colorectal carcinogenesis. Dig Dis 30:310–315
Tóth K, Sipos F, Kalmár A, Patai AV, Wichmann B, Stoehr R, Golcher H, Schellerer V, Tulassay Z, Molnár B (2012) Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One 7:e46000
Tóth K, Wasserkort R, Sipos F, Kalmár A, Wichmann B, Leiszter K, Valcz G, Juhász M, Miheller P, Patai ÁV, Tulassay Z, Molnár B (2014) Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS One 9:e115415
Burgess NG, Tutticci NJ, Pellise M, Bourke MJ (2014) Sessile serrated adenomas/polyps with cytologic dysplasia: a triple threat for interval cancer. Gastrointest Endosc 80:307–310
Erichsen R, Baron JA, Hamilton-Dutoit SJ, Snover DC, Torlakovic EE, Pedersen L, Frøslev T, Vyberg M, Hamilton SR, Sørensen HT (2016) Increased risk of colorectal cancer development among patients with serrated polyps. Gastroenterology 150:895–902.e5
Regula J, Rupinski M, Kraszewska E, Polkowski M, Pachlewski J, Orlowska J, Nowacki MP, Butruk E (2006) Colonoscopy in colorectal-cancer screening for detection of advanced neoplasia. N Engl J Med 355:1863–1872
Rothwell PM, Wilson M, Elwin CE, Norrving B, Algra A, Warlow CP, Meade TW (2010) Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet 376:1741–1750
Heigh RI, Yab TC, Taylor WR, Hussain FT, Smyrk TC, Mahoney DW, Domanico MJ, Berger BM, Lidgard GP, Ahlquist DA (2014) Detection of colorectal serrated polyps by stool DNA testing: comparison with fecal immunochemical testing for occult blood (FIT). PLoS One 9:e85659
Park SJ, Rashid A, Lee JH, Kim SG, Hamilton SR, Wu TT (2003) Frequent CpG island methylation in serrated adenomas of the colorectum. Am J Pathol 162:815–822
Wynter CV, Walsh MD, Higuchi T, Leggett BA, Young J, Jass JR (2004) Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut 53:573–580
O'Brien MJ, Yang S, Mack C, Xu H, Huang CS, Mulcahy E, Amorosino M, Farraye FA (2006) Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol 30:1491–1501
Kim YH, Kakar S, Cun L, Deng G, Kim YS (2008) Distinct CpG island methylation profiles and BRAF mutation status in serrated and adenomatous colorectal polyps. Int J Cancer 123:2587–2593
Subramaniam MM, Chan JY, Soong R, Ito K, Yeoh KG, Wong R, Guenther T, Will O, Chen CL, Kumarasinghe MP, Ito Y, Salto-Tellez M (2009) RUNX3 inactivation in colorectal polyps arising through different pathways of colonic carcinogenesis. Am J Gastroenterol 104:426–436
Kim KM, Lee EJ, Ha S, Kang SY, Jang KT, Park CK, Kim JY, Kim YH, Chang DK, Odze RD (2011) Molecular features of colorectal hyperplastic polyps and sessile serrated adenoma/polyps from Korea. Am J Surg Pathol 35:1274–1286
Kaji E, Uraoka T, Kato J, Hiraoka S, Suzuki H, Akita M, Saito S, Tanaka T, Ohara N, Yamamoto K (2012) Externalization of saw-tooth architecture in small serrated polyps implies the presence of methylation of IGFBP7. Dig Dis Sci 57:1261–1270
Fu X, Li L, Peng Y (2012) Wnt signalling pathway in the serrated neoplastic pathway of the colorectum: possible roles and epigenetic regulatory mechanisms. J Clin Pathol 65:675–679
Beggs AD, Jones A, Shepherd N, Arnaout A, Finlayson C, Abulafi AM, Morton DG, Matthews GM, Hodgson SV, Tomlinson IP (2013) Loss of expression and promoter methylation of SLIT2 are associated with sessile serrated adenoma formation. PLoS Genet 9:e1003488
Muto Y, Maeda T, Suzuki K, Kato T, Watanabe F, Kamiyama H, Saito M, Koizumi K, Miyaki Y, Konishi F, Alonso S, Perucho M, Rikiyama T (2014) DNA methylation alterations of AXIN2 in serrated adenomas and colon carcinomas with microsatellite instability. BMC Cancer 14:466
Dhir M, Yachida S, Van Neste L, Glöckner SC, Jeschke J, Pappou EP, Montgomery EA, Herman JG, Baylin SB, Iacobuzio-Donahue C, Ahuja N (2011) Sessile serrated adenomas and classical adenomas: an epigenetic perspective on premalignant neoplastic lesions of the gastrointestinal tract. Int J Cancer 129:1889–1898
Gaiser T, Meinhardt S, Hirsch D, Killian JK, Gaedcke J, Jo P, Ponsa I, Miró R, Rüschoff J, Seitz G, Hu Y, Camps J, Ried T (2013) Molecular patterns in the evolution of serrated lesion of the colorectum. Int J Cancer 132:1800–1810
Patai ÁV, Valcz G, Hollósi P, Kalmár A, Péterfia B, Patai Á, Wichmann B, Spisák S, Barták BK, Leiszter K, Tóth K, Sipos F, Kovalszky I, Péter Z, Miheller P, Tulassay Z, Molnár B (2015) Comprehensive DNA methylation analysis reveals a common ten-gene methylation signature in colorectal adenomas and carcinomas. PLoS One 10:e0133836
Holemon H, Korshunova Y, Ordway JM, Bedell JA, Citek RW, Lakey N, Leon J, Finney M, McPherson JD, Jeddeloh JA (2007) MethylScreen: DNA methylation density monitoring using quantitative PCR. BioTechniques 43:683–693
Péterfia B, Kalmár A, Patai AV, Bodor A, Micsik T, Hollósi P, Egedi K, Wichmann B, Tulassay Z, Molnár B (2016) Construction of a multiplex mutation hot spot PCR panel: the first step towards colorectal cancer genotyping on the GS Junior platform. J Cancer. doi:10.7150/jca.16037
Valcz G, Patai ÁV, Kalmár A, Péterfia B, Fűri I, Wichmann B, Műzes G, Sipos F, Krenács T, Mihály E, Spisák S, Molnár B, Tulassay Z (2014) Myofibroblast-derived SFRP1 as potential inhibitor of colorectal carcinoma field effect. PLoS One 9:e106143
Lee EJ, Chun SM, Kim MJ, Jang SJ, Kim do S, Lee DH, Youk EG (2016) Reappraisal of hMLH1 promoter methylation and protein expression status in the serrated neoplasia pathway. Histopathology 69:198–210
Hazewinkel Y, de Wijkerslooth TR, Stoop EM, Bossuyt PM, Biermann K, van de Vijver MJ, Fockens P, van Leerdam ME, Kuipers EJ, Dekker E (2014) Prevalence of serrated polyps and association with synchronous advanced neoplasia in screening colonoscopy. Endoscopy 46:219–224
Sipos F, Műzes G, Patai AV, Fűri I, Péterfia B, Hollósi P, Molnár B, Tulassay Z (2013) Genome-wide screening for understanding the role of DNA methylation in colorectal cancer. Epigenomics 5:569–581
Acknowledgements
Authors are deeply indebted to colleagues at the Endoscopy Unit and also to Gabriella Farkas Kónyáné (2nd Department of Medicine, Semmelweis University) for her skilled technical work and assistance in the immunohistochemical analyses.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Financial Support
This study was supported by the National Office for Research and Technology, Hungary, (Grant number: TECH_08-A1/2–2008-0114) and by the Hungarian Scientific Research Fund (OTKA, Grant number: K 111743). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional information
Árpád V. Patai, Barbara Kinga Barták and Bálint Péterfia contributed equally to this work.
Electronic supplementary material
Supplementary Fig. 1
Characteristic histopathological image of SSA. SSA: sessile serrated adenoma (JPEG 11 kb)
Supplementary Fig. 2
Principle of Methyl-Profiler DNA methylation analysis (EpiTect Methyl qPCR Array System, figure provided by Qiagen, with permission) (JPEG 4 kb)
Supplementary Fig. 3
Principle of methylation-sensitive restriction enzyme analysis. Mock reaction contains no enzymes, double reaction contains both enzymes. Methylation-sensitive restriction enzyme cleaves at unmethylated CpG sites, whereas methylation-dependent restriction enzyme cleaves at methylated CpG sites. Empty circle unmethylated cytosine, full circle: methylated cytosine (JPEG 7 kb)
Supplementary Table 1
(DOC 79 kb)
Rights and permissions
About this article
Cite this article
Patai, Á.V., Barták, B.K., Péterfia, B. et al. Comprehensive DNA Methylation and Mutation Analyses Reveal a Methylation Signature in Colorectal Sessile Serrated Adenomas. Pathol. Oncol. Res. 23, 589–594 (2017). https://doi.org/10.1007/s12253-016-0154-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-016-0154-6