Abstract
Coxsackievirus A10 (CVA10) is one of the major etiological agents of hand, foot, and mouth disease. There are no vaccine and antiviral drugs for controlling CVA10 infection. Reverse genetic tools for CVA10 will benefit its mechanistic study and development of vaccines and antivirals. Here, two infectious clones for the prototype and a Myc-tagged CVA10 were constructed. Viable CVA10 viruses were harvested by transfecting the viral mRNA into human rhabdomyosarcoma (RD) cells. Rescued CVA10 was further confirmed by next generation sequencing and characterized experimentally. We also constructed the vectors for CVA10 subgenomic replicon with luciferase reporter and viral capsid with EGFP reporter, respectively. Co-transfection of the viral replicon RNA and capsid expresser in human embryonic kidney 293T (HEK293T) cells led to the production of single round infectious particles (SRIPs). Based on CVA10 replicon RNA, SRIPs with either the enterovirus A71 (EVA71) capsid or the CVA10 capsid were generated. Infection by EVA71 SRIPs required SCARB2, while CVA10 SRIPs did not. Finally, we showed great improvement of the replicon activity and SRIPs production by insertion of a cis-active hammerhead ribozyme (HHRib) before the 5′-untranslated region (UTR). In summary, reverse genetic tools for prototype strain of CVA10, including both the infectious clone and the SRIPs system, were successfully established. These tools will facilitate the basic and translational study of CVA10.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human Enterovirus Genus includes A, B, C and D (EV-A, B, C and D) species, and consists of many important human pathogens, such as the Coxsackievirus B3 of EV-B, Poliovirus of EV-C and Enterovirus D68 of EV-D (Tapparel et al. 2013). Although considered with low pathogenicity, EV-As are found to be the leading causes of large outbreaks of hand, foot, and mouth disease (HFMD), posing threat for public health, especially in Asia–Pacific region (Aswathyraj et al. 2016). Among the EV-A species, EVA71 and CVA16 are the major pathogens for HFMD and the severest cases are mainly caused by EVA71 infection (Yang et al. 2017). One major breakthrough in the field is the development of prophylactic vaccines for EVA71. However, this vaccination can only prevent diseases caused by EVA71 but not other EV-As (Li et al. 2014; Zhu et al. 2014). Recently, epidemiological surveys showed that CVA6 and CVA10 became more prevalent, replacing the EVA71 and CVA16 as the dominant pathogens for HFMD (Fu et al. 2019). The clinical manifestations of CVA6 and CVA10 infections are unique. CVA6 often causes an atypical HFMD accompanied by a varicella rash (Bian et al. 2015), while CVA10 has been associated with HFMD/herpangina and sometimes with post-HFMD onychomadesis (Bian et al. 2019; Davia et al. 2011), though the underlying mechanisms for the peculiar pathogenesis are not well understood.
Reverse genetic tools are crucial for studying viruses. So far, among 25 serotypes of EV-A, infectious clone of EVA71 (Tan et al. 2016), CVA16 (Deng et al. 2015; Wang et al. 2016) and CVA6 (Yang et al. 2015) have been previously established, while the others are not available yet. Very recently, an infectious clone of clinically isolated CVA10 from China has been generated (Liu et al. 2019). Here, we reported the construction of both the infectious clone and single-round-infectious particles (SRIPs) for the CVA10 prototype strain (Kowalik strain, GenBank Accession: AY421767.1).
Materials and Methods
Materials and Reagents
Human embryonic kidney 293T (HEK293T) cells, human rhabdomyosarcoma (RD) cells, African green monkey kidney Vero cells, human neuroblastoma SK-N-SH cells, human cervical epithelial Hela cells, human lung fibroblast MRC-5 cells, and Human oral epidermoid carcinoma KB cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 µg/mL streptomycin. All cell lines were from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). EVA71 (FJ08089 strain), CVA10 (SHAPHC798F/SH/CHN/2010) and the rescued CVA10 (AY421767.1) were propagated in RD cells. Viral titer was determined in RD cells by plaque assay.
Mouse monoclonal antibody against EV71 capsid protein VP1 was from Abcam (Cambridge, UK). The secondary antibody conjugated to Allophycocyanin (APC) was from BD Biosciences (Franklin Lakes, CA, USA). AlexaFluor594-Conjugated AffiniPure Goat Anti-Mouse IgG (H + L) was from ZSGB-BIO (Beijing, China). Fluoroshield Mounting Medium with DAPI was from Abcam. Mouse anti-Flag monoclonal antibody was from Sigma (M2 mAb, St Louis, MO, USA). Rabbit anti-GAPDH and mouse anti-3A monoclonal antibody were from Youke (Shanghai, China). Mouse anti-dsRNA antibody was from SCICONS (J2 mAb, Budapest, Hungary). Rabbit anti-Myc monoclonal antibody was from Cell Signaling Technologies (Danvers, MA, USA). Luciferase assay system was from Promega (E1500, Madison, WI, USA).
Cloning Strategy
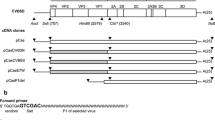
Three segments (1–2436, 2437–5064, 5065–7409) covering the CVA10 (AY421767.1) genome were synthesized and cloned in the pUC57 vector. The cloning backbone PL451 and the three segments of CVA10 carrying overlapping end (15–20 bp) were amplified and seamlessly linked together in the transformed E. coli by recombination according to HieffClone™MultiOneStep Cloning kit (YEASEN, Shanghai, China). The CVA10 capsid segments were seamlessly cloned into pcDNA6.0-EGFP to form the CVA10 capsid expresser. The construct of CVA10 replicon was derived from the plasmid of EVA71 replicon. Firstly, the original EVA71 5′-untranslated region (UTR) was replaced by CVA10 5′-UTR. Secondly, the non-structural gene segments of EVA71 (from 2A to 3′UTR) were replaced by that of CVA10. The cloning strategy and vector maps were illustrated in Supplementary Figure S1.
In Vitro Transcription
The plasmids of CVA10 infectious clone and replicon were linearized by SalI (NEB, Ipswich, MA, USA), and then purified by TIANquick Midi Purification Kit (TIANGEN, Beijing, China). The linearized plasmids were transcribed by T7 polymerase following the manual of MEGAscript T7 in vitro transcription kit (Cat. AM1334, Invitrogen, USA). The RNA quality was confirmed by agarose gel electrophoresis.
RNA Transfection
CVA10 mRNA was introduced into RD cells using Lipofectamine 3000 (Life Technologies, Carlsbad, USA) according to the manufacturer’s instruction. RD cells were seeded at 3 × 105 cells/well in 12-wells plates. Next day, the mixture containing 0.5 μg of in vitro transcribed RNA and transfection reagents was added into RD monolayers (90% confluence) and incubated for 6 h at 37 °C. The medium was then changed and cells were maintained in fresh culture medium.
Virus Next Generation Sequencing (NGS)
Virus NGS was conducted at Shanghai Tanpu Biotechnology Co., Ltd. Briefly, viral RNA was extracted from culture supernatants using the TIANamp RNA kit (TIANGEN, Beijing, China), and then the first strand cDNA synthesis was conducted using NEBNext Ultra RNA Prep Kit with random primers. Second Strand Master Mix was added into the original reaction mixture to synthesize the second strand. NGS libraries were prepared using the Nextera DNA sample preparation kit (Illumina, CA, USA) for Illumina sequencing according to the manufacture’s instructions. After quality control, trimming and adapter clipping, data filtering and de-novo assembly, whole genome sequence was obtained and analyzed as described previously (Watson et al. 2013).
Viral Infection and Plaque Assay
The plaque assay was performed using 6 or 12-wells plates containing RD cell monolayers. Ten-fold series of viral dilutions were added and the plate was shaken every 15 min for 1 h. Then the inoculums were removed and 2 or 1 mL of DMEM containing 2% FBS and 1% low melting point agarose (Promega, Madison, USA) was added to each well, before incubation at 37 °C. After cultured for 3–5 days, the plates were stained with 0.1% crystal violet (Sigma, St. Louis, USA) containing 10% formaldehyde and plaques were counted to measure the viral titer.
Transmission Electron Microscopy (TEM)
The CVA10 culture supernatant was harvested and the cell debris was removed by passage through a 0.45 µm filter (Pall, Puerto Rico). The virus sample was precipitated by incubation with 8% polyethylene glycol (PEG) 8,000 in PBS (pH 7.4) at 4 °C for 12 h. After one more centrifugation, the virus resuspended in PBS and subsequently loaded onto a 10%–50% continuous sucrose gradient for ultracentrifugation in Beckman SW41 Ti rotors at 175,000 g for 3 h. The fractions (60 mL per fraction) at 20% to 40% sucrose were collected and individually concentrated by diafiltration using an Amicon 100 K tube (Millipore, Belerica, MA USA) at 4000 g for 30 min. The concentrated virus was stored at 4 °C. Characterization of virus particles was analyzed by negative staining electron microscopy. Briefly, virus was inactivated by 1/4000 (v/v) formalin at 37 °C for 3 days and then absorbed onto 200-mesh carbon-coated copper grid for 20 min at room temperature. The grids were washed twice with ddH2O and subsequently negatively stained with 2% phosphotungstic acid (pH 6.4) for 2 min. The stained grid was dried for 3 days and observed under a FEI Talos F200 transmission electron microscope (ThermoFisher, USA).
Immunofluorescence
RD cells were seeded in 12-well plate (3 × 105 cells/well) with coverslips. The next day, RD cells were incubated with EV71, CVA10 or CVA10-Myc (MOI = 1) for 6 h. Then, RD cells were washed with cold PBS, fixed with 4% paraformaldehyde (Sigma, St Louis, USA) for 15 min, permeabilized with 0.05% Triton X-100 in 2% FBS/PBS, and then stained with mouse anti-dsRNA antibody (1:250 dilution, J2 clone) (SCICONS, Budapest, Hungary), mouse anti-3A antibody (1:1000 dilution, YOUKE) or rabbit anti-Myc antibody (1:250 dilution, CST) for 1 h at room temperature. Three washes with PBS were followed by 30 min-incubation with the secondary antibodies. After washes with PBS, coverslips were stained by mounting medium with DAPI. Immunofluorescent imaging was taken on EVOS® FL Color Imaging Systems (Life technology, Grand Island, NY, USA).
Viral RNA Quantification
Viral RNA was extracted from infected cell lysates using the TIANamp RNA kit (Cat.SD101, TIANGEN, Beijing, China). Quantification of viral RNA was performed by quantitative real-time RT-PCR assay using specific primers for VP1. To make a standard curve, a fragment of CVA10 capsid from nt 2007 to 2145 was cloned into pcDNA6.0. The plasmid was linearized with SalI (NEB), in vitro transcribed using the MEGAscript T7 transcription kit (Invitrogen, USA), and DNase treated to generate the RNA standard.
Western Blot
Cells were harvested and lysed in Radioimmunoprecipitation assay buffer with a mixture of proteinase inhibitors (MedChem Express, Monmouth Junction, NJ, USA). The concentration of proteins was quantified by bicinchoninic acid protein assay (Biosharp, China) and equal amounts of proteins were loaded and separated by SDS-PAGE, then transferred to a nitrocellulose membrane. The membrane was blocked with blocking buffer (0.1% Tween-20 in PBS containing 5% BSA) and then was incubated overnight with primary antibody diluted in blocking buffer at 4 °C. The membrane was then washed three times in 0.1% Tween-20/PBS and incubated with anti-mouse IgG conjugated to AlexaFluor790 (Jackson ImmunoResearch, West Grove, PA, USA) for 1 h at room temperature. The immunoblots were visualized using an Odyssey Fc Imager (Lincoln, NE, USA).
Luciferase Measurement
SRIPs of CVA10, CVA10 HHRib and EVA71 were generated by sequential transfection of HEK293T cells with the capsid expresser and the CVA10 replicon mRNA. The capsid plasmid (1 μg) was firstly transfected into HEK293T cells at 60%–80% confluence. 24 h later, 1.5 μg of replicon RNA was then transfected using lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). The pseudovirus was harvested 24 h post-RNA transfection with two rounds of freeze–thaw cycle. Then 200 μL of pseudovirus mixed with 800 μL of fresh DMEM was applied to RD cells in 12-well plates. The SRIPs were scored by assay of luciferase activity after 12 h infection at 37 °C. Briefly, the inoculums were removed after 12 h incubation, and then the cells were washed twice by PBS and lysed directly on the plates by the addition of 150 μL of cell culture lysis reagent. Ten microliters of the cell lysate were then used to assay luciferase activity by using a luciferase assay kit (E1500, Promega, Madison, WI, USA). Luciferase activity was expressed as relative luciferase units (RLU).
Statistical Analyses
Significance of differences was evaluated using Student’s t test. Statistical analysis was performed with GraphPad Prism version 6.0 (La Jolla, CA, USA). The P values (P) less than 0.05 were considered statistically significant.
Results
Construction of Infectious Clone of Coxsackievirus A10
To create the infectious clone of prototype CVA10, three segments covering the full length of CVA10’s genome were synthesized and seamlessly cloned together, with a T7 promoter and poly(A) tail at the 5′ and 3′ end respectively (Fig. 1A and Supplementary Fig. S1). The CVA10 mRNA was generated by in vitro transcription using the linearized plasmid as the template (Fig. 1B). After introducing the CVA10 mRNA into RD cells, cytopathic effect (CPE) was observed 24 h later, indicating the successful rescue of prototype CVA10 virus (Fig. 1C). The morphological characterization of the rescued CVA10 viral particles was revealed by TEM analysis. The prototype CVA10 and another clinical isolated CVA10 strain (SHAPHC798F/SH/CHN/2010, abbreviated as 798F) (Fig. 1D) held similar spherical shape with diameters of ~ 30 nm, as other enteroviruses of the piconaviridae family. After 3 passages in RD cells, the virus titer was determined to be 2.0 × 108 PFU/mL (Fig. 1E). Viral infection in cells was confirmed by dsRNA staining (Fig. 1F). Finally, rescued viral genome sequence was confirmed without any mutation by next generation sequencing (data not shown).
Construction of an infectious clone for prototype CVA10. A The cartoon shows the structure of the CVA10 infectious clone which is led by a T7 promotor. P1, P2 and P3 represent VP4-VP2-VP3-VP1, 2A-2B-2C and 3A-3B-3C-3D polyproteins, respectively. B The linearized PL451-EVA71 and PL451-CVA10 were transcribed by T7 polymerase. The purified mRNAs for EVA71 and CVA10 were analyzed by electrophoresis on agarose gel. Lane M, ssRNA ladder marker. C RD cells were transfected with 0.5 μg CVA10 mRNA per well. The cytopathic effect (CPE) was detected 24 h after transfection. D The electron microscopic characterizations of the CVA10 particles. Scale bar, 100 nm. E The rescued CVA10 from initial mRNA transfection was passaged 3 times and harvested as the stock. The viral titer of the CVA10 stock was determined by plaque assay. F RD cells were infected by EVA71 and CVA10 at an MOI of 1. At 6 h post-infection, cells were fixed and stained with anti-dsRNA J2 antibody, followed by Alexa Flour 594 conjugated secondary antibody. DAPI was used to visualize the nuclei. (Red, dsRNA; Blue, nuclei).
Virological Characteristics of the Rescued Coxsackievirus A10
To further characterize the rescued CVA10 viruses, we examined the growth curve of CVA10 in different cell lines, including RD (human rhabdomyosarcoma cells), Vero (African green monkey kidney cells), SK-N-SH (human neuroblastoma cells), Hela (human cervical epithelial cells), MRC-5 (human lung fibroblasts) and KB (Human oral carcinoma) cells. Both the prototype and 798F CVA10 could infect all six cell lines and cause CPE. Although the growth of rescued CVA10 was restricted in MRC-5 and SK-N-SH cells, the 798F CVA10 continued growing in MRC-5 cells. Meanwhile, 798F CVA10 showed higher growth levels than rescued CVA10 in Vero and Hela cells. The growth rates of two CVA10 strains are similar in RD and KB cells (Fig. 2A).
Characterization of the rescued CVA10. A The growth curves of the two CVA10 strains on different cell lines. The pictures above showed the CPE and the charts below depicted the viral growth pattern by quantitative RNA levels. B, C RD SCARB2 WT and KO cells were infected with EVA71 or CVA10 at an MOI of 1. The infectivity was monitored by VP1 (EVA71) or 3A (CVA10) staining at 6 h post-infection (B). CPE was detected at 24 h post-infection (C). *P < 0.05; **P < 0.01.
Among Enterovirus A species, EVA71 and CVA16 use human scavenger receptor class B, member 2 (SCARB2) protein as their receptor. However, CVA6 and CVA10 infections are SCARB2-independent (Staring et al. 2018). Here, to confirm the differential requirement for SCARB2 by EVA71 and CVA10, the SCARB2 null (SCARB2-KO) cells were created by CRISPR/Cas9 techniques (Supplementary Fig. S2). We found that SCARB2 KO cells were not infected by EVA71 (Fig. 2B) and showed no CPE (Fig. 2C). However, CVA10 infected both control and SCARB2 KO cells, and caused CPE in both cell lines (Fig. 2B, 2C). These results indicated that the rescued CVA10 infection was SCARB2-independent as expected.
The CVA10 Infectious Clone Allows Insertion of Short Tag Sequence in the 3A Region
Protein tag is a useful tool for characterizing the viral proteins, especially when proper antibody is not available. Previous study showed that the N-terminal of enterovirus 3A protein could tolerate insertion of short sequences, including poliovirus (Li et al. 2016) and Coxsackievirus B3 (CVB3) (van der Schaar et al. 2016). Previously, our lab confirmed that FLAG-tag can be inserted after the second amino acid of 3A protein in the genome of EVA71, and the modified EVA71 is viable, with the 3A protein detected by anti-FLAG antibody (unpublished data). Here, we inserted the Myc-tag after the second amino acid of 3A protein in the CVA10 infectious clone (Fig. 3A). In comparative plaque assay, we found that the rescued CVA10-Myc maintained comparable titers as CVA10 after passaging in RD cells (Fig. 3B). Thus, we concluded that the Myc-insertion did not interfere the viral activity. To detect the expression of 3A-Myc, we infected RD cells with CVA10-Myc and performed immunofluorescence and Western blot. The results showed the Myc-tag could be detected and was co-localized with viral 3A protein (Fig. 3C, 3D). However, for unknown causes, we failed to detect the FLAG-tagged or HA-tagged protein in CVA10-FLAG or CVA10-HA infected cells by Western blot, despite that the rescued CVA10-FLAG or CVA10-HA viruses contained the inserted FLAG or HA tag correctly (Supplementary Fig. S3).
Construction of CVA10-3A-Myc infectious clone. A The cartoon shows that a Myc-tag sequence is inserted into the CVA10 infectious clone after the second amino acid (proline) of 3A. B The rescued CVA10 with a Myc-tag was harvested after four continuous passages and the titer was measured by plaque assay. C, D RD cells were infected by CVA10 and CVA10-Myc at an MOI of 1. At 6 h post-infection, the expression of Myc was detected by immunofluorescence (C) and Western blot (D). For immunofluorescent assay, cells were co-stained with mouse anti-3A and rabbit anti-Myc antibodies, followed by anti-mouse IgG conjugated with Alexa Fluor 488 and anti-rabbit IgG conjugated with Alexa Flour 594 secondary antibodies, respectively. DAPI was used to visualize the nuclei. (3A, green; Myc, red; nuclei, blue). For Western blot, cells were harvested and lysed for SDS-PAGE. Then the blot was probed with anti-3A or anti-Myc antibodies. GAPDH was used as an internal control.
Subgenomic Replicon and Single Round Infectious Particles of Coxsackievirus A10
Firstly discovered in polio virus, and now confirmed in other +ssRNA viruses as well that the genomic replication of these viruses, including enteroviruses, usually do not require their structural genes (Kaplan and Racaniello 1988). Hence, subgenomic replicon with viral structural gene replaced by a reporter gene, can efficiently replicate when introduced into the host cells, so it can be used as a powerful research tool. Here, we made the vector for subgenomic replicon of CVA10 by replacing the capsid coding region of the infectious clone with the gene for luciferase (Fig. 4A). The in vitro derived CVA10 replicon RNA could replicate after transfection into RD cells, although its activity was lower compared with the EVA71 replicon (Fig. 4B). The replication inhibitors, GnHCl (2C inhibitor) and Rupintrivir (3C inhibitor), significantly suppressed the activity of both replicons.
The establishment of CVA10 replicon and pseudovirus. A The constructs of CVA10 capsid with an EGFP reporter and CVA10 subgenomic replicon with a luciferase reporter. The viral 2A protease recognition site AITTL was inserted between the reporter and the viral replicon or capsid. B RD cells were transfected with 0.5 μg EVA71 or CVA10 replicon mRNA per well, and viral replication was monitored by luciferase activity. GnHCl and Rupintrivir were used to inhibit viral replication. C The schematic diagram of single round infection assay. HEK293T cell monolayers were co-transfected with viral capsid and replicon mRNA sequentially. Progeny pseudovirus recovered from the transfection were defined as passage 1 (P1). Progeny viruses recovered from subsequent infection of cells with P1 virus were defined as passage 2 (P2) virus. Passage 3 (P3) virus was collected with the same method. EVA71 and CVA10 pseudoviruses of P1, P2 and P3 were used to infect RD cells, and luciferase activity was measured at 12 h post-infection. D CVA10 replicon based SRIPs with either EVA71 or CVA10 capsid were generated. The SRIPs infectivity in RD SCARB2 WT or KO cells were examined by luciferase activity. E Predicted structure of the cis-active hammerhead ribozyme (HHRib) attached to the 5′end of the CVA10 replicon. F RD cells were transfected by CVA10 HHRib replicon and CVA10 replicon RNAs, or infected by their SRIPs. Luciferase activity was measured. Means of three independent experiments with SD were shown. *P < 0.05; **P < 0.01.
Previous study showed that replicable subgenomic replicon can be assembled into viral particles when viral structural proteins were provided in-trans, and this strategy has been adopted to make single round infectious particles (SRIPs) of many +ssRNA viruses, for example the SRIPs of EVA71 (Chen et al. 2012). The infection tropism of SRIPs is determined by their trans-complemented capsids. Moreover, SRIPs only cause single round of infection. Thus, SRIPs are safe and useful to study viral tropism and entry mechanisms. To confirm that SRIPs can only replicate at one round, plasmids expressing viral capsid gene and viral subgenomic replicon RNAs were transfected into HEK293T cells sequentially. Then the pseudoviruses of passage 1 (P1), P2 and P3 were harvested and the viral infectivity was measured (Fig. 4C). We found high luciferase activity of P1 virus but minimal activities of those from the P2 and P3 viruses, indicating the single round infection by SRIPs. As expected, SRIPs with EVA71 capsid could infect RD cells but failed to infect SCARB2-KO RD cells, while SRIPs with CVA10 capsid could infect both RD and KO cells (Fig. 4D).
A number of positive- and negative-strand RNA viruses require exact termini for successful replication (Herold and Andino 2000). The synthetic transcripts transcribed by T7 polymerase contain several extra nucleotides at the 5′ end, which may affect the replication activity. To generate authentic 5′ end, a cDNA copy of a cis-active hammerhead ribozyme (HHRib), was cloned between the T7 RNA polymerase promoter and the 5′-terminal sequences of the replicon cDNA (Fig. 4E). Indeed, due to removal of extra sequences by HHRib, the replication activity of the CVA10 replicon was improved significantly. In addition, we determined that the titers of the EVA71 SRIPs were 2.5 × 106 TCID50/mL, the CVA10 SRIPs were 2.14 × 104 TCID50/mL, while the CVA10 HHRib SRIPs were increased to 1.35 × 106 TCID50/mL (Fig. 4F).
Discussion
Probably due to its relatively low incidence and mild clinical significance in western world, EV-A species are less studied compared to EV-B and EV-C species. However, viruses of EV-A species are causing large outbreaks of HFMD in the Aisa-Pacific regions in recent years and spiked a major research interest ever since. Much effort had been put into the study of two major agents, the EVA71 and CVA16, while other viruses were relatively omitted. Recently, enteroviruses other than EVA71 and CVA16 are emerging as the new threats, such as CVA6 and CVA10 (Fu et al. 2019). Among the EV-A serotypes, CV-A6 and CV-A10 form a distinct cluster different from the EVA71 and CVA16 cluster (Liu et al. 2016). The HFMD caused by CVA6 and CVA10 also have distinct clinical features showing as varicella rash caused by CVA6 (Bian et al. 2015) and onychomadesis by CVA10 (Davia et al. 2011). So far, there is no vaccine for infections by non-EVA71 enteroviruses (Klein 2015). Development of the reverse genetic tools for CVA10 will greatly facilitate the future studies of this virus and other EV-A strains. For example, the newly established infectious clone can be further modified to create reporter viruses, like luciferase or EGFP viruses (Caine and Osorio 2017) which are useful in high-throughput screening or animal modeling. It can also be applied to rapidly produce vaccine candidates (Yang et al. 2016). Finally, the CVA10 infectious clone and replicon are valuable tools for studying its biology, such as probing the genetic basis for its molecular function (Li et al. 2016), and studying the driving forces for viral adaptation, recombination and evolution (Volle et al. 2019). Recently, Liu et al. (2019) reported the creation of a CVA10 infectious clone based on a clinical isolate. However, different strains of CVA10 hold strain-specific properties and are often not universally available. In contrast, the Kowalik strain in current study is the CVA10 prototype, which have been widely studied and referred as the standard by researchers worldwide. Thus, creation of the infectious clone based on CVA10 prototype provides a valuable tool for the whole research community.
It is possible to insert additional sequences at the 3A site in human enteroviruses. This modification was originally attempted in poliovirus (Teterina et al. 2011). Later it was utilized to create a split-GFP tagged CVB3 (van der Schaar et al. 2016). Now, we showed that it was also possible to insert foreign sequences into this particular site in other EV-A strains. The inserted Myc-tag sequence was stable after subsequent passages in the CVA10 infectious clone. In addition, the Myc-tagged 3A protein can be detected by immunofluorescence and Western blot using anti-Myc antibody, which facilitated the detection of viral infection. This approach makes it possible for monitoring CVA10 infection and furthering our understanding of virus-host interaction by the tagged protein.
In the current study, we have made both the virus infectious clone and the SRIPs for CVA10. Viral subgenomic-replicon can be used in RNA replication studies and it can inform us whether the RNA replicon is replication-competent, and it can be combined with various viral capsids to form unique SRIPs or chimeric SRIPs with new properties. The SRIPs are safer than the infectious virions and are useful for trans-encapsidation mechanism or the viral entry (Yuan et al. 2018). Finally, since a successful infectious clone requires both functional replicon and capsid, the SRIPs help to reveal which part of the cloned genome is involved in a failed infectious clone. For example, we recently identified the notably lower protein levels of EVA89 capsid might be responsible for the failure rescuing of EVA89 viruses from the infectious clone, although the EVA89 replicon was capable of high levels of replication (unpublished data). Finally, based on prior knowledge of the requirement on authentic 5′ end for optimized viral replication (Herold and Andino 2000), we inserted a cis-active hammerhead ribozyme in front of the 5′-end of replicon plasmid. The viral RNAs derived from these plasmids showed significantly improved viral replication in RD cells and led to significantly enhanced production of SRIPs.
In summary, we for the first time established the infectious clone and the SRIP system for CVA10 prototype strain, another important member in Enterovirus A species. These tools will largely facilitate basic and translational studies of CVA10 and related viruses in the future.
References
Aswathyraj S, Arunkumar G, Alidjinou EK, Hober D (2016) Hand, foot and mouth disease (HFMD): emerging epidemiology and the need for a vaccine strategy. Med Microbiol Immunol 205:397–407
Bian L, Wang Y, Yao X, Mao Q, Xu M, Liang Z (2015) Coxsackievirus A6: a new emerging pathogen causing hand, foot and mouth disease outbreaks worldwide. Expert Rev Anti Infect Ther 13:1061–1071
Bian L et al (2019) Hand, foot, and mouth disease associated with coxsackievirus A10: more serious than it seems. Expert Rev Anti Infect Ther 17:233–242. https://doi.org/10.1080/14787210.2019.1585242
Caine EA, Osorio JE (2017) In vivo imaging with bioluminescent enterovirus 71 allows for real-time visualization of tissue tropism and viral spread. J Virol 91:e01759–16
Chen P et al (2012) Molecular determinants of enterovirus 71 viral entry: cleft around GLN-172 on VP1 protein interacts with variable region on scavenge receptor B2. J Biol Chem 287:6406–6420
Davia JL et al (2011) Onychomadesis outbreak in Valencia, Spain associated with hand, foot, and mouth disease caused by enteroviruses. Pediatr Dermatol 28:1–5
Deng C, Li X, Liu S, Xu L, Ye H, Qin CF, Zhang B (2015) Development and characterization of a clinical strain of Coxsackievirus A16 and an eGFP infectious clone. Virol Sin 30:269–276
Fu X, Wan Z, Li Y, Hu Y, Jin X, Zhang C (2019) National epidemiology and evolutionary history of four hand, foot and mouth disease-related enteroviruses in China from 2008 to 2016. Virol Sin. https://doi.org/10.1007/s12250-019-00169-2
Herold J, Andino R (2000) Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J Virol 74:6394–6400
Kaplan G, Racaniello VR (1988) Construction and characterization of poliovirus subgenomic replicons. J Virol 62:1687–1696
Klein MH (2015) EV71 vaccines: a first step towards multivalent hand, foot and mouth disease vaccines. Expert Rev Vaccines 14:337–340
Li R et al (2014) An inactivated enterovirus 71 vaccine in healthy children. N Engl J Med 370:829–837
Li Z et al (2016) Identification of a nucleotide in 5′ untranslated region contributing to virus replication and virulence of Coxsackievirus A16. Sci Rep 6:20839
Liu CC et al (2016) Immunological and biochemical characterizations of coxsackievirus A6 and A10 viral particles. Antiviral Res 129:58–66
Liu Q et al (2019) Construction and characterization of an infectious cDNA clone of coxsackievirus A10. Virol J 16:98
Staring J, van den Hengel LG, Raaben M, Blomen VA, Carette JE, Brummelkamp TR (2018) KREMEN1 is a host entry receptor for a major group of enteroviruses. Cell Host Microbe 23:636–643
Tan CW, Tee HK, Lee MH, Sam IC, Chan YF (2016) Enterovirus A71 DNA-launched infectious clone as a robust reverse genetic tool. PLoS One 11:e0162771
Tapparel C, Siegrist F, Petty TJ, Kaiser L (2013) Picornavirus and enterovirus diversity with associated human diseases. Infect Genet Evol 14:282–293
Teterina NL, Pinto Y, Weaver JD, Jensen KS, Ehrenfeld E (2011) Analysis of poliovirus protein 3A interactions with viral and cellular proteins in infected cells. J Virol 85:4284–4296
van der Schaar HM et al (2016) Illuminating the sites of enterovirus replication in living cells by using a split-GFP-tagged viral protein. mSphere 1:e00104–16
Volle R et al (2019) High permissiveness for genetic exchanges between enteroviruses of species A, including enterovirus 71, favors evolution through intertypic recombination in Madagascar. J Virol. https://doi.org/10.1128/JVI.01667-18
Wang X, Shen C, Chen T, Lan K, Huang Z, Zhang Y, Liu Q (2016) Improved plasmid-based recovery of coxsackievirus A16 infectious clone driven by human RNA polymerase I promoter. Virol Sin 31:339–341
Watson SJ, Welkers MR, Depledge DP, Coulter E, Breuer JM, de Jong MD, Kellam P (2013) Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos Trans R Soc Lond B Biol Sci 368:20120205
Yang B et al. (2017) Epidemiology of hand, foot and mouth disease in China, 2008 to 2015 prior to the introduction of EV-A71 vaccine Euro Surveill 22
Yang L et al (2015) Construction and characterization of an infectious clone of coxsackievirus A6 that showed high virulence in neonatal mice. Virus Res 210:165–168
Yang YT et al (2016) Development of a full-length cDNA-derived enterovirus A71 vaccine candidate using reverse genetics technology. Antiviral Res 132:225–232
Yuan M et al (2018) Enhanced human enterovirus 71 infection by endocytosis inhibitors reveals multiple entry pathways by enterovirus causing hand-foot-and-mouth diseases. Virol J 15:1
Zhu F et al (2014) Efficacy, safety, and immunogenicity of an enterovirus 71 vaccine in China. N Engl J Med 370:818–828
Acknowledgements
We want to thank Prof. Wenhui Li at National Institute of Biomedical Sciences (NIBS) for providing us the infectious clone and subgenomic replicon of EVA71. This work was supported by the Shanghai Public Health Clinical Center (KY-GW-2018-17) and Shanghai Municipal Commission of Health and Family Planning (20174Y0099).
Author information
Authors and Affiliations
Contributions
MW and JY designed and finished most of experiments, analyzed data and drafted the manuscript; LZ, MW, LL, RY, MC, JX and YZ have performed part of the experiments; ZY involved in experiment design, data analysis and manuscript preparation; SZ played major roles in experiment design and data analysis, and finalized the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Animal and human rights statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, M., Yan, J., Zhu, L. et al. The Establishment of Infectious Clone and Single Round Infectious Particles for Coxsackievirus A10. Virol. Sin. 35, 426–435 (2020). https://doi.org/10.1007/s12250-020-00198-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12250-020-00198-2