Abstract
Dengue virus (DENV) and Zika virus (ZIKV) have spread throughout many countries in the developing world and infect millions of people every year, causing severe harm to human health and the economy. Unfortunately, there are few effective vaccines and therapies available against these viruses. Therefore, the discovery of new antiviral agents is critical. Herein, a scorpion venom peptide (Smp76) characterized from Scorpio maurus palmatus was successfully expressed and purified in Escherichia coli BL21(DE3). The recombinant Smp76 (rSmp76) was found to effectively inhibit DENV and ZIKV infections in a dose-dependent manner in both cultured cell lines and primary mouse macrophages. Interestingly, rSmp76 did not inactivate the viral particles directly but suppressed the established viral infection, similar to the effect of interferon (IFN)-β. Mechanistically, rSmp76 was revealed to upregulate the expression of IFN-β by activating interferon regulatory transcription factor 3 (IRF3) phosphorylation, enhancing the type-I IFN response and inhibiting viral infection. This mechanism is significantly different from traditional virucidal antimicrobial peptides (AMPs). Overall, the scorpion venom peptide Smp76 is a potential new antiviral agent with a unique mechanism involving type-I IFN responses, demonstrating that natural AMPs can enhance immunity by functioning as immunomodulators.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dengue virus (DENV) and Zika virus (ZIKV), belonging to the Flaviviridae family, are a global threat to human health and result in huge economic losses. DENV and ZIKV are enveloped, positive single-stranded RNA viruses (Chan and Choi 2016) that are mosquito-borne. Due to urbanization and global warming (McMichael et al.2006), the two viruses have spread more widely in tropical and subtropical areas. The incidence of dengue has increased 30-fold over the last 50 years, with 396 million individuals infected every year, an 18% hospitalization rate, and approximately 13,600 deaths (Halstead 2007; Shepard et al.2016). DENV has four serotypes (DENV1–4) that can all result in life-threatening dengue hemorrhagic fever and dengue shock syndrome (Cui et al.2018; Heymann et al.2016; Ubol et al.2008). According to the World Health Organization (WHO), ZIKV is endemic to over 84 countries, territories, or sub-national areas including Africa, the Asia–Pacific regions, and the Americas—particularly South America (Bogoch et al.2016) (http://www.who.int/emergencies/zika-virus-tmp/en/). Infection with ZIKV during pregnancy can lead to congenital Zika syndrome (Alesha Grant et al.2016; Cui et al.2018; Heymann et al.2016), which is a serious health burden to society. Currently, a ZIKV vaccine is not available. Moreover, the tetravalent dengue vaccine (i.e., Dengvaxia) exhibits limited efficacy and can cause severe dengue in naive individuals, thus, it is only licensed in 9–11 countries. There are also no specific therapeutics against DENV and ZIKV (Screaton et al.2015; Yu et al.2017b). Therefore, the discovery of new antiviral agents is urgently needed.
Scorpion venom is a rich source of peptides with a variety of pharmacological functions (Carballar-Lejarazu et al.2008). Antimicrobial peptides (AMPs) are important components of scorpion venom, and some scorpion venom AMPs have been shown to have activity against multiple viral pathogens through different mechanisms (Hong et al.2013; Zeng et al.2018; Zhao et al.2012). For example, Ctry2459 and its derived peptides can inactivate hepatitis C virus (HCV) particles directly (Hong et al.2013), while Eval418 and its derived peptides can suppress herpes simplex virus (HSV)-1 infection by inactivating HSV-1 and inhibiting its attachment to host cells (Zeng et al.2018). Mucroporin-M1 can inhibit hepatitis B virus (HBV) replication by activating the MAPK pathway and downregulating hepatocyte nuclear factor (HNF4)-α in vitro and in vitro (Zhao et al.2012). Meanwhile, scorpine was shown to possess anti-DENV activity in vitro (Carballar-Lejarazu et al.2008).
Innate immune recognition of viral infection triggers antiviral immune responses. DENV and ZIKV are recognized by pattern recognition receptors (PRRs), particularly retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5), that are expressed on innate immune cells (Loo and Gale 2011; Morrison et al.2013). Interferon (IFN)-mediated antiviral responses are important against viral infection and pathogenesis (Schoggins et al.2011; Seth et al.2006; Muller et al.1994). IFN-α/β receptors receive stimulation from type-I IFN, after which hundreds of cellular genes are upregulated, leading to antiviral responses (Jones et al.2005; Morrison et al.2013). Although AMPs are effectors of the innate immune response and can kill bacteria, enveloped viruses, fungi, and tumor cells (Reddy et al.2004), natural AMPs are rarely explored for their regulation of the IFN system, which includes type-I and II IFN immune responses (Biragyn et al.2002; Lande et al.2007).
Smp76, from the venom of the scorpion Scorpio maurus palmatus, possesses 76 amino acid residues and is approximately 8397 Da in size (Abdel-Rahman et al.2013). Its N-terminal amino acid sequence is similar to cecropins, whereas its C-terminal region has three pairs of disulfide bridges, sharing the same CSαβ motif with defensins (Renaud Conde et al.2000). Sequence similarity showed that Smp76 belongs to the family of β-KTx-like scorpine from Pandinus imperator and HS-1 from Heterometrus laoticus (Renaud Conde et al.2000; Uawonggul et al.2014). Considering the high sequence similarity of Smp76 with scorpine and HS-1 (Fig. 1A), we examined the antiviral activity of Smp76, a venom peptide previously characterized from the scorpion Scorpio maurus palmatus (Abdel-Rahman et al.2013). In the present study, we implemented a fusion expression and purification strategy in Escherichia coli BL21(DE3) to obtain the recombinant venom peptide (rSmp76) and then tested it on cultured cell lines and mouse macrophages to examine its antiviral effects and mechanisms.
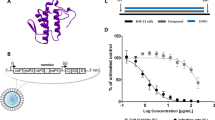
Expression, purification, and identification of the recombinant peptide Smp76 (rSmp76). A Sequence alignment of the three scorpine-like peptides. Highly conserved cysteine residues are marked in yellow font, forming three pairs of disulfide bonds. B The cDNA coding sequence of Smp76 mature peptide was cloned into the expression vector pET-32a at the Kpn I-Xho I restriction sites for cloning and expression of rSmp76 in E. coli BL21(DE3). C SDS-PAGE of rSmp76 purified from E. coli BL21(DE3). Lane 1, protein marker; lane 2, E. coli containing pET-32a-rSmp76 without induction; lane 3, E. coli induced with IPTG; lane 4, ultrasonicated supernatant from induced E. coli; lane 5, ultrasonicated sediment of induced E. coli; lane 6, purified His-tag fusion protein after affinity chromatography; lane 7, fusion protein cleaved by enterokinase; lane 8, rSmp76 after HPLC purification. D HPLC profile of rSmp76 after cleavage by enterokinase. E HPLC purity analysis of rSmp76 with an analytical column. F MALDI-TOF-MS mass spectrum of the purified rSmp76.
Materials and Methods
Cells and Viruses
The human lung adenocarcinoma cell line A549, human hepatoma cell line Huh7, and African green monkey kidney cell line Vero were cultured in Dulbecco’s modified Eagle’s medium (Gibco DMEM; Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (Gibco FBS; Thermo Fisher Scientific) and 1% penicillin/streptomycin at 37 °C in a humidified 5% CO2 incubator. The human monocytic cell line THP-1 was cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific) supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C in a humidified 5% CO2 incubator. THP-1 cells were differentiated to macrophages with 60 nmol/L phorbol-12-myristate-13-acetate (TPA) for 12–14 h and then cultured for 24 h without TPA. The Aedes albopictus mosquito cell line C6/36 was cultured at 28 °C without CO2 in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Peritoneal macrophages were harvested from mice by washing with PFE (PBS supplemented with 5% FBS and 2 mmol/L EDTA) 3 days after injecting the mice intraperitoneally with 4% thioglycollate medium.
DENV serotype-2 TSV01 strain (DENV-2 TSV01) was kindly provided by Dr. Bo Zhang from the Wuhan Institute of Virology, Chinese Academy of Sciences. DENV serotype-2 New Guinea C strain (DENV-2 NGC) was kindly gifted by Prof. Jianguo Wu from Wuhan University. The HSV-1 F strain was stored in our laboratory. ZIKV Puerto Rico strain (PRVABC59) cDNA plasmid (pBR322-Z2) was kindly provided by Dr. Ren Sun and Dr. Danyang Gong from the University of California, Los Angeles. ZIKV mRNA was obtained by transcription of recombinant plasmid in vitro, after which ZIKV mRNA was transfected into Vero cells. Zika virus was harvested 6 days later.
Mice
Ifnar1-/- mice with a C57BL6/J genetic background were kindly provided by Professor Bo Zhong from Wuhan University. Wild-type C57BL6/J mice were purchased from the Hubei Provincial Center for Disease Control and Prevention, China. Mice were maintained in the specific pathogen-free facility of Wuhan University.
Antibodies and Reagents
Antibodies used were as follows, DENV type 1–4 mouse monoclonal antibody (sc-57,800; Santa Cruz Biotechnology, Dallas, TX), IRF3 rabbit monoclonal antibody (D83B9; Cell Signaling Technologies, Danvers, MA), p-IRF3 rabbit monoclonal antibody (S396; Cell Signaling Technologies), anti-GAPDH mouse antibody (60004-1-Ig; Proteintech, Rosemont, IL), and goat HRP conjugated anti-mouse IgG (GM01H; NovoGene, Beijing, China). Bestar® SybrGreen qPCR master mix reagent was purchased from (DBI® Bioscience, Ludwigshafen, German). Loading sample buffer (5 ×; BL502A) was obtained from Biosharp, Hefei, China. Protein markers used were PageRuler Prestained Protein Ladder (26617; Thermo Fisher Scientific) and Protein Marker Low Range (66–4.1 kDa; Kerun Biology, Chongqing, China). The protease and phosphatase inhibitor cocktails MCE HY-K0010 and MCE HY-K0022, respectively, were purchased from MedChemExpress (Monmouth Junction, NJ).
Peptide Expression, Purification, and Characterization
The amino acid sequence of Smp76 peptide was obtained from published data (Abdel-Rahman et al.2013). Smp76 cDNA was reverse translated from the protein sequence. We used overlapping PCR to insert the DNA fragment into the expression plasmid pET-32a with Kpn I and Xho I restriction enzymes and an enterokinase cleavage site (DDDDK; Fig. 1B). The recombinant vector was then transformed into the chemically competent Escherichia coli BL21(DE3). Next, recombinant E. coli BL21(DE3) was cultured in LB medium with 100 µg/mL ampicillin at 37 °C and when cell density reached OD600 = 0.5, isopropyl β-D-thiogalactoside (IPTG) was added at a final concentration of 1 mmol/L. The cells were then lysed by ultrasonication at 400 Hz for 99 cycles and the supernatant containing the fusion peptide was collected. The obtained supernatant was subjected to Ni-chelating affinity chromatography and the fusion peptide was dialyzed against 1enterokinase buffer for 5 h. Next, the fusion peptide was cleaved with an enterokinase (Morebio, Wuhan, China) overnight at 23 °C and separated by reverse-phased high-performance liquid chromatography (RP-HPLC) using a C18 column (10 × 250 mm, 5 μm; Elite HPLC, Dalian, China). Finally, the target peptide was collected by detecting at a wavelength of 230 nm and immediately freeze-dried. The purity and molecular mass of rSmp76 were confirmed by RP-HPLC and matrix-assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF–MS), respectively.
RNA Extraction and Quantitative Reverse Transcription (qRT)-PCR
Total RNA of cells or tissues was extracted by TRIzol reagent (TaKaRa, Kusatsu, Japan) and then reverse transcribed to synthesize first-strand cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). cDNA was quantified by qRT-PCR using the Bestar® SybrGreen qPCR master mix reagent on an ABI 7500 Fast Instrument (Applied Biosystems, Foster City, CA) using standard cycling conditions. The detailed primers for qRT-PCR were in Supplementary Table S1.
Western Blot Analysis
Cells were lysed with RIPA lysis buffer plus the protease and phosphatase inhibitor cocktails on ice. The lysate was centrifuged for 15 min at 14,400 ×g and the supernatant was collected to quantify the obtained proteins with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). An equal amount (20 μg) of proteins was loaded and separated by SDS-PAGE and then transferred to a nitrocellulose membrane (Merck Millipore, Burlington, MA). The membrane was blocked in 5% bovine serum albumin (BSA) for 2 h at room temperature and then incubated with primary antibodies (1;1000; anti-E protein, p-IRF3, and IRF3 antibodies) at 4 °C overnight. The membrane was then exposed to HRP-conjugated anti-mouse or anti-rabbit secondary antibodies (1:10,000) at room temperature for 2 h and the results visualized with an enhanced chemiluminescence kit (Super RX-N–C; Yestar Healthcare Holdings Company, Guangxi, China).
Plaque assay
A549 or Vero cells were seeded in 12-well plates (2 × 105 cells/well) with DMEM high glucose, 10% FBS, and 1% penicillin/streptomycin at 37 °C. When the cells were confluent, the medium was removed, and cells were washed with PBS. Then, viruses in DMEM supplemented with 1% FBS and 1% penicillin/streptomycin were added for 1 h. The unadsorbed viruses were washed off and cells were overlaid with 1 mL 1% agarose containing 2% FBS and 1% penicillin/streptomycin. The plates were cultured at 37 °C for 3 and 5 days for Vero and A549 cells, respectively. Finally, cells were stained with 1 mL 1% crystal violet solution in 10% methanol for 3 h, after which the stain was removed with tap water and the plaques were counted. The virus titter was expressed as plaque forming unites (PFU) per milliliter.
MTT Assay
Cells were seeded in 96-well plates (7,000–10,000 cells/well) and cultured at 37 °C for 24 h. A series of peptides at different concentrations were added into the medium and then cells were cultured at 37 °C for 48 h. Next, 20 µL MTT (Invitrogen, Carlsbad, CA) solution (5 mg/mL in PBS buffer) was added to each well and the plate was further incubated at 37 °C for 4 h. After removing the medium, 100 µL DMSO was added and the plate was incubated for 20 min at room temperature with shaking to completely dissolve the crystal purple formazan. Finally, absorbance was measured at 570 nm.
Peptide Antiviral Assay
As mentioned, cells were seeded in 12-well plates (2 × 105 cells/well) and cultured overnight, after which the medium was removed and the cells rinsed with PBS. Next, the cells were incubated with virus (multiplicity of infection [MOI] = 0.1; 2 × 104 PFU) and peptides in FBS-free DMEM. After 1 h of incubation, cells were rinsed and replenished with medium containing peptides throughout the experiment. DENV RNA was then subjected to qRT-PCR after 48 h.
Viral Attachment Assay
A549 cells were incubated with virus (2 × 104 PFU) and peptides at 4 °C for 1 h and then rinsed with PBS and shifted to 37 °C for viral entry. After 1 h of incubation, cells were rinsed and replenished with fresh medium. DENV RNA was then subjected to qRT-PCR after 48 h.
Viral Entry Assay
Cells were infected with virus (2 × 104 PFU) for 1 h at 4 °C for viral attachment to cells, and then cells were rinsed before adding peptides. The incubation temperature was then shifted to 37 °C for viral entry. After 1 h of incubation, cells were rinsed and replenished with fresh medium. DENV RNA was then subjected to qRT-PCR after 48 h.
Viral Inactivation Assay
rSmp76 (40 µmol/L) was incubated with virus (2 × 104 PFU) for 1 h at 37 °C and then the mixture was diluted 50× and added to the cells for 1 h. After inoculation, the cells were rinsed with PBS and replenished with fresh medium. DENV RNA was then subjected to qRT-PCR after 48 h.
RNase Digestion Assay
Released genomic RNA from DENV was detected by RNase digestion assays and qRT-PCR as described by Yu et al. (2016). Briefly, DENV (2 × 104 PFU) was incubated with rSmp76 at room temperature for 1 h. Released genomic RNA from treated DENV was then digested with RNase A (D6950-01; Omega, Norcross, GA) at 37 °C for 1 h. After inactivation of residual RNase A, undigested genomic RNA in the intact viral particles was extracted using TRIzol reagent, reversed transcribed, and quantified as mentioned above.
Post-infection Assay
Cells were infected with virus (2 × 104 PFU), which was then removed after 1 h at 37 °C. The cells were rinsed and replenished with medium containing peptides throughout the experiment and then DENV RNA was subjected to qRT-PCR after 48 h.
Cell-Stimulation Assay
Cells were incubated with peptides for 1 h at 37 °C and then peptide-treated cells were rinsed before virus infection (2 × 104 PFU) for 1 h at 37 °C. Next, the cells were rinsed and replenished with fresh medium and then DENV RNA was subjected to qRT-PCR after 48 h.
qRT-PCR and Enzyme-Linked Immunosorbent Assay (ELISA) of IFN-β and Interferon-Stimulated Genes (ISGs)
Cells were seeded in 12-well plates at 2 × 105 cells/well. After overnight culturing, the medium was removed, and the cells were rinsed and replenished with FBS-free medium. Four different treatments were performed as follows, the mock and Smp76 groups were left uninfected, while the DENV and Smp76 + DENV groups were infected with DENV (MOI = 1; 2 × 105 PFU) for 1 h, after which the medium was removed and cells were rinsed with PBS. Mock and DENV groups were replenished with fresh medium, while Smp76 and Smp76 + DENV groups were replenished with fresh medium containing rSmp76 peptides. After 12 h, RNA and supernatants were harvested for qRT-PCR and ELISA, respectively. ELISA was performed using the VeriKine Human IFN βeta ELISA Kit (catalog no. 41410; PBL Assay Science, Piscataway, NJ) according to manufacturer’s instructions.
Statistical Analysis
Data were analyzed using GraphPad Prism software (GraphPad Software Inc., La Jolla, CA) and expressed as the mean ± standard deviation (SD) of at least three experiments.
Results
Prokaryotic Expression, Purification, and Identification of rSmp76
Scorpions are small arachnids that are mostly 5–6 cm in length and secrete a low amount of scorpion venom; a single scorpion specimen can normally produce 1–3 µL venom by mild electrical stimulation. It is thus difficult to isolate, purify, and identify the monomeric peptide component from natural scorpion venom. It is even more difficult to prepare a sufficient amount of scorpion venom peptide to investigate its biological functions and related mechanisms. Thus, developing a strategy for producing sufficient amounts of a single scorpion venom peptide is very important. To address these shortcomings, we constructed a prokaryotic expression and purification system for the scorpion venom peptide Smp76, which was previously characterized from Scorpio maurus palmatus, by an integrated strategy of fusion proteins, affinity chromatography, and peptide release by enzymatic cleavage (Fig. 1B–1F). High-purity rSmp76 was prepared as described in the materials and methods section. SDS-PAGE and HPLC analysis of rSmp76 demonstrated that the collected component was a single peptide (Fig. 1C–1E), while MALDI-TOF-MS (Fig. 1F) revealed that the experimental molecular mass of the peptide containing three disulfide bonds (8397.24 Da) was similar to its theoretical molecular mass (8397.07 Da; Fig. 1A).
Smp76 Inhibits DENV/ZIKV RNA Infection and Protein Synthesis
Smp76 shares high sequence similarity with two scorpion venom peptides, scorpine and HS-1, that possess antibacterial activities against multiple microorganisms (Fig. 1A) (Carballar-Lejarazu et al.2008; Uawonggul et al.2014). In addition, scorpine exhibits the inhibitory activity to DENV (Carballar-Lejarazu et al.2008). Thus, we tested the antiviral activity of rSmp76 and found that rSmp76 can inhibit DENV-2 (TSV01) RNA replication (Fig. 2A) and protein synthesis (Fig. 2B) in A549 cells in a dose-dependent manner. Inhibitory rates of DENV-2 (TSV01) infection were 10.4, 26.4, 44.1 and 75.7% by rSmp76 under concentrations 1, 2, 5 and 10 µmol/L, respectively. The IC50 value of rSmp76 against DENV-2 (TSV01) was 6.21 µmol/L in A549 cells. Viral infectious particles in the supernatant were also reduced significantly (Fig. 2C, Fig. 2D). Additionally, the more virulent DENV-2 strain NGC was also used to test the antiviral activity of rSmp76, which led to similar results as for DENV-2 strain TSV01 (Fig. 2E–2H). Furthermore, DENV-2 (TSV01) infection was effectively inhibited by rSmp76 in other cells lines, including Huh7, THP-1, and primary mouse macrophages (Fig. 2I–2K), and the half maximal inhibitory concentrations (IC50) for rSmp76 against DENV-2 (TSV01) were revealed to be 6.21, 6.52, 4.69, and 5.17 µmol/L in A549, Huh7, and THP-1 cells as well as primary mouse macrophages, respectively. Meanwhile, cellular infection with ZIKV (Fig. 2L–2N) and HCV (data not shown) were also suppressed by the rSmp76 peptide. The inhibitory rates of rSmp76 against ZIKV were 17.3%, 34.9%, 38.8% and 73.8%, respectively, under the concentrations of 1, 2, 5 and 10 μmol/L. The IC50 value of rSmp76 against ZIKV was 6.63 μmol/L (Fig. 2L–2N). In contrast, rSmp76 did not affect HSV-1 infection, even under different concentrations (Supplementary Fig. S1 A–C). Altogether, the scorpion venom peptide Smp76 was found to effectively inhibit cellular infection by various viruses under non-cytotoxic concentrations (Supplementary Fig. S2).
Dose-dependent inhibitory activity of rSmp76 against DENV and ZIKV in vitro. A–D rSmp76 dose-dependent inhibition of DENV-2 (TSV01) infected-A549 cells. E–H rSmp76 dose-dependent inhibition of DENV-2 (NGC) infected-A549 cells. A, E RNA levels were analyzed by qRT-PCR. B, F Intracellular DENV-2 E protein levels were analyzed by western blotting. C, D, G, H Extracellular DENV-2 particles were diluted 100 × and quantified by plaque formation. I–K rSmp76 dose-dependent inhibition of DENV-2 (TSV01) infected-Huh7, THP-1, and mouse peritoneal macrophages (PM) analyzed by qRTPCR. L Dose-dependent inhibition of rSmp76 to ZIKV (PRVABC59) infection in A549 cells. (M and N) Plaque assay. Extracellular ZIKV (PRVABC59) particles were diluted 100 × and quantified by plaque formation. Data represent the mean ± SD of three independent experiments. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 (Student’s t-test). Con, control. PM, mouse peritoneal macrophages. The internal controls of subfigure A, E and I–L are GAPDH.
rSmp76 Plays Antiviral Roles at the Post-infection Stage of the Viral Life Cycle
rSmp76 inhibited DENV infection in multiple cell types (Fig. 2A, 2I–2K). To investigate the antiviral mechanism of rSmp76, we incubated the viruses and/or cells with rSmp76 at different stages of the viral life cycle (Fig. 3A) and analyzed its antiviral activity (Fig. 3B–3G). We found that rSmp76 did not inactivate viral particles directly (Fig. 3B), as it did not affect virus infectivity after co-incubating with the peptide before infecting cells with the virus. Furthermore, rSmp76 also did not inhibit virus attachment to cells (Fig. 3D) nor viral entry (Fig. 3E). Interestingly, rSmp76 exhibited significant antiviral activity when pre-incubated with cells before infection (Fig. 3C) and after establishment of DENV infections (Fig. 3F). Thus, our findings suggest that rSmp76 mainly plays antiviral roles at the post-infection stage of the viral life cycle.
Analysis of scorpion venom peptide rSmp76 effects during the viral life cycle. A Schematic diagram summarizing cells and/or virus treatment with rSmp76 (10 μmol/L). B Viral inactivation assay. C Cell stimulation assay. D Viral attachment assay. E Viral entry assay. F Post-infection assay. G RNase digestion assay. Data represent the mean ± SD of three independent experiments. **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001 (Student’s t-test). Con, control. The internal controls of subfigure B–F are GAPDH.
rSmp76 Upregulates IFN-β by Activating IRF3 Phosphorylation
IFN-mediated antiviral responses are critical against viral infection and pathogenesis (Schoggins et al.2011; Seth et al.2006; Muller et al.1994). Therefore, we examined whether the antiviral function of rSmp76 is associated with IFNs. The following IFNs screening revealed that rSmp76 antiviral activity was associated with IFN-β regulation (Fig. 4). When rSmp76 or IFN-β (1 ng/mL) were added to cells 1 h post-infection, both exhibited antiviral activity (Fig. 4A). In contrast, when rSmp76 or IFN-β (1 ng/mL) were added to cells 2 days post-infection, neither exhibited any antiviral activity (Fig. 4B). The consistency in antiviral activity between rSmp76 and IFN-β indicated a relationship. In subsequent experiments, we observed that rSmp76 promotes IFN-β expression at the transcriptional and translational levels in both uninfected and infected cells (Fig. 4C, 4D). When virus-infected cells were treated with rSmp76, IFN-β expression was upregulated in a dose-dependent manner (Fig. 4E). For further confirmation, we tested the antiviral activity of rSmp76 in peritoneal macrophages from IFN-α/β receptor-deficient (Ifnar1-/-) and wild-type mice. Indeed, rSmp76 was found to inhibit DENV infection in peritoneal macrophages from wild-type mice (Fig. 4F) but not in those from Ifnar–/– mice (Fig. 4G), suggesting that the antiviral activity of rSmp76 relies on the type-I IFN system. rSmp76 was also found to upregulate IFN-β expression by enhancing IRF3 phosphorylation (Fig. 4H). Thus, we concluded that rSmp76 inhibits DENV by enhancing IRF3 phosphorylation, which triggers IFN-β upregulation. Furthermore, it was previously reported that IFN-β does not inhibit HSV-1 infection efficiently in vitro as HSV-1 is not sensitive to IFN-β (Sainz and Halford 2002); this may partly explain why rSmp76 did not affect HSV-1 infection under our experimental conditions (Supplementary Fig. S1 A–C).
rSmp76 upregulates IFN-β by activating IRF3 phosphorylation. A Comparison of the antiviral activity of rSmp76 (10 μmol/L) and IFN-β (1 ng/mL) against DENV at 1 h post-infection. B Comparison of the antiviral activity of rSmp76 (10 μmol/L) and IFN-β (1 ng/mL) against DENV at 48 h post-infection. C qRT-PCR expression levels of IFN-β. D ELISA of IFN-β protein levels in supernatants. E rSmp76 dose-dependent enhancement of IFN-β expression detected in the supernatant. F and G Antiviral effect of rSmp76 on peritoneal macrophages from wild type C57BL6/J (F) and Ifnar1-/- mice (G). DENV RNA levels were normalized to GAPDH. H Effect of rSmp76 on IRF3 phosphorylation by western blot analysis. The A549 cells were infected by DENV-2 (TSV01) with MOI = 1 for 12 h. Data represent the mean ± SD of three independent experiments. *P ≤ 0.05; ***P ≤ 0.001; ****P ≤ 0.0001 (Student’s t-test). Con, control. The internal controls of subfigure A, B and C are GAPDH.
Effects of rSmp76 on Downstream ISGs
When IFN-α/β receptors receive stimulation from IFN-β, hundreds of cellular genes are upregulated, leading to antiviral responses (Jones et al.2005; Morrison et al.2013). ISG15 and OAS2 are the major downstream ISGs of IFN-β, and both possess anti-DENV functions (Hishiki et al.2014; Dai et al.2011; Lin et al.2009). After exposing A549 cells to four different treatments (see “Materials and Methods” section), qRT-PCR was employed to detect ISG15 and OAS2 mRNA levels. As a result, ISG15 and OAS2 were found upregulated in response to rSmp76 treatment of both uninfected and infected cells (Fig. 5A, 5C). When cells were infected with DENV, ISG15 and OAS2 were also upregulated in a dose-dependent manner (Fig. 5B, 5D). However, ISG15 level of DENV-infected Smp76-treated cells was lower than that of uninfected Smp76-treated cells, which may be related to the fact that DENV can degrade STAT2, downregulating transcription and translation of ISG15 (Morrison et al.2013). Overall, our findings indicate that rSmp76 upregulates IFN-β, which increases expression levels of ISG15 and OAS2 and results in viral infection inhibition.
rSmp76 upregulates ISGs downstream of the type-I IFN response. A qRT-PCR expression levels of ISG15. B Dose-dependent enhancement of ISG15 expression levels in DENV-infected cells treated with rSmp76. C qRT-PCR expression levels of OAS2. D Dose-dependent enhancement of OAS2 expression levels in DENV-infected cells treated with rSmp76. Data represent the mean ± SD of three independent experiments. *P ≤ 0.05; **P ≤ 0.01 (Student’s t-test). Con, control. The internal controls of subfigure A–D are GAPDH.
Discussion
AMPs is an important self-protective factor against pathogenic microorganisms. In the present study, rSmp76 was found to inhibit DENV and ZIKV in vitro in a dose-dependent manner. Moreover, we found that Smp76 is not a virucidal peptide because it does not inactivate virus particles directly (Fig. 3B) or destroy viral envelope proteins tested by RNase digestion assays (Fig. 3G), but rather stimulates cells to upregulate IFN-β, achieving antiviral function. In previous studies, most peptides, including natural (Carballar-Lejarazu et al.2008; Holthausen et al.2017; Hong et al.2014; Jiang et al.1993; Yasin et al.2004) and synthetic (Hong et al.2013; Schmidt et al.2010; Yu et al.2017b) peptides, were revealed to play antiviral roles by inactivating viral particles or inhibiting viral cellular entry. In addition to direct virucidal activity, some AMP members were also reported to possess immunomodulatory functions (Easton et al.2009; Hancock and Finlay 2004; Hilchie et al.2013; Scott et al.2007). For example, the peptide Mucroporin-M1 can activate the MAPK pathway and downregulate HNF4α to inhibit HBV replication in vitro and in vivo (Zhao et al.2012). In fact, the greatest difficulty during treatment of viral diseases is clearance of established infections. Our study demonstrated that the scorpion venom peptide Smp76 can upregulate IFN-β to suppress the established viral infections, which is a more potent and efficient way than traditional AMPs to treat viral infections.
Treatment of viral infections with direct-acting antivirals (DAAs) may exhibit decreased efficacy due to the high variability of viruses (Theofilopoulos et al.2005). Therefore, an alternative treatment may enhance the protective effects of host innate immunity, such as IFN activation, against DENV and ZIKV progression. Such a strategy can potentially avoid drug resistance and the effects of genetic variability in the viral genome (Yu et al.2017a). Numerous studies have demonstrated that type-I IFN is involved in the pathogenesis of DENV infection and the progression from undifferentiated febrile illness (DF) to life-threatening infection (Pichlmair and Reis e Sousa 2007; Ubol et al.2008). Therefore, the Smp76-induced IFN response may function as a potential anti-DENV factor by exerting cytoprotective effects against DENV-associated cell damage and life-threatening symptoms associated with dengue hemorrhagic fever and dengue shock syndrome (Yu et al.2017a).
Smp76, a scorpine-like peptide from the venom of the scorpion Scorpio maurus palmatus, was successfully expressed and purified in an E. coli BL21(DE3) system by an integrated strategy of fusion proteins, affinity chromatography, and enzymatic cleavage. Smp76 was found to inhibit DENV and ZIKV infections in cultured human cell lines and primary mouse macrophages in a concentration-dependent manner. Mechanistically, Smp76 does not inactivate viral particles directly but suppresses established viral infection by upregulating IFN-β expression through IRF3 phosphorylation, which enhances type-I IFN responses to inhibit viral infection. However, it is necessary to elucidate the detailed molecular mechanisms of Smp76 that promotes IFN-β expression during viral infection in future studies.
References
Abdel-Rahman MA, Quintero-Hernandez V, Possani LD (2013) Venom proteomic and venomous glands transcriptomic analysis of the Egyptian scorpion Scorpio maurus palmatus (Arachnida: Scorpionidae). Toxicon 74:193–207
Alesha Grant SS, Tripathi Shashank, Balasubramaniam Vinod, Lisa Miorin MS, Schwarz Megan C, Sánchez-Seco Mari Paz, Matthew J, Evans SM, García-Sastre A (2016) Zika virus impairs growth in human neurospheres and brain organoids. Cell Host Microbe 19:882–890
Biragyn A, Ruffini PA, Leifer CA, Klyushnenkova E, Shakhov A, Chertov O, Shirakawa AK, Farber JM, Segal DM, Oppenheim JJ, Kwak LW (2002) Toll-like receptor 4-dependent activation of dendritic cells by β-Denfensin 2. Science 298:1025–1029
Bogoch II, Brady OJ, Kraemer MU, German M, Creatore MI, Brent S, Watts AG, Hay SI, Kulkarni MA, Brownstein JS, Khan K (2016) Potential for Zika virus introduction and transmission in resource-limited countries in Africa and the Asia-Pacific region: a modelling study. Lancet Infect Dis 16:1237–1245
Carballar-Lejarazu R, Rodriguez MH, Hernandez-Hernandez FC, Ramos-Castaneda J, Possani LD, Zurita-Ortega M, Reynaud-Garza E, Hernandez-Rivas R, Loukeris T, Lycett G, Lanz-Mendoza H (2008) Recombinant scorpine: a multifunctional antimicrobial peptide with activity against different pathogens. Cell Mol Life Sci 65:3081–3092
Chan JF, Choi GK (2016) Zika fever and congenital Zika syndrome: an unexpected emerging arboviral disease. J Infect Dis 220:449
Cui X, Wu Y, Fan D, Gao N, Ming Y, Wang P, An J (2018) Peptides P4 and P7 derived from E protein inhibit entry of dengue virus serotype 2 via interacting with β3 integrin. Antivir Res 155:20–27
Dai JF, Pan W, Wang PH (2011) ISG15 facilitates cellular antiviral response to dengue and west nile virus infection in vitro. Virol J 8:486–492
Easton DM, Nijnik A, Mayer ML, Hancock RE (2009) Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends Biotechnol 27:582–590
Halstead SB (2007) Dengue. Lancet 370:1644–1652
Hancock RE, Finlay BB (2004) Can innate immunity be enhanced to treat microbial infections. Nat Rev Microbiol 2:497–504
Heymann DL, Hodgson A, Sall AA, Freedman DO, Staples JE, Althabe F, Baruah K, Mahmud G, Kandun N, Vasconcelos FC, Bino S, Menon KU (2016) Zika virus and microcephaly: why is this situation a PHEIC? Lancet 387:719–721
Hilchie AL, Wuerth K, Hancock RE (2013) Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat Chem Biol 9:761–768
Hishiki T, Han QE, Arimoto K, Shimotohno K, Igarashi T, Vasudevan SG, Suzuki Y, Yamamoto N (2014) Interferon-mediated ISG15 conjugation restricts dengue virus 2 replication. Biochem Biophys Res Commun 448:95–100
Holthausen DJ, Lee SH, Kumar VT, Bouvier NM, Krammer F, Ellebedy AH, Wrammert J, Lowen AC, George S, Pillai MR, Jacob J (2017) An amphibian host defense peptide is virucidal for human H1 hemagglutinin-bearing influenza viruses. Immunity 46:587–595
Hong W, Zhang R, Di Z, He Y, Zhao Z, Hu J, Wu Y, Li W, Cao Z (2013) Design of histidine-rich peptides with enhanced bioavailability and inhibitory activity against hepatitis C virus. Biomaterials 34:3511–3522
Hong W, Li T, Song Y, Zhang R, Zeng Z, Han S, Zhang X, Wu Y, Li W, Cao Z (2014) Inhibitory activity and mechanism of two scorpion venom peptides against herpes simplex virus type 1. Antivir Res 102:1–10
Jiang SB, Lin K, Strick N, Neurath AR (1993) HIV-1 inhibition by a peptide. Nature 365:113
Jones M, Davidson A, Hibbert L, Gruenwald P, Schlaak J, Ball S, Foster GR, Jacobs M (2005) Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J Virol 79:5414–5420
Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Wang YH, Su B, Nestle FO, Zal T, Mellman I, Schroder JM, Liu YJ, Gilliet M (2007) Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 449:564–569
Lin RJ, Yu HP, Chang BL, Tang WC, Liao CL, Lin YL (2009) Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J Immunol 183:8035–8043
Loo YM, Gale M Jr (2011) Immune signaling by RIG-I-like receptors. Immunity 34:680–692
McMichael AJ, Woodruff RE, Hales S (2006) Climate change and human health: present and future risks. Lancet 367:859–869
Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, Mulder LC, Fernandez-Sesma A, Garcia-Sastre A (2013) Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog 9:e1003265
Muller U, Steinhoff U, Reis LF, Hemmi S, Paviovic J, Zinkernagel RM, Aguet M (1994) Functional role of type I and type II Interferons in antiviral defense. Science 264:1918–1921
Pichlmair A, Reis e Sousa C (2007) Innate recognition of viruses. Immunity 27:370–383
Reddy KV, Yedery RD, Aranha C (2004) Antimicrobial peptides: premises and promises international. J Antimicrob Agents 24:536–547
Renaud Conde FZ, Rodriguez Mario H, Possani Lourival D (2000) Scorpine, an anti-malaria and anti-bacterial agent purified from scorpion venom. FEBS Lett 471:165–168
Sainz B, Halford WP (2002) Alpha/beta interferon and gamma interferon synergize to inhibit the replication of herpes simplex virus type 1. J Virol 76:11541–11550
Schmidt AG, Yang PL, Harrison SC (2010) Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Pathog 6:e1000851
Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485
Scott MG, Dullaghan E, Mookherjee N, Glavas N, Waldbrook M, Thompson A, Wang A, Lee K, Doria S, Hamill P, Yu JJ, Li Y, Donini O, Guarna MM, Finlay BB, North JR, Hancock RE (2007) An anti-infective peptide that selectively modulates the innate immune response. Nat Biotechnol 25:465–472
Screaton G, Mongkolsapaya J, Yacoub S, Roberts C (2015) New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol 15:745–759
Seth RB, Sun L, Chen ZJ (2006) Antiviral innate immunity pathways. Cell Res 16:141–147
Shepard DS, Undurraga EA, Halasa YA, Stanaway JD (2016) The global economic burden of dengue: a systematic analysis. Lancet Infect Dis 16:935–941
Theofilopoulos AN, Baccala R, Beutler B, Kono DH (2005) Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol 23:307–336
Uawonggul N, Sukprasert S, Incamnoi P, Patramanon R, Thammasirirak S, Preecharram S, Bunyatratchata W, Kuaprasert B, Daduang J, Daduang S (2014) Bacterial overexpression ofrecombinant heteroscorpine-1 (rHS-1), a toxin from Heterometrus laoticus scorpion venom: trends forantibacterial application and antivenom production. Biochem Genet 52:459–473
Ubol S, Masrinoul P, Chaijaruwanich J, Kalayanarooj S, Charoensirisuthikul T, Kasisith J (2008) Differences in global gene expression in peripheral blood mononuclear cells indicate a significant role of the innate responses in progression of dengue fever but not dengue hemorrhagic fever. J Infect Dis 197:1459–1467
Yasin B, Wang W, Pang M, Cheshenko N, Hong T, Waring AJ, Herold BC, Wagar EA, Lehrer RI (2004) θ Defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. J Virol 78:5147–5156
Yu JS, Tseng CK, Lin CK, Hsu YC, Wu YH, Hsieh CL, Lee JC (2017a) Celastrol inhibits dengue virus replication via up-regulating type I interferon and downstream interferon-stimulated responses. Antivir Res 137:49–57
Yu Y, Deng Y, Zou P, Wang Q, Dai Y, Yu F, Du L, Zhang N, Tian M, Hao J, Meng Y, Li Y, Zhou X, Chan JF, Yuen KY, Qin CF, Jiang SB, Lu L (2017b) A peptide-based viral inactivator inhibits Zika virus infection in pregnant mice and fetuses. Nat Commun 8:15672
Zeng Z, Zhang R, Hong W, Cheng Y, Wang H, Lang Y, Ji Z, Wu Y, Li W, Xie Y, Cao Z (2018) Histidine-rich modification of a scorpion-derived peptide improves bioavailability and inhibitory activity against HSV-1. Theranostics 8:199–211
Zhao Z, Hong W, Zeng Z, Wu Y, Hu K, Tian X, Li W, Cao Z (2012) Mucroporin-M1 inhibits hepatitis B virus replication by activating the mitogen-activated protein kinase (MAPK) pathway and down-regulating HNF4alpha in vitro and in vivo. J Biol Chem 287:30181–30190
Acknowledgements
We are indebted to Dr. Bo Zhang from the Wuhan Institute of Virology, Chinese Academy of Sciences for kindly providing DENV serotype-2 TSV01 strain. We thank Dr. Ren Sun and Dr. Danyang Gong from the University of California, Los Angeles for sharing pBR322-Z2 plasmid. We gratefully acknowledge Prof. Wu Jianguo and Prof. Bo Zhong from Wuhan University for their kindly providing DENV-2 (NGC) and Ifnar1-/- mice, respectively. This work was supported by grants from National Science Fund of China (Nos. 31572289, 31872239 and 81630091), International S&T Cooperation Program of China (No. S2016G3110), Hubei Science Fund (Nos. 2015CFA042 and 2016CFA018), China-Kazakhstan Cooperation Program (No. CK-07-09), and Fundamental Research Funds for the Central Universities in China (Nos. 2042017kf0242 and 2042017kf0199). SAA is grateful for financial support from Higher Education Commission (HEC) of Pakistan.
Author information
Authors and Affiliations
Contributions
FFL, ZQX, XCG and ZLJ designed the experiments and analyzed the data. ZLJ, FS and MJG performed most of the experiments. ZLJ and YTC wrote the manuscript. SAA, WXL, YLW and ZJC revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Animal and Human Rights Statement
All animal experiments were in accordance with and were approved by the Institutional Animal Care and Use Committee of Wuhan University (Wuhan, China).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ji, Z., Li, F., Xia, Z. et al. The Scorpion Venom Peptide Smp76 Inhibits Viral Infection by Regulating Type-I Interferon Response. Virol. Sin. 33, 545–556 (2018). https://doi.org/10.1007/s12250-018-0068-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12250-018-0068-4