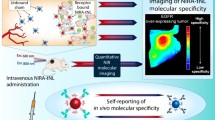
Abstract
Purpose
Abundance of receptors on tumor vasculature presents a prominent target for theranostic applications. The alphavbeta3 integrin receptors expressed on vascular endothelial cells during angiogenesis were therefore considered targets for imaging. Non-invasive visualization of tumor growth and/or delivery systems can appreciate tumor localization and disposition kinetics of carriers, respectively. Herein, we report near-infrared fluorescence imaging (NIRFI) of solid tumors using targeted fluorescence nanoliposomes in vivo.
Methods
Fluorescence nanoliposomes surface modified with cRGD-peptide were injected into CD1 athymic (nu/nu) mice bearing C6 glioblastoma xenografts (300 mm3). At different time points, mice were subjected to NIRFI for visualization of tumor xenografts and nanocarrier tracing in vivo.
Results
NIRFI showed tumor localization of 1,1′-dioctadecyl-3,3,3′,3′-tetramethyl indotricarbocyanine iodide (DiR18)-incorporated-targeted liposomes with maximum tumor-to-tissue occurring at 24-h post-liposome administration. Interaction of integrin receptors with targeted liposomes had contributed to an intense NIRF signal. Molecular studies showed an elevated expression of alphavbeta3 integrin receptors in tumor xenografts.
Conclusion
From the studies, it can be concluded that non-invasive localization of tumors and tracing of liposome carriers had been achieved using receptor targeting and NIRFI approaches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Angiogenesis, tumor growth, and metastasis are hallmark events of cancer [1, 2]. Specifically, angiogenesis is critical for growth and survival of tumors. When tumors reach 2 mm3, increased interstitial pressure within the tumor inhibits diffusion of metabolites and nutrients necessary for tumor growth [3, 4]. During this period, cellular hypoxia causes sprouting of new blood vessels (angiogenesis) [5]. New tumor sprouts “turn-on” angiogenic signals leading to activation of signaling molecules, (e.g., integrins, cadherins, selectins, and immunoglobulin families) which participate in different steps of cancer (e.g., benign-to-malignant tumor formation, tumor growth and progression, invasion, and metastasis) [6]. Cairns et al. showed upregulation of αvβ3 integrin receptors on proliferating tumor vessels [1].
Integrins are divalent cation-dependent heterodimeric membrane glycoproteins, which serve as cell adhesion receptors for extracellular matrix (ECM) proteins (e.g., fibrinogen, fibronectin, laminin, and vitronectin) [6]. Combination of α- and β-subunits determines ligand specificity, signaling, and functional properties of a given integrin. Overexpression of αvβ3 integrins is found in tumors and neo-vasculatures. Gasparini et al. correlated integrin expression levels with aggressiveness of cancer [7]. Most integrins recognize and bind to their respective proteins through short peptide sequences, such as Arg-Gly-Asp (RGD) [8]. This dimeric protein complex is involved in regulating tumor growth, angiogenesis, and metastasis [6]. Abundance of integrins on tumor vascular cell surfaces thus presents a RGD-based targeting for tumor imaging and drug delivery. Cyclic RGD compounds, such as cyclic pentapeptide (cRGDfK), have been shown to exhibit significant binding affinity and selectivity for αvβ3 integrin receptors [9].
Synthetic peptides have been investigated for targeting integrin receptors [8, 10]. We report liposome nanoparticles’ surface modified with lipid-peptide grafts for targeting tumor-expressed integrin receptors. Nanoliposomes offer distinct advantages [11]: (a) biodegradability, (b) structural ease to conjugate-targeting ligands, and (c) ability to incorporate tracer dyes for diagnostic applications. Saul et al. used post-incorporation method for intercalation of targeting ligands into lipid bilayers of doxorubicin-loaded liposomes [12]. It facilitated modulation of intercalation of targeting ligands in lipid bilayers and improved tumor targeting efficiency. Herein, we used post-incorporation method for surface modification of fluorophore-loaded liposomes with targeting ligands.
Various in vivo molecular imaging modalities were used for tumor localization and as management guidance for chemotherapy [13, 14]. Aiding to imaging instrumentation, novel contrast agents are essential for achieving imaging objectives. Relaxation of tissue-bound water protons for computed tomography (CT), radioisotopes (e.g., 123I, 99mTc, 111In, and 18F) for scintigraphy (positron emission tomography, PET/single-photon emission computed tomography, SPECT), supermagnetic or paramagnetic metals or complexes (e.g., Gd+3, Mn+2, and Fe+3) for magnetic resonance imaging (MRI), microbubbles (e.g., gas-filled lipid microbubbles) for ultrasound (US), and visible and near-infrared (NIR) dyes for optical imaging are examples. Each imaging method is unique in achieving imaging objectives, such as sensitivity, spatial and temporal resolution, depth of tissue penetration, and cost of the procedure. MRI, CT, and US methods possess high spatial and temporal resolution, but have low sensitivity in ligand detection [14]. The scintigraphic method has very high sensitivity for ligand detection, but suffers from poor spatial and temporal resolution. Optical imaging has high sensitivity and sub-cellular resolution for probe/tracer detection. On the other side, it has limited tissue penetration.
Using specific near-infrared fluorophores, optical imaging has been utilized for detection of tumors [15,16,17]. These near-infrared fluorophores emit light with tissue penetration approaching 10–15 cm [18]. Biological tissues exhibit a high photon absorbance in visible (350–700 nm) and infrared (> 900 nm) ranges with exception in the NIR region (700–900 nm). A significant advantage of NIRF-based optical imaging is absorbance spectra of biomolecules (e.g., oxyhemoglobin) reaching minima in the NIR region [10, 19]. In this study, NIRF tracer dye, 1,1′-dioctadecyl-3,3,3′,3′-tetramethyl indotricarbocyanine iodide (DiR18), was incorporated in lipid bilayers of liposomes for high-resolution NIRFI in vivo.
In the present study, optimized composition of targeted fluorescent nanoliposomes (targeted liposomes, (Fig. 1)) was administered to CD1 athymic (nu/nu) female mice bearing C6 glioblastoma xenografts, for NIRFI of solid tumors. NIRFI demonstrated localization of liposomes in tumor xenografts. Disposition kinetics of NIRF signals was considered as a function of temporal and spatial association of liposomes with integrin receptors on tumors. NIRFI signal intensities were correlated to αvβ3 integrin receptor levels in vivo. Also, we demonstrated αvβ3 integrin receptor specificity of liposomes in vitro. Plain fluorescent nanoliposomes (plain liposomes) served as control in various studies.
Illustration of targeted DiR18 fluorescence liposomes. Green- and black-colored twirls represent DSPE-PEG2000 and DSPE-PEG2000-Mal (functionalized lipid) moieties, respectively; cyclic pentane ring-shaped structures represent cRGD-peptide comprised of five amino acids (arginine-glycine-(d)-aspartic acid-phenylalanine-cysteine). Orange dots in lipid bilayers of liposomes represent DiR18, NIRF dye molecules. Also illustrated are functional sites of targeting ligand (cRGD-peptide). Arginine-glycine-(d)-aspartic acid of cyclopentane ring represents integrin receptor-binding site; cysteine represents conjugation site between peptide and lipid moieties; phenylalanine acts as linker amino acid between tri-peptide (arginine-glycine-(d)-aspartic acid) and cysteine
Methods
Materials
Hydrogenated soy phosphatidylcholine (HSPC), cholesterol (CHOL), distearoyl phosphatidylethanolamine-polyethylene glycol2000 (DSPE-PEG2000), and distearoyl phosphatidylethanolamine-polyethylene glycol2000-maleimide (DSPE-PEG2000-Mal) were obtained from Avanti Lipids (Birmingham, AL). DiR18 dye was obtained from Invitrogen (Boston, MA). Cyclic RGD-peptide was purchased from Peptide International (Louisville, KY). 125I-echistatin with a specific activity of 2000 μCi/mmol was purchased from Amersham Biosciences (Piscataway, NJ). Echistatin was purchased from Sigma (St. Louis, MO). All other chemicals were purchased from Sigma or Fisher Scientific (Suwannee, GA).
Design of Experiments for Optimization of Liposomes
I-optimal design was used to model and optimize targeted liposome composition and characteristics. Four parameters, HSPC/CHOL ratio, DSPE-PEG2000 concentration, DSPE-PEG2000-Mal-cRGD-peptide conjugate concentration, and incubation temperature, were identified as critical independent variables affecting response factors, ligand grafting efficiency, and mean diameter of liposomes. The I-optimal model was used for optimization runs. As shown in Table S1 (supplementary material 2), three levels of independent variables were selected to evaluate interaction effects. The + 1 and − 1 represent upper and lower limits of the ranges, respectively. Zero corresponds to the central point of levels. Independent variables were kept within the range, while response variables were either maximized or matched with those of the targets. Based on the I-optimal model, a total of 25 random runs were performed. Response variables could be related to independent variables through second-order polynomial function (Eq. 1) to estimate linear, quadratic, and interactions terms.
Where Yi represents predicted response, Xi and Xj are codes for independent variables, βo is intercept coefficient of second-order polynomial, βi is linear coefficient, βij is interaction coefficient. Statistical significance of response variables was determined by analysis of variance (ANOVA). A p value less than 0.05 was considered statistically significant. The model fit was checked accounting for R 2 and adjusted R 2 (adj-R 2) values. Statistical optimization runs were performed using JMP 10 software (SAS Inc., NC).
Preparation of Liposomes
Liposomes were prepared by methods similar to those described elsewhere [20]. Briefly, liposomes were prepared with an optimized composition of HSPC/CHOL/DSPE-PEG2000 in 54: 44: 2 M ratios. Organic solution comprised of lipids, cholesterol, and DiR18 fluorescent dye (10 μM) was vacuum dried at 45–48 °C to form a thin dried layer. This film was hydrated with (N-[2-hydroxyethyl]piperazine-N′-[2-ethanesulfonic acid]) (HEPES) buffer (50 mM, pH 6.8) at 65 °C to form multi-lamellar vesicles (MLVs). These MLVs were extruded (Northern Lipids, Canada) through 200/100-nm sandwiched polycarbonate nucleopore membrane filters (Whatman, MA) to obtain liposomes in the size range, 100–150 nm. The DiR18 entrapment efficiency was calculated using Eq. 2.
where A2 was the amount of DiR18 in lipid bilayers and A1 was the initial amount of DiR18 used for loading per 1 mL of liposomes; entrapment efficiency was expressed as %w/v.
Preparation and Characterization of Targeting Ligand Grafts
DSPE-PEG2000-Mal grafts were synthesized by addition reaction between maleimide and thiol groups of functionalized lipid (78.2 mg) and cRGD-peptide (15.5 mg), respectively. Lipid-peptide grafts were purified (for removal of free DSPE-PEG2000-Mal, cRGD-peptide, and TCEP HCl components) by dialysis (3000 Da MWCO, Pierce, Rockford, IL) against HEPES buffer, pH 6.5 at 4 °C for 12 h (h). The purified product was freeze-dried to obtain solid form. DSPE-PEG2000-Mal graft was characterized by MALDI-TOF (unpublished data).
Surface Modification of Liposomes with Ligand Grafts
DSPE-PEG2000-Mal-cRGD conjugates (46.8 mg; MW 3458.29 Da) were grafted onto the surface of liposomes (1 mL) at 55 °C (above phase transition temperature (Tm) of HSPC) [21]. After surface modification of liposomes with targeting ligand grafts, dialysis was performed to remove ungrafted conjugate. The cRGD-peptide (grafted as DSPE-PEG2000-Mal-cRGD conjugate on the liposome surface) and lipid concentration were quantified using HPLC [22] and colorimetric [23] methods, respectively. The percent targeting ligand grafting efficiency was calculated using Eq. 3.
where G2 was the amount of ligands grafted on the liposome surface and G1 was the initial amount of ligands used for grafting of 1 mL of liposomes; grafting efficiency was expressed in terms of percentage.
Physico-chemical Characterization of Liposomes
The mean diameter and zeta potential of liposomes were determined by dynamic light scattering (DLS) studies using zetasizer® (Malvern Inc., Westborough, MA). The morphology was determined by negative staining of liposome specimens on carbon support films. Aqueous solution (2% w/v) of uranyl acetate (pH 6.5) was employed to stain the specimens. Specimens were studied using a transmission electron microscope, functioning at 200 kV (JEOL-FX 2000).
Cell Lines
C6 glioblastoma cell line was obtained from the American Type Culture Collection (Manassas, VA) and maintained at 37 °C in 5% CO2-humidified atmosphere. Cells were cultured in modified Dulbecco’s medium (GIBCO, Carlsbad, CA) and 5% fetal bovine serum (FBS). C6 cell line was used to induce angiogenesis in vascular endothelial cells. Human umbilical vascular endothelial cells (HUVECs) were obtained from Cascade Biologics (Portland, OR) and cultured in medium 200 according to supplier’s instructions.
In Vitro Receptor-Binding Studies
Affinity of cRGD-peptide, conjugate, targeted liposomes, echistatin, and cRAD-peptide for αvβ3 integrin receptors on the surface of HUVECs was determined by competitive binding experiments using 125I-echistatin as the radioactive ligand. Receptor-binding assays were carried as described previously [24]. Briefly, HUVECs were harvested, washed with PBS, and resuspended (2 × 106 cells/mL) in a binding buffer (Tris-HCl, pH 7.4; 0.1% w/v BSA). The binding buffer containing HUVECs was aliquoted into micro-titer 96-well plates (filter pore size 0.65 μm). These plates were incubated with 125I-echistatin (0.05 nM/L) in the presence of increasing concentrations of cRAD-peptide (0.1 nM–5 μM/L), cRGD-peptide, or conjugate or targeted liposomes (containing equivalent to 0.1 nM–5 μM/L of cRGD-peptide). The total incubation volume was adjusted to 200 μL/well. After the cells were incubated at room temperature for 4 h, plates were washed with cold binding buffer. Micro-titer plate filters were collected and washed with 2 M NaOH (boiling), and radioactivity was determined using gamma counter (Packard, CT). The best fit IC50 values were calculated by fitting data to non-linear regression analysis (JMP 7, NC).
In Vivo Tumor Model
Animal procedures were performed according to a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Tennessee Health Science Center, Memphis. The CD1 athymic (nu/nu) female mice (8–9 weeks) were obtained from Harlan laboratories (Indianapolis, IN). The C6 glioblastoma cells were trypsinized, washed, centrifuged, and resuspended to a concentration, 4 × 106 cells/0.1 mL in PBS, and injected subcutaneously into flanks (right hind leg) of mice. When tumors reached 300 mm3 in diameter (10 days after implantation), tumor-bearing mice were administered liposome formulations via retro-orbital plexus method (targeted liposomes or plain liposomes; 100 μL/mice) and subjected to NIRFI studies. For the studies, n = 5 mice per time point were used.
Expression of αvβ3 Integrins
Reverse transcriptase polymerase chain reaction was performed to estimate expression levels of αv and β3 subunits at the mRNA level in vivo. Following isolation of tumor tissue, cells were lysed with TRIzol® reagent and total cellular RNA was extracted according to manufacturer’s instructions (Invitrogen, CA). RNA was reverse transcribed into cDNA synthesis with oligo (dT) primer using Superscript III First Strand Synthesis System (Invitrogen, CA). Primer sequences listed in Table 1 were designed using Primer3 software. The concentration of primers was 200 nM for forward and reverse primers. PCR amplification conditions include the following: 50 °C for 30 min, 1 cycle; 94 °C for 5 min, 1 cycle; 35 cycles of 94 °C for 45 s, 55–58 °C for 45 s, and 72 °C for 2 min; and 68 °C for 7 min, 1 cycle. Ten microliters of PCR amplification product and a 123-bp DNA ladder were separated using agarose (1.5%) gel electrophoresis. Gels were stained with ethidium bromide and photographed under ultraviolet transillumination. Results were expressed as relative intensity of each gene with respect to corresponding β-actin band (internal control). The band intensities were analyzed using ImageJ software (NIH, MD). A standard Western blot analysis was performed to estimate protein expression levels of αv and β3 subunits.
Imaging Studies
In vivo NIRFI studies were conducted in a custom-assembled photometric Chemipro imaging station with fluorescence capabilities (Roper Scientific, Trenton, NJ). In brief, solid tumor-implanted nude mice were anesthetized. Following systemic administration of liposomes, real-time NIRF images were acquired at 0.5 (earlier), 0.08, 3, 6, 12, 24, and 48 h. Images acquired 0.5 h (earlier) mean control images were acquired 0.5 h earlier to the administration of fluorescence liposomes. Animals were placed inside a black color-coated light-tight box and images were acquired in excitation/emission module. For whole-body imaging, NIR and white light were supplied by fiber-optic light system connected to a 175-W Xenon source, which was used to guide multiple excitation filter wheels (Sutter Instruments Co., Novato, CA). Fluorescence excitation light was filtered through 710-nm band-pass filter (Chrom Technology, Rockingham, VT). Filtered excitation light was passed through NIRF emission filter, 780 nm (Andover, Salem, NH). Thereafter, NIRF images were acquired with liquid nitrogen-cooled charge-coupled device (CCD) camera (Roper Scientific, NJ). Image acquisition and analysis were performed with Metamorph software (Universal Imaging, PA). A pseudo-color image was generated for each captured image. For image quantitative analysis, a defined standard region of interest (ROI) was selected over organ or tissue image and intensity/area estimation was performed. Background images were acquired at every time point to calculate signal-to-noise (S/N) ratios. Images were integrated as mean fluorescence intensities (MFI) and then were converted to S/N ratios. S/N ratio at a definite time point was calculated using Eq. 4:
Blood and Tumor Distribution Studies
Following image acquisition, mice (n = 4 mice per time point) were sacrificed for collection of blood and tumor tissue at defined time points, 0.08, 3, 6, 12, 24, and 48 h. Frozen tissues were minced and extracted for DiR18 fluorophore in methanol. As described previously [25], with a modification, DiR18 concentration in serum and methanol extracts of tissues was analyzed by RP-HPLC method. In brief, HPLC analysis was carried using mobile phase composition, solvent A: 70% water/0.07% trifluoroacetic acid and solvent B: 30% acetonitrile/0.03% trifluoroacetic acid. Fluorescence quantification of DiR18 was performed at 750 nm (λ max), and retention time was 15.8 min.
Statistical Analysis
All in vivo data were reported in arithmetic means. Error bars represent one standard error of mean (SEM) or standard deviation (SD). Difference between the means of indicated groups was analyzed by one-way ANOVA. The p values less than 0.05 (p < 0.05) were considered significance.
Results
I-optimal Design of Formulation Composition
Multi-linear regression analysis and ANOVA were performed to fit the I-optimal model and examine statistical significance of the terms. The I-optimal design emphasizes prediction of response rather than coefficients [26]. This design determines the optimum region (e.g., composition, process conditions) in the design space where response falls within the acceptable range. Also, the I-optimal design minimizes average prediction variance inside the region of independent variable limits [27, 28]. At the center of the design space, average variance is 0.1667 compared to 0.25 of the D-optimal design, which means shorter confidence intervals for precise prediction of response variables. Table S2 (supplementary material 2) presents estimated regression coefficients of independent variables affecting response variables, R 2 values, adj-R 2 values, and p values. The R 2 values for ligand graft efficiency and mean diameter of liposomes were 0.974 and 0.979, respectively, indicating that the I-optimal model was suitable to predict response variables. Also, adj-R 2 values were used to evaluate the fit model. The adj-R 2 values for graft efficiency and mean diameter were 0.94 and 0.95, respectively, indicating that non-significant terms were not included in the model.
The I-optimal design evaluated linear, quadratic, and interaction effects of independent variables on response variables. Fig. 2 a, b demonstrates effects of HSPC/CHOL ratio and conjugate concentration and their interaction effects on graft efficiency and mean diameter, respectively. Only HSPC/CHOL ratio showed significant effect (p < 0.0001) on ligand graft efficiency, whereas HSPC/CHOL ratio (p < 0.0001) and conjugate concentration (p < 0.005) influenced mean diameter of liposomes to a greater degree. A non-linear relationship between HSPC/CHOL ratio and ligand graft efficiency was observed (i.e., graft efficiency increased with an increase of HSPC/CHOL ratio and decreased at the upper limit of HSPC/CHOL ratio), while a linear relationship between HSPC/CHOL ratio and mean diameter of liposomes was observed. Conjugate concentration showed an inverse relation on mean diameter of liposomes. It has no effect on ligand graft efficiency. Cholesterol appeared to show varied effects on the stability of liposomal membranes. Higher cholesterol concentration had generated vesicles in the micellar size range, 30–80 nm, which reduced graft efficiency. Lower cholesterol concentration destabilized membranes leading to large and polydispersed liposomes.
Response surface plots demonstrating the effect of independent variables on response variables of targeted nanoliposomes. Response variables: targeting ligand grafting efficiency and mean diameter of nanoliposomes as a function of independent variables: HSPC/CHOL ratio and conjugate concentration (a, b); HSPC/CHOL ratio and incubation temperature (c, d). Values in brackets of axis titles represent low and high levels used for experimental and prediction purposes. e, f Correlation between actual and predicted values of ligand graft efficiency and mean diameter, respectively. Number in brackets of HSPC/CHOL ratio represents molar ratio of HSPC lipid to cholesterol. Conjugate concentration in brackets represents molar concentration (expressed in mM) of lipid-peptide conjugate
Figure 2c, d shows effects of HSCP/CHOL ratio and incubation temperature on graft efficiency and mean diameter of liposomes, respectively. Incubation temperature showed a significant effect (p < 0.0001) on graft efficiency and mean diameter of liposomes. A linear relationship was observed between incubation temperature and graft efficiency, i.e., graft efficiency increased with an increase of incubation temperature. And, an inverse relation was observed between incubation temperature and mean diameter. Above Tm of lipids, lipid layers exhibit a liquid-crystalline nature. These liquid-crystalline domains intercalated lipid-peptide ligand grafts, resulting in higher targeting ligand graft efficiency.
From Table S2 (supplementary material 2), it can be observed that HSPC/CHOL ratio and incubation temperature exhibited significant quadratic effects (p < 0.0001) on graft efficiency and mean diameter of liposomes. Quadratic surface effects tend to optimize probabilities of lower or higher responses from different runs of the same independent factor. These effects optimize towards maximization of favorable responses. The I-optimal design encompassed six interactions for four independent factors. X1X4 (p < 0.0027) and X2X3 (p < 0.004) interactions showed significant effects on ligand graft efficiency, whereas X1X4 (p < 0.0001) and X3X4 (p < 0.0046) interactions exhibited significant effects on mean diameter of liposomes. Higher HSPC/CHOL ratio and incubation temperature destabilized liposome membranes. Below Tm, HSPC exists in a rigid crystalline state (tight lipid molecule domains), which posed unfavorable condition for ligand grafting. Mean diameter of liposomes remained stable in the crystalline state of phospholipids (i.e., < 45 °C).
Figure 2e, f represents correlation of predicted and experimental R 2 values for graft efficiency and mean diameter of liposomes. Higher R 2 values indicated that experimental values were close to predicted values. Optimized independent variables were found to generate stable liposomes with ligand graft efficiency, 85.6% and mean diameter, 132 nm. From Tables 2 and 3, optimum formulation conditions indicated that predicted response variables were reliable.
Characterization of Nanoliposomes
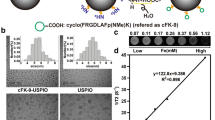
DSPE-PEG2000-Mal-cRGD graft was formed by Michael addition reaction between maleimide group of DSPE-PEG2000-Mal and sulfhydryl group (cysteine amino acid) of cRGD-peptide, respectively. MALDI-TOF analysis correlated theoretical molecular weight of the conjugate, 3458.19 Da with estimated molecular weight (m/z = 3459.19 Da for [M + H]+) (unpublished data). The yield of the conjugate was 92% (gravimetric basis after purification). For receptor targeting, liposomes were surface modified with targeting ligand (DSPE-PEG2000-Mal-cRGD conjugate). Approximately, 85% grafting efficiency was observed. Total number of DSPE-PEG2000-Mal-cRGD grafts was determined on the basis of liposome number (Nvol) (supplementary material 1). Approximately, 150 targeting ligands per liposome were intercalated into lipid bilayers of liposomes.
Due to lipophilic nature, DiR18 fluorophore was incorporated into lipid bilayers of liposomes. DiR18 entrapment in targeted and plain liposomes was found to be 96.8% w/v and 98.1% w/v, respectively. Total lipid concentration was 74.7 and 74.5 mM, respectively. As shown in Fig. 3a, b, mean diameter of targeted and plain liposomes was 131 ± 7 and 114 ± 12 nm, respectively. Polydispersity index was ≤ 0.1, which indicates unimodal liposomal particle size distribution. The zeta potential of targeted and plain liposomes was − 17.4 ± 4.8 and − 22.1 ± 7.9 mV, respectively. Negative charge of liposomes was due to negatively charged HSPC in the lipid bilayers of liposomes. Short-term (1 week) and long-term (6 months) colloidal stability studies were conducted at three different thermal conditions, 2–8, 15, and 25 °C. These stability studies indicated that the liposome formulations were stable at 2–8 °C for 6 months. Accordingly, liposome formulations were stored at 2–8 °C. Zeta potential and polydispersity indices were used to determine colloidal stability of formulations.
Affinity Studies
Integrin receptor-binding characteristics of cRGD-peptide domain of DSPE-PEG2000-Mal-cRGD conjugate and targeted liposomes were determined in competitive receptor-binding experiments. As shown in Fig. 4, we measured IC50 values by competitive binding measurements between 125I-echistatin and RGD-peptide. The binding of 125I-echistatin to αvβ3 integrin receptors on HUVECs was inhibited by cRGD-peptide, DSPE-PEG2000-Mal-cRGD conjugate, and targeted liposomes in a log-concentration-dependent manner. The measured IC50 values of competing ligands, cRGD-peptide, DSPE-PEG2000-Mal-cRGD conjugate, and targeted liposomes were 6.3 ± 2.5, 50.1 ± 10.2, and 316.3 ± 21.3 nmol/L, respectively. Targeted liposomes required high concentration for affinity to integrin receptors. Low binding affinity of cRGD-peptide domain of targeted liposomes could be due to steric hindrance of PEG chains. Non-specific peptide, cRAD-peptide, showed no competitive binding to αvβ3 integrin receptors.
Competition of specific binding of 125I-labeled echistatin with ligands. Ligands include cRGD-peptide, lipid-peptide conjugate (DSPE-PEG2000-Mal-cRGD conjugate), and targeted liposomes to αvβ3 integrin receptors. Receptor bound by 125I-echistatin was displaced by cRGD-peptide, conjugate, and targeted liposomes. Specific radioactivity of 125I-echistatin bound to integrin receptors in the absence of competing ligands was considered as 100%. cRAD-peptide was considered a non-specific control. IC50 values were reported as equivalent to cRGD concentration. Compared to cRGD-peptide and conjugate, targeted liposomes exhibited high IC50 values. Steric hindrance was assumed to be responsible for high IC50 values of targeted liposomes. Each point represents mean ± SEM of three experiments. The black circle, black up-pointing triangle, black square, and white square represent cRGD-peptide, lipid-peptide conjugate, targeted liposomes, and cRAD-peptide, respectively
αvβ3 Integrin Receptor Expression
In tumor tissue, mRNA and protein expression of αv and β3 integrin subunits was detected by RT-PCR and Western blot techniques, respectively. Figure 5a shows a representative agarose gel demonstrating mRNA expression of αv and β3 integrin subunits. Compared to normal muscle tissue, tumors showed an elevated mRNA expression of αv and β3 subunits (Fig. 5b) (10 days post-tumor implantation). Figure 5c represents Western blot analysis of αv and β3 subunits in tumor tissue. As shown in Fig. 5d, protein levels of αv and β3 subunits increased to 1.7- (p < 0.05) and 1.9- (p < 0.05) folds, respectively. From data, it can be observed that tumor formation resulted in the upregulation of αvβ3 integrin receptors.
RT-PCR data. a Expression of αv and β3 integrin subunits in solid tumor implanted on right plank of athymic nu/nu mice. Gel photographs show corresponding RT-PCR-amplified integrin subunits from tumor tissue, where lane 1 represents non-tumor plank tissue (negative for integrin receptor subunits), and lanes 2, 3, 4, 5, and 6 represent tumor tissues from five mice, respectively. b Histograms show densitometry analysis of αv and β3 integrin subunits amplification relative to the amount of corresponding β-actin amplification, expressed as percent of control where control is 100%. Data are from pooled tumor tissues of n = 5 mice. Values are a mean ± SEM of three auto-radiograms data; asterisks indicate statistical significance at p < 0.0001 from non-tumor tissue. c Total tissue expression of αv and β3 integrin subunits was determined by Western blot method. Lane 1 represents non-tumor tissue expression of integrin subunits, whereas lanes 2, 3, 4, 5, and 6 represent tumor tissues from five mice, respectively. Equal amount (25 μg) of total protein was loaded onto wells. d Relative expression of integrin subunits was determined by densitometry analysis of stained blot. Data are from pooled tumor tissues of n = 5 mice. Values are a mean ± SEM of three blots data
Real-Time NIRFI
NIRFI was used for non-invasive visualization and quantitation of targeted liposomal uptake in tumor xenografts. Figure 6a, b shows NIRF images of C6 glioblastoma xenograft-bearing mice groups administered with targeted and plain liposome formulations. In both the groups, NIRF images of fluorescent liposome deposition in tumor could be observed. Compared to plain liposomes, targeted liposomes showed significant uptake in tumors. As shown in Fig. 6c, tumor sites exhibited peak NIRF levels, 12- and 24-h post-administration of plain and targeted liposomes, respectively. The intensity of NIRF image in targeted liposome administered group was 2.8-folds (p < 0.05) higher than that of control mice. At the end of 72 h, fluorescence levels were still detectable in targeted liposome group (unpublished data). Compared to plain liposomes, targeted liposomes exhibited significant reticuloendothelial system (RES) uptake of liposomes. The RES uptake of targeted liposomes continued to increase till 24 h and decreased thereafter.
CD1 athymic mice were implanted with GFP (green fluorescence protein)-transfected C6 glioma cells. GFP tumor and NIRF background indicate image acquisition of tumor with fluoro-isothiocyanate (FITC) (500–600 nm) and NIRF (700–800 nm) filters, before injection of a targeted liposomes and b plain liposomes (control). Post-administration (retro-orbital plexus injection) of 100 μL of liposomes, NIRF images were acquired at definite time intervals, 0.08, 3, 6, 12, 24, and 48 h. All NIRF images were acquired with a 5-s exposure time. Fluorescence signal from DiR18 dye was pseudo-colored, and representative images demonstrate tumor and organ uptake of liposomes (inset shows the liposome injection site, lungs, diaphragm, RES (liver and spleen), and tumor regions after administration of liposomes), c quantification of NIRF signal (emitted from tumor site) as a function of time after administration of targeted (black square) and plain (black circle) liposomes in tumor-bearing mice. Mean fluorescence intensity of NIRF signals was expressed in p/s/cm2/sr units. Data points are a mean ± SEM of samples from n = 3 mice/time point in tumor or control groups, d blocking of integrin receptors with cRGD-peptide (unlabeled; 30 μg/kg; retro-orbital plexus injection) prior to administration of targeted liposomes. DI. GFP tumor illuminated with FITC, DII. NIRF background and DIII. NIRF image acquisition 24-h post-administration of targeted liposomes. Herein, MFI of NIRF signal (competitive binding) is 1254.8 ± p/s/cm2/sr compared to 4527.1 ± 129.3 p/s/cm2/sr of plain liposomes and 17,577.3 ± 258.5 p/s/cm2/sr of targeted liposomes signals
To further determine the presence of integrin receptors, mice were injected with cRGD-peptide prior to injection of targeted liposomes. At 24 h, NIRF signal emitted from tumor was suppressed compared to that of mice administered with targeted liposomes alone (Fig. 6DIII). cRGD-peptide in vivo was consistent with in vitro data, i.e., cRGD-peptide competed with targeted liposomes for interacting with integrin receptors. NIRF signal was not detected in skeletal muscle, which can serve as negative control for the presence of integrin receptors. From the above data, it can be observed that targeted DiR18 liposomes detected integrin receptors expressed on solid tumors.
Tumor and Blood Distribution Kinetics of Liposomes
In vivo distribution kinetic studies of targeted and plain liposomes were performed in mice bearing subcutaneous C6 glioblastoma xenografts. Figure 7a demonstrates tumor-to-blood (T/B) mean fluorescence intensity (MFI) ratios of targeted and plain liposome formulations in tumor-bearing mice. Corroborating NIRF signal intensities, tumor tissue-extracted DiR18 concentration of targeted liposomes was maximum 24-h post-administration. And thereafter, fluorescence decreased. Targeted and plain liposome groups showed maximum T/B values, 8.3-folds (p < 0.01) and 3.1-folds (p < 0.05) at 24- and 12-h post-administration of liposomes, respectively. Higher tumor (T)-to-blood (B) ratio was due to lower blood activity and higher deposition of targeted liposomes in tumor site. At 48 h, T/B value in mice administered with targeted liposomes was 4.5-folds (p < 0.01). For calculation purpose, MFI of tumor was normalized to 1 mm3 and MFI of blood was normalized to 15 μL (approximately, 1% of total blood volume of mouse).
Quantification of tumor (1 mm3)-to-blood (15 μL) NIRF signals after administration of targeted (black square) and plain (black circle) liposomes in tumor-bearing mice. NIRF signals (mean fluorescence intensity) were expressed in p/s/cm2/sr units. Data points are a mean ± SEM of samples from n = 4 mice/time point
Discussion
Molecular imaging technologies (e.g., CT, MRI, and PET) have been used for diagnosis of tumors in pre-clinical and clinical models [10, 29,30,31]. Here, we applied NIRF optical imaging for non-invasive detection of tumors and tracing of injectable carriers. However, NIRFI suffers from limited specificity and sensitivity, which are dependent on the following: (a) density of target molecule in a disease site, (b) affinity of targeted ligand for the receptor, (c) payload (e.g., tracer dye or drug) of the targeted carrier, and (d) permeation characteristics of the targeted carrier. To impart specificity and sensitivity, we developed cRGD-peptide-conjugated liposomes for targeting integrins on tumor vasculature in C6 glioblastoma xenograft rodent model. Liposomes were surface modified with targeting ligands and incorporated with DiR18 fluorescent dye. These NIRF imaging probes possess greater photon penetration into living tissues, which may be desirable for imaging deep-seated organs [32]. Desu HR et al. demonstrated emission of fluorescence from deep-seated tissues such as the lungs [33]. In similar terms, non-invasive detection of deep-seated tumors can be observed using NIRF imaging.
NIRFI station acquired an overlay of pseudo-color fluorescence image superimposed on gray-scale photographic image. Since fluorescence intensity and contrast ratios of NIRF images were dependent on viewing angles and optical path length between excitation light and imaging animal, a fixed angle (60° mounting angle of mice) and path length were maintained for image acquisition [34]. At 10-μM DiR18 concentration (of liposome formulations), NIRFI visualized tumor tissue of mice. Higher DiR18 concentration (> 50 μM) caused saturation of acquired images. The tumor-to-normal tissue contrast ratio measured by in vivo imaging was less than an isolated tumor tissue (exogenous tissue). Non-invasive whole-body imaging in mice (nu/nu) resulted in fluorescence attenuation (compared to imaging of isolated tumor tissue), which could be due to loss of emission and scattering of light occurred during transmission across the skin.
In comparison to normal tissues, tumors possess defective vasculatures with substantial location-dependent heterogeneity; the net effect has lower blood flow, which impedes exposure of targeted nanoparticles to tumors. On the other hand, tumor vasculature has a discontinuous endothelium, which can be more permeable (100- to 780-nm pore size) compared to normal vasculature (< 6 nm in postcapillary venules; 50–150 nm in hepatic/splenic sinusoidal endothelium) [35]. Along with mean diameter, ligand density influences targeting properties of nano-sized carriers.
The use of design of experiments (DoE) for preparing targeted liposomes is relatively a new analytical tool in the field of advanced drug delivery. In contrast to the popular method for preparation of targeted liposomes, the authors adopted a post-incorporation method for intercalation of targeting ligands into lipid bilayers of fluorescent liposomes. Earlier, Saul et al. demonstrated that post-incorporation method had improved targeting efficiency of liposomes [12]. In these experimental studies, authors utilized the same post-incorporation method for surface modification of liposomes with targeting ligands. Such a method involves numerous composition and process variables for optimization of targeted liposomes. Henceforth, the authors considered DoE as an analytical approach to minimize the number of experiments to be performed for preparing optimized targeted liposome formulation. In practice, authors had conducted fewer experiments (25 in number) that otherwise require over 150 experiments. Authors had validated the optimum engraftment efficiency using lower and higher (saturated concentration) ligand concentrations for surface modification of liposomes with targeting ligands. Over the saturated targeting ligand concentration, liposome bilayers ruptured and exhibited multimodal distribution in size. In fact, DoE studies were performed to optimize formulation composition that favors tumor-targeting properties of liposomes. Optimized liposomes with 150 targeting ligands/liposome and mean diameter in the range 100–150 nm were used for targeting purpose.
Angiogenesis is an important phenomenon in proliferation and growth of primary and secondary (metastasis) tumors [2, 36]. During angiogenesis, integrin receptors on sprouting capillary vessels mediate tumor cell adhesion to vessels, and thus play a key role in the survival of tumor cells [37]. Therefore, integrin receptors on tumor vasculature were considered prominent targets for NIRFI. Molecular studies showed upregulation of αvβ3 integrin receptors in tumor tissues. Mice administered with targeted liposomes showed an intense NIRF signal, which may be due to interaction of a large population of targeted liposomes with upregulated integrins on tumor vasculature.
Compared to passive liposomes, targeted liposomes were deposited in tumor areas to a greater degree. Tumors and reticulo-endothelial system have constitutive expression of α3βv integrin receptors [6, 38, 39]. And thereby, tumor and RES-expressed integrins could have triggered adherence of cRGD-peptide-anchored nano-sized liposomes to the RES area. This phenomenon might have prompted deposition of targeted liposomes in tumor xenografts and the RES region. In contrast to plain liposomes (3-h post-administration), an immediate uptake of targeted liposomes (0.08-h post-administration) in tumors was observed. Intensity of NIRF signal in targeted liposome group continued to increase till 24 h and decreased thereafter. Besides fluorescence intensity, the area of NIRF signal in targeted liposome group was larger than that in plain liposome group. The difference could be accounted for the interaction of a large population of targeted DiR18 liposomes with integrin receptors. In vitro receptor-binding studies and in vivo integrin receptor-blocking studies showed significant interaction of targeted liposomes with αvβ3 integrins. In addition, enhanced permeation retention (EPR) of liposomes in tumor vasculature was assumed to contribute to liposome localization in tumors. Corroborating NIRF image signals, tissue-extracted DiR18 concentration (i.e., T/B value) of targeted liposomes in tumor tissue was significantly (p < 0.01) higher. Plain liposomes also exhibited localization in tumor tissue, which could be due to EPR effect of leaky tumor vasculature, i.e., passive influx of nanoliposomes (< 120 nm) in tumor tissue.
Clearance of liposomes is dependent on various factors, such as formulation composition, size, targeted receptor, targeting ligand, targeted organ, receptor-ligand interactions, and species. Here, differences in PK profiles (MFI kinetics) could be due to receptors on tissues (including tumor tissues) and their interactions with targeting ligands. Receptor-mediated uptake of ligands/targeted carriers could have facilitated rapid influx of targeted carriers. This receptor-mediated uptake of targeted carriers was assumed for fast blood clearance into tissues. In contrast, passive carriers undergo slower blood clearance into tissues through intercellular channels. And, tissue clearance of liposomes is largely dependent on receptor-ligand interactions and tissue vasculature properties. Integrin-RGD peptide interactions and EPR effect might have slowed down clearance of targeted liposomes from tumors. The abovementioned two events could be the causative factors for slower tissue clearance and significant tumor deposition of targeted liposomes. In contrast, these could also be the reasons for faster tumor clearance and lower tumor deposition of passive liposomes.
Elimination of liposomes can be explained by two clearance processes, one is the degradation of liposomes in blood and the other is uptake by RES, especially the liver. Degradation process can be approximated by the first-order degradation rate constant which is suggested to be governed by the affinity of liposomes to complement activation factors. Again, the affinity of liposomes to complement activation factors could be influenced by size of liposomes. It has been shown that the percentage of liposomes recognized by complement activation factors was lower in the small liposomes (≤ 0.2 μm) [38,39,40]. In our studies, we used liposomes less than 0.2 μm in size. Therefore, size-dependent degradation of liposomes can be minimal so forth. And, size-dependent differences in liposome recognition can be attributed to different curvatures. It has been recognized that the lipid bilayer composition of liposomes (e.g., lipids, cholesterol, PEGylated lipids) could alter degradation of liposomes in blood [40]. Liposomes with composition HSPC/CHOL/DSPE-PEG2000 were used in vivo. PEGylated lipids could impart stealth factor to liposomes. This stealthness could have protected liposomes from degradation due to complement activation factors. On the other hand, RES uptake was observed with surface-modified liposomes. Compared to passive liposomes, surface-modified liposomes (or RGD-peptide-conjugated liposomes) exhibited significant RES uptake. Probably, an integrin receptor-positive liver could be a causative factor for RES uptake of targeted liposomes rather than a complement activation-triggered elimination mechanism.
Advantages of NIRFI probe are cost-effective, biodegradable, and safe. Preparation of targeted liposomes involved intercalation of targeting ligands into lipid bilayers of liposomes by post-incorporation method. With this method, density of targeting ligands can be modulated to improve targeting efficiency. Fluorescent dye concentration can be changed without altering ligand density of liposomes to improve sensitivity of the NIRF method. Also, targeted fluorescent liposomes can be utilized for delivery of encapsulated or entrapped drug moieties to tumor sites. On the other side, liposomes have the tendency to undergo RES uptake due to their particulate nature. Disposition kinetics of fluorescent liposomes can be determined by disposition of liposomes themselves rather than fluorescent dye.
Compared to CT, MRI, and PET, acquisition of images through near-infrared fluorescence method is a complex procedure. Due to diffusion of fluorescence in visible light, image acquisition requires dark backgrounds. Also, it requires image reconstruction with sophisticated image-processing algorithms that create images, akin to tomographic images. Rasmussen et al. successfully utilized NIRFI for rapid and economical assessment of lymphatic contribution to edema that was associated with venous disease in humans [41]. This study has demonstrated reduction of fluorescence diffusion using indocyanine green (ICG) and Food and Drug Administration (FDA)- and Institutional Review Board (IRB)-approved use of indocyanine green (ICG) for lymphatic imaging of human subjects. In near future, it could be possible that NIRFI could be utilized for non-invasive detection and imaging of tumors and tumor vasculature in humans.
Conclusion
Targeted fluorescent liposomes were evaluated for imaging of solid tumors. In vitro, receptor-binding studies demonstrated binding of cRGD-peptide domain of targeted liposomes to integrin receptors. Rapid uptake and prolonged retention of targeted liposomes in tumor tissue were observed. The strong NIRF signal from tumors was assumed due to interaction between αvβ3 integrin receptors and cRGD-peptide of targeted liposomes. From the studies, it can be inferred that non-invasive localization of tumors and tracing of liposome carriers were possible using receptor targeting and NIRFI approaches.
References
Cairns RA, Khokha R, Hill RP. Molecular mechanisms of tumor invasion and metastasis: an integrated view. Curr Mol Med. 2003;3:659–71.
Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–57.
Jain RK. Transport of molecules in the tumor interstitium: a review. Cancer Res. 1987;47:3039–51.
Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat Med. 2000;6:389–95.
Fang J, Shing Y, Weidershain D. Matrix metalloproteinase-2 is required for the switch to the angiogenic phenotype in a tumor model. Proc Natl Acad Sci U S A. 2000;97:3884–9.
Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nature Review Cancer. 2010;10:9–22.
Gasparini G, Brooks PC, Biganzoli E. Vascular integrin avb3: a new prognostic indicator in breast cancer. Clin Cancer Res. 1998;4:2625–34.
Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715.
Ye Y, Xu B, Nikiforovich GV, Bloch S, Achilefu S. Exploring new near-infrared fluorescent disulfide-based cyclic RGD peptide analogs for potential integrin-targeted optical imaging. Bioorg Med Chem Lett. 2011;21:2116–20.
Chen X, Conti PS, Moats RA. In vivo near-infrared fluorescence imaging of integrin avb3 in brain tumor xenografts. Cancer Res. 2004;64:8009–14.
Torchilin VP. Recent advances with liposomes as pharmaceutical carriers. Nat Rev Drug Discov. 2005;4(2):145–60.
Saul JM, Annapragada A, Natarajan JV, Bellamkonda RV. Controlled targeting of liposomal doxorubicin via the folate receptor in vitro. J Control Release. 2003;92:49–67.
Folkman J, Beckner K. Angiogenesis imaging. Acad Radiol. 2000;7:783–5.
Joshi BP, Wang TD. Exogenous molecular probes for targeted imaging in cancer: focus on multi-modal imaging. Cancers. 2010;2:1251–87.
Murata M, Tahara K, Takeuchi H. Real-time in vivo imaging of surface-modified liposomes to evaluate their behavior after pulmonary administration. Eur J Pharm Biopharm. 2014;86(1):115–9.
Khemthongcharoen N, Jolivot R, Rattanavarin S, Piyawattanametha W. Advances in imaging probes and optical microendoscopic imaging techniques for early in vivo cancer assessment. Adv Drug Deliv Rev. 2014;74(Jul):53–74.
Portoy E, Lecht S, Lazarovici P, Danino D, Magdassi S. Cetuximab-labeled liposomes containing near-infra probe for in vivo imaging. Nanomedicine. 2011;7(4):480–8.
Mahmood U, Weissleder R. Near-infrared optical imaging of proteases in cancer. Mol Cancer Ther. 2003;2:489–96.
Becker A, Hessenius C, Licha K. Receptor-targeted optical imaging of tumors with near-infrared fluorescent ligands. Nat Biotechnol. 2001;19:327–33.
Johnson SM, Bangham AD, Hill MW, Korn ED. Single bilayer liposomes. Biochim Biophys Acta. 1971;233(3):820–6.
Manca ML, Sinico C, Maccioni AM, Diez O, Fadda AM, Manconi M. Composition influence on pulmonary delivery of rifampicin liposomes. Pharmaceutics. 2012;4:590–606.
Nallamothu R, Wood GC, Pattillo CB, Scott RC, Kiani MF, Moore BM, et al. A tumor vasculature targeted liposome delivery system for combretastatin A4: design, characterization, and in vitro evaluation. AAPS PharmSciTech. 2006;7(2):E7–E16.
Fiske CH, Subbarow Y. The colorimetric determination of phosphorus. J Biol Chem. 1925;66:375–400.
Kumar CC, Nie H, Rogers CP, Malkowski M, Maxwell E, Catino JJ, et al. Biochemical characterization of the binding of echistatin to integrin avb3 receptor. J Pharmacol Exp Ther. 1997;283:843–53.
Cho H, Kwon GS. Polymeric micelles for neoadjuvant cancer therapy and tumor primed optical imaging. ACS Nano. 2011;5:8721–9.
Myers RH, Montgomery DC, Anderon-Cook CM, editors. Response surface methodology: process and product optimization using designed experiments. New York: Wiley; 2009.
Haines LM. The application of the annealing algorithm to the construction of exact optimal designs for linear regression models. Technometrics. 1989;29:439–47.
Hardin RH, Sloane NJA. A new approach to the construction of optimal designs. J Stat Plann Interference. 1993;37:339–69.
Blasberg RG, Gelovani J. Molecular-genetic imaging: a nuclear medicine-based perspective. Mol Imaging. 2002;1:280–300.
Choudhury RP, Fuster V, Fayad ZA. Molecular, cellular and functional imaging of atherothrombosis. Nat Rev Drug Discov. 2004;3:913–25.
Citrin D, Lee AK, Scott T, Sproull M, Menard C, Tofilon PJ, et al. In vivo tumor imaging in mice with near-infrared labeled endostatin. Mol Cancer Ther. 2004;3(4):481–8.
Quaresima V, Matcher SJ, Ferrari M. Identification and quantification of intrinsic optical contrast for near-infrared mammography. Photochem Photobiol. 1998;67(1):4–14.
Desu HR, Wood GC, Thoma LA. Noninvasive detection of lung inflammation by near-infrared fluorescence imaging using bimodal liposomes. J Fluoresc. 2016;26(1):241–53.
Zaheer A, Lenkinski RE, Mahmood A, Jones AG, Cantley LC, Frangioni JV. In vivo near-infrared fluorescence imaging of osteoblastic activity. Nat Biotechnol. 2001;19:1148–54.
Wang J, Lu Z, Gao Y, Wientjes MG, Au JL-S. Improving delivery and efficacy of nanomedicines in solid tumors. Nanomedicine. 2011;6(9):1605–20.
Eichler AF, Chung E, Kodack DP, Loeffler JS, Fukumura D, Jain RK. The biology of brain metastases—translation to new therapies. Nat Rev Clin Oncol. 2011;8:344–56.
Wolburg H, Noell S, Mack A, Wolburg-Buchholz K, Fallier-Becker P. Brain endothelial cells and the glio-vascular complex. Cell Tissue Res. 2009;335:75–96.
Allen TM, Everest JM. Effect of liposome size and drug release properties on pharmacokinetics of encapsulated drug in rats. J Pharmacol Exp Ther. 1983;226:539–44.
Funato K, Yoda R, Kiwada H. Contribution of complement system on destabilization of liposomes composed of hydrogenated egg phosphatidylcholine in rat fresh plasma. Biochim Biophys Acta. 1992;1103:198–204.
Harashima H, Hiraiwa T, Ochi Y, Kiwada H. Size dependent liposome degradation in blood: in vivo/in vitro correlation by kinetic modeling. J Drug Target. 1995;3:253–61.
Rasmussen JC, Aldrich MB, Guilliod R, Fife CF, O'Donnell TF Jr, Sevick-Muraca EM. Near-infrared fluorescence lymphatic imaging in a patient treated for venous occlusion. Journal of Vasc Surg Cases. 2015;1:201–4.
Acknowledgements
Experimental studies were performed in the Department of Pharmaceutical Sciences, University of Tennessee Health Science Center, Memphis, TN, USA. The authors thank Anuj Shukla for his support during imaging studies.
Funding
This project received financial support from Parenteral Medications Laboratories, University of Tennessee Health Science Center, Memphis, TN, USA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Animal procedures were performed according to a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Tennessee Health Science Center, Memphis.
Competing Interests
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Desu, H.R., Thoma, L.A. & Wood, G.C. αvβ3 Integrin Receptor Targeting and Near-Infrared Imaging of Solid Tumors Using Surface-Modified Nanoliposomes. J Pharm Innov 13, 1–14 (2018). https://doi.org/10.1007/s12247-017-9296-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12247-017-9296-7