Abstract
Feruloyl esterases (FAEs) are a crucial component of the hemicellulose-degrading enzyme family that facilitates the degradation of lignocellulose while releasing hydroxycinnamic acids such as ferulic acid with high added value. Currently, the low enzyme yield of FAEs is one of the primary factors limiting its application. Therefore, in this paper, we optimized the fermentation conditions for the expression of FAE BpFaeT132C−D143C with excellent thermal stability in Escherichia coli by experimental design. Firstly, we explored the effects of 11 factors such as medium type, isopropyl-β-d-thiogalactopyranoside (IPTG) concentration, and inoculum size on BpFaeT132C−D143C activity separately by the single factor design. Then, the significance of the effects of seven factors, such as post-induction temperature, shaker rotational speed, and inoculum size on BpFaeT132C−D143C activity, was analyzed by Plackett–Burman design. We identified the main factors affecting the fermentation conditions of E. coli expressing BpFaeT132C−D143C as post-induction temperature, pre-induction period, and post-induction period. Finally, we used the steepest ascent path design and response surface method to optimize the levels of these three factors further. Under the optimal conditions, the activity of BpFaeT132C−D143C was 3.58 U/ml, which was a significant 6.6-fold increase compared to the pre-optimization (0.47 U/ml), demonstrating the effectiveness of this optimization process. Moreover, BpFaeT132C−D143C activity was 1.52 U/ml in a 3-l fermenter under the abovementioned optimal conditions. It was determined that the expression of BpFaeT132C−D143C in E. coli was predominantly intracellular in the cytoplasm. This study lays the foundation for further research on BpFaeT132C−D143C in degrading agricultural waste transformation applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Feruloyl esterases (FAEs; EC 3.1.1.73), also called cinnamate esterases, are a specific group of enzymes known for their ability to break ester bonds in lignocellulose, ferulic acid esters, oligosaccharide ferulic acid esters, and polysaccharide ferulic acid esters, leading to the release of ferulic acid (FA) and other hydroxycinnamic acids (Oliveira et al. 2019). FAEs can break down the ester bond of FA and other hydroxycinnamic acids connected to hemicellulose and lignin. This process disrupts the dense network structure of the plant cell wall, resulting in improved efficiency of lignocellulose degradation and releasing FA or FA dehydrodimers with high added value from it (Kroon et al. 1999; Schulz et al. 2018). FAEs play a crucial role in accelerating lignocellulose breakdown and maximizing the use of agricultural wastes. Thus, more studies on FAEs and their improvement may help achieve carbon peaking and carbon neutrality targets.
Currently, research on FAEs is limited, with just around a hundred different types researched and published. Existing research shows that the yield of FAEs is often low and has poor heat resistance, which is the key obstacle to deploying FAEs in industrial production (Oliveira et al. 2019). Current FAEs may be cloned, produced, and molecularly changed to overcome these concerns by utilizing molecular biology methods. For instance, Yin et al. (2015) selected two amino acid positions, Ser33 and Asn92, in AuFaeA, for iterative saturation mutagenesis based on B-factor, ΔΔG values, and structural analysis. Consequently, they were able to produce a mutant enzyme that had much higher heat stability. Our research team cloned and expressed the FAE BpFae from Burkholderia pyrrocinia in Escherichia coli in another study (Fu et al. 2020). By optimizing the fermentation conditions for expression using single factor design, we achieved a final enzyme activity of 2.5 units (U)/ml, which is more than a 90-fold increase in enzyme production compared to the natural strain (Fu et al. 2020).
The FAE BpFae, originating from B. pyrrocinia, exhibits notable acid resistance and an optimal reaction pH of 4.5. Additionally, they demonstrate exceptional hydrolysis properties and efficiency, rendering them conducive for synergistic utilization with xylanase in the effective degradation of wheat bran to produce xylooligosaccharide (XOS) and FA. Nevertheless, like most FAEs, BpFae displays inadequate heat resistance, as evidenced by a residual enzyme activity of merely 49% after a 30-min exposure to 50 ℃, constraining its potential applications (Fu et al. 2022). A mutant enzyme, BpFaeT132C−D143C, was obtained through rational design to enhance the heat resistance of the enzyme. The residual enzyme activity of BpFaeT132C−D143C was found to be 76% after 30 min of exposure to 50 ℃, 1.5 times higher than that of the wild-type enzyme BpFae, and it may have a better application prospect (the data have not been published). To investigate the potential utilization of the mutant enzyme in the breakdown of agricultural waste to produce XOS or FA, this paper used an experimental design to optimize the fermentation conditions of the recombinant bacterium pGEX-4T-1-BpFaeT132C−D143C, which is capable of producing the BpFaeT132C−D143C, in order to enhance its expression levels. Past research has consistently shown that statistical approaches successfully optimize microbial fermentation settings. These methods enable rapid and precise determine of the ideal conditions, improving fermentation outcomes. After undergoing optimization through various methods such as single factor design, Plackett–Burman (PB) design, steepest ascent design, and the response surface methodology (RSM), the activity of FAE BpFae in E. coli induced by lactose was measured to be 7.43 U/ml (Fan et al. 2020b). Similarly, following a similar optimization process, the activity of prodigiosin synthetase in the optimal medium was found to be 2.4 times higher than when using the initial medium (You et al. 2018). Therefore, this paper will also use single factor design, PB design, steepest ascent design, and RSM to systematically optimize the fermentation conditions for the expression of FAE BpFaeT132C−D143C in E. coli.
Materials and methods
Materials, reagents, and media preparation
The recombinant bacterium pGEX-4T-1-BpFaeT132C−D143C was constructed and preserved in our laboratory. The antibiotic ampicillin (Amp), FA standard, ferulic acid methyl ester, acrylamide, and bisacrylamide (N,N-methyl bidentate bisacrylamide) were purchased from Beijing Mreda. Acetonitrile and methanol were of domestic chromatographic purity; all other routine reagents were of domestic analytical purity unless otherwise specified. Luria–Bertani (LB) medium, terrific broth (TB) medium, LB medium with MgCl2 (LBBM), LB medium with MgCl2 and extra NaCl (LBBNM), LB medium with MgCl2 and glycerol (LBBMG), LB medium with MgCl2, glycerol, and sorbitol (LBBSMG), terrific broth medium with glycerol and NaCl (TB-GN), super optimal broth (SOB), and medium for expression (MX) were prepared as reported previously (Chen et al. 2011; Zamani et al. 2015; Golotin et al. 2016).
Enzyme production by fermentation
The recombinant bacterium pGEX-4T-1-BpFaeT132C−D143C was conserved and inoculated into an LB medium containing 40 µg/ml Amp, with an inoculum of 0.2%. It was then cultured at 37 ℃ and 200 rpm for 12 h. After that, the cultivated seed was added to a new LB medium containing 40 µg/ml Amp, at an inoculum of 0.2%, and cultured at 37 ℃ and 200 rpm for another 4 h (the optical density at 600 nm (OD600) is 0.6–0.8). The expression of BpFaeT132C−D143C was induced by adding 0.5 mmol/l isopropyl-β-d-thiogalactopyranoside (IPTG) at 20 ℃ and 200 rpm for 16 h.
Preparation of crude enzyme solution
The crude enzyme solution was prepared based on the method described by Fu et al. (2020). The fermentation broth was centrifuged at 4 ℃ and 10,000 rpm for 5 min to collect the bacterial cells. The bacteria were then resuspended in 50 mmol/l citric acid-sodium citrate buffer (pH 5.5) and disrupted by ultrasonication in an ice water bath for 10 min with a 2-s on/3-s off program (130 W, 20 kHz). The suspension was harvested by centrifugation at 4 ℃ and 10,000 rpm for 10 min. The supernatant which was the crude enzyme solution was collected.
FAE activity assay
FAE activity was determined using our team’s pre-established methods (Fu et al. 2020). The procedure involved mixing 20 µl of 25 mmol/l ferulic acid methyl ester solution (prepared with 50% DMSO) with 430 µl of citric acid-sodium citrate buffer (50 mmol/l, pH 5.5) and preheating it at 50 ℃ for 3 min. Next, 50 µl of suitably diluted enzyme solution was added, and the reaction was mixed and left at 50 ℃ to react for 10 min. After that, 500 µl of acetonitrile was added to terminate the reaction, and the reaction was completed. The FA content produced in the system was detected by high-performance liquid chromatography (HPLC), with the buffer solution used as a blank control test instead of the enzyme solution. The amount of enzyme required to release 1 µmol of FA within 1 min under the above conditions was defined as one enzyme activity unit.
Experimental design to optimize enzyme production conditions
Single factor design
The fermentation conditions for IPTG-induced production of BpFaeT132C−D143C were optimized using single factor design (Table 1), which included the medium types, IPTG concentration, inoculum size, initial pH, post-induction temperature, shaker rotational speed, pre-induction period, post-induction period, medium volume, surfactant species, and Tween-80 concentration.
PB design
Based on the results of single factor design, a PB design was carried out by Minitab 17.1 software for seven factors, including post-induction temperature (X1), shaker rotational speed (X2), inoculum size (X3), IPTG concentration (X4), initial pH (X5), post-induction period (X6), and pre-induction period (X7), and each factor was selected as a high level (+ 1) and a low level (− 1), respectively, with the BpFaeT132C−D143C activity as the response value, and regression models were established based on the experimental data.
Steepest ascent path design
Based on the regression model obtained from the PB design, we screened out the factors that had significant effects on BpFaeT132C−D143C activity, and set the direction of change and step size according to the effects of these factors on BpFaeT132C−D143C activity and experimental experience, and designed the steepest ascent path to further optimize the conditions for the expression of BpFaeT132C−D143C.
RSM
After approaching the region of highest BpFaeT132C−D143C activity produced by the expression through the steepest ascent path design, the experimental Box-Behnken design (BBD; Design expert 11.0) of the response surface methodology was used to optimize the three factors identified by the PB experiment that had the most significant effect on enzyme production (post-induction temperature (A), pre-induction period (B), and post-induction period (C) were optimized). The centroid of each factor in the experiment was the level of each factor determined by the highest value in the steepest ascent path design.
Culture amplification using a bioreactor
The optimal fermentation conditions obtained from the optimization process were used to scale production in a 3-l fermenter. The initial pH of the fermentation medium was adjusted to 8.0. The recombinant BL21 (DE3) E. coli cells harboring a pGEX-4T-1-BpFaeT132C−D143C construct were inoculated in 50 ml of LB medium with 40 µg/ml Amp, and the flask was incubated at 37 ℃ and 180 rpm for 12 h. The cultivated seed was added to 1.8 l working volume at an inoculation ratio of 3.2% (v/v). The cultivation temperature was 37 ℃, the rotation speed of the bioreactor was 150 rpm, and the airflow rate was 0.75, the vessel volume per minute (vvm; airflow rate/operating volume) at the pre-induction stage. After the incubation for 3.5 h, the temperature was adjusted to 21 ℃, and the final concentration of IPTG was added to the culture medium for 48 h. The samples were taken at 6-h intervals, and the wet weight of the bacterial organisms, OD600, pH of fermentation broth, culture medium supernatant, and intracellular BpFaeT132C−D143C activity were measured.
Cellular distribution of FAEs
The distribution of BpFaeT132C−D143C produced by E. coli in cells was analyzed using the methods of Zhu et al. (2022) and Li et al. (2017). The specific steps were as follows: Fermentation was carried out under the optimized conditions, as described above, and 50 ml of the fermentation broth was centrifuged at 10,000 rpm for 5 min to collect the centrifuged supernatant 1, which was recorded as the extracellular crude enzyme broth. The centrifuged bacterium was resuspended in 50 mmol/l citrate buffer (pH 5.5) with 20 g/l sucrose, placed in an ice bath for 2 h, and then centrifuged at 10,000 rpm for 5 min to collect supernatant 2. The centrifuged organisms were resuspended again in 50 ml citrate buffer, placed in an ice bath for 30 min, and centrifuged at 10,000 rpm for 5 min to collect supernatant 3. The combined supernatants of two centrifugations were recorded as periplasmic crude enzyme solution. The precipitated portion was resuspended again and collected as an extracellular crude enzyme solution. The precipitated part was resuspended in citrate buffer again. Ultrasonic waves broke the cells according to the above conditions, and the supernatant taken by centrifugation was recorded as the cytoplasmic crude enzyme solution. One percent (w/v) sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) sampling buffer was added to the remaining cell debris and boiled in a boiling water bath for 10 min. The supernatant, after centrifugation, was recorded as the insoluble inclusion bodies.
Determination of FA
The determination of FA content was carried out by HPLC under the following conditions (Xu et al. 2019): the column was a ZORBAX Eclipse Plus C-18 column, the detector was an ultraviolet detector with a detection wavelength of 320 nm, the column temperature was 20 ℃, the mobile phase was an isocratic mixture of methanol and 0.7% acetic acid at a flow rate of 0.5 ml/min, and the injection volume was 10 µl.
SDS-PAGE
SDS-PAGE was carried out according to the method of Laemmli (1970) using a 10% separating gel and a 4.5% concentrating gel, and the gel was stained with 0.25% Kaumas Brilliant Blue R-250 at the end of electrophoresis.
Statistical analysis
Statistical differences in the assessed strategies were analyzed using a one-way ANOVA (P < 0.05) with Tukey’s test. Assays have been conducted at least in triplicate, and the reported values correspond to the mean value and its standard deviation. Minitab 17.1 (Minitab, Inc.), SPSS 24.0 (IBM Corp., New York, USA), OriginPro 9.1 (OriginLab, Northampton, MA, USA), Design-Expert 11 (Stat-Ease, Inc., USA), and Excel 2016 (Microsoft, USA) were used to analyze the data.
Results and discussion
Shake flask level optimization of FAE BpFae T132C−D143C fermentation conditions
Single factor design
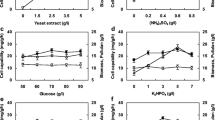
The composition of the medium and the content of each component may vary from one type to another; consequently, the effect on the expression of exogenous proteins in E. coli will also vary. In order to select a suitable medium for the expression of BpFaeT132C−D143C, the effects of nine standard culture media, including LBBMG, LB, TB, SOB, MX, LBBM, TB-GN, LBBSMG, and LBBNM, on the expression of the genes were compared. The results revealed that different media have significantly varying effects on the expression of BpFaeT132C−D143C, as shown in Fig. 1a. LB and MX were the most favorable media for its expression, while LBBM, LBBMG, and LBBNM were highly unfavorable for its expression. Comparative analysis revealed that media containing both Na+ and Mg+ were unfavorable for the expression of BpFaeT132C−D143C, which was verified by the fact that the expression was also deficient in the LBBSMG medium. It was hypothesized that the simultaneous presence of Na+ and Mg+ could inhibit the expression of certain enzyme activities or related factors during the transcription or translation process of the expression system of E. coli, which could decrease the expression of BpFaeT132C−D143C. LB medium was selected as the base fermentation medium for subsequent optimization of fermentation conditions, combining the effects of LB and MX on BpFaeT132C−D143C activity as well as cost. It is worth noting that the optimal medium type for BpFaeT132C−D143C expression was different from BpFae (SOB medium) (Fu et al. 2020). This may be due to the expression of the enzyme genes also changing when they are mutated.
Effects of medium types (LBBMG, LB, TB, SOB, MX, LBBM, TB-GN, LBBSMG, and LBBNM) (a), IPTG concentrations (0.025 mmol/l, 0.05 mmol/l, 0.1 mmol/l, 0.2 mmol/l, 0.4 mmol/l, and 0.8 mmol/l) (b), inoculum size (0.1%, 0.2%, 0.4%, 0.8%, 1.6%, 3.2%, and 6.4%, v/v) (c), pH (4.0, 5.0, 6.0, 7.0, 8.0, and 9.0) (d), post-induction temperature (16 ℃, 20 ℃, 24 ℃, 28 ℃, and 32 ℃) (e), shaker rotational speed and medium volume (140 rpm, 160 rpm, 180 rpm, 200 rpm, and 220 rpm, and 12.5 ml, 25 ml, 50 ml, 75 ml, 100 ml, and 125 ml/250 ml) (f), pre-induction period (2 h, 4 h, 6 h, 8 h, and 10 h) (g), post-induction period (12 h, 18 h, 24 h, 30 h, and 36 h) (h), surfactant types (control, glycerol, Tween-20, Tween-40, Tween-60, Tween-80, Triton X-100, and Triton X-114; the addition amount of each surfactant is 1g/l) (i), and Tween-80 concentration (0 g/l, 0.5 g/l, 1 g/l, 2 g/l, 4 g/l, and 8 g/l) (j) on BpFaeT132C−D143C activity
The pGEX-4T-1 plasmid contains a lactose manipulator, and the lactose analog, IPTG, induces the expression of proteins corresponding to the genes downstream of the lactose manipulator, and it is a common and highly efficient inducer of exogenous protein expression (You et al. 2018; Xiao et al. 2023). The concentration of IPTG influences the efficiency of the recombinant E. coli in the expression of exogenous proteins. As demonstrated in Fig. 1b, with the increase in IPTG concentration, the enzyme activity of BpFaeT132C−D143C expressed by E. coli showed a tendency to increase and then decrease. BpFaeT132C−D143C activity was highest when the IPTG concentration was 0.05 mmol/l, which was 0.37 U/ml, and was in line with the required IPTG concentration of BpFae (Fu et al. 2020). Studies have shown that a lower IPTG concentration is not strong enough to induce E. coli, and the expression of exogenous proteins is low. A too high concentration of IPTG produces a certain toxicity effect on the cell, inhibiting cell growth. Furthermore, it will also result in much faster protein expression than folding and transit, which will create a lot of exogenous protein inclusions and destabilize plasmids, both of which will lower enzyme function (Choi et al. 2000; Duan et al. 2013; Ko et al. 2009; Kosinski et al. 1992; Yang et al. 2017).
For the E. coli expression system, the inoculum size affects the biomass of E. coli at the time of induction, and subsequently, the expression of the target protein under the same pre-induction period conditions (Fu et al. 2020). According to the result (Fig. 1c), BpFaeT132C−D143C activity increased with the increase in inoculum size, and the highest activity of BpFaeT132C−D143C was observed at 0.38 U/ml when the inoculum size was 3.2%. However, no significant change in the activity of BpFaeT132C−D143C was observed when the inoculum size was further increased. A suitable inoculum size has been shown to reduce the bacterial growth hysteresis period and accumulate the right amount of biomass for the production of foreign proteins (Huang et al. 2012; Shu et al. 2016). A large inoculum size, however, may force the strain to develop too quickly, which might lead to a lack of nutrients during the enzyme-producing stage and an increase in toxic metabolites. This, in turn, can inhibit the expression of exogenous proteins or lead to cell death, which can decrease in the amount of expression.
The expression of cell membranes and periplasmic proteins, the secretion and activity of associated enzymes during cellular metabolism, and the stability of metabolites are all impacted by the pH of the environment in which microorganisms thrive (Choi et al. 2004). An adapted pH can promote the expression of associated enzymes and the accumulation of microbial metabolites. Although it varies, the ideal pH needed for various foreign proteins in the E. coli expression system is typically between 4.4 and 9.2 (Stancik et al. 2002). For this reason, the expression of BpFaeT132C−D143C in E. coli under the initial pH condition of 4–9 was explored. The results (Fig. 1d) showed that with the increase of pH, the activity of BpFaeT132C−D143C showed an initial increase followed by a decrease. The enzyme activity was highest at an initial pH of 8, reaching 0.45 U/ml, which differed significantly from the optimal pH conditions required for the expression of BpFae (pH 5.0). The results suggest that the mutation of the exogenous genes affects the growth of E. coli or the expression of the exogenous genes and it is also possible that the different exogenous proteins expressed may have different effects on the growth and metabolism of E. coli (Fan et al. 2020b; Fu et al. 2020).
The temperature has a significant effect on bacterial growth and reproduction, plasmid stability, exogenous protein expression, folding and translocation, and enzyme activity during metabolism (Gadgil et al. 2005; Zhou et al. 2018; Liu et al. 2015). It is a critical factor that affects the soluble expression of exogenous proteins in E. coli. The results showed that with an increase in the post-induction temperature, the activity of BpFaeT132C−D143C first increases and then decreases and is highest at a post-induction temperature of 24 ℃ (Fig. 1e), which is consistent with the results of most studies. The optimal post-induction temperature is between 20–25 ℃, at which the synthesis rate of the target protein BpFaeT132C−D143C is appropriate, and the polypeptide chain can be correctly folded (Krause et al. 2016; Zhou et al. 2018). If the temperature is below this range, the normal metabolism of E. coli is inhibited, resulting in a low synthesis rate of exogenous proteins. Although, theoretically, above this temperature range, such as 32 ℃, E. coli’s cell reproduction and the number of exogenous proteins produced would grow, the overexpressed exogenous proteins cannot be adequately folded or may be harmful to cells, which, in turn, leads to lower enzyme activity (Choi et al. 2000; Li et al. 2014; Su et al. 2015, 2019; Voulgaris et al. 2015; Vuillemin et al. 2014; Xiao et al. 2023).
The growth of E. coli is affected by the content of dissolved oxygen, which also plays a crucial role in exogenous protein expression. Studies showed that transient anaerobiosis can alter metabolic pathways in E. coli by initiating the expression of anaerobic genes and inhibiting the expression of exogenous genes (Sandoval-Basurto et al. 2005). The medium volume and shaker rotational speed are the two primary parameters that influence the quantity of dissolved oxygen in the fermentation system under shaking flask culture conditions. It was found (Fig. 1f) that the activity of BpFaeT132C−D143C increased with an increase in shaker rotational speed and was the highest at 180 rpm and 200 rpm. Continuing to improve the rotational speed increases the amount of dissolved oxygen but, at the same time, increases the shear force on the E. coli cells, resulting in abnormal cell growth and decreased BpFaeT132C−D143C activity (Ukkonen et al. 2013; Yang et al. 2024). Additionally, as the medium volume increased, the activity of BpFaeT132C−D143C decreased, indicating that the expression of this enzyme in E. coli requires a higher amount of dissolved oxygen. Contrary to BpFae, our experimental observations imply that unanticipated differences in the necessary conditions may occur even when highly similar enzymes are produced in the same expression vector. It is hypothesized that slight differences in the exogenous proteins may impose a very different burden on the expressing cells (Fan et al. 2020b).
A proper pre-induction period is crucial for the expression of heterologous proteins in E. coli. Generally, inducers are added at mid to late logarithmic growth in E. coli to favor heterologous protein expression. As demonstrated in Fig. 1g, the optimal induction of BpFaeT132C−D143C occurred at 6 h, which is during the middle to late logarithmic stage of growth. Premature induction of the strain triggers untimely expression of BpFaeT132C−D143C, yet this approach is hampered by the toxicity of IPTG towards the strain and the negative consequences of premature heterologous protein expression on cellular growth (Fan et al. 2020b; Zhou et al. 2018). Consequently, this leads to diminished biomass production and plasmid instability in E. coli, ultimately resulting in low expression levels of BpFaeT132C−D143C. This is evident from the near-zero activity observed for BpFaeT132C−D143C when induced with IPTG at the 2-h mark. On the contrary, delayed induction can be attributed to the significant depletion of nutrients in the medium by the strain during the pre-induction growth phase. Specifically, the exhaustion of crucial nutrients like proteins, which are indispensable for enzyme synthesis and expression during induction, coupled with the possible build-up of inhibitory metabolites that hinder the expression of exogenous proteins during growth, culminates in diminished enzyme activity (Fan et al. 2020b).
The post-induction period is one of the crucial factors that affect the soluble expression of target proteins. It is advantageous for the expression of foreign proteins and the management of production costs to have an appropriate post-induction period (Asther et al. 2002; Liu et al. 2011). The optimal post-induction period required for the expression of different target proteins varies (Sadeghian-Rizi et al. 2020). As illustrated in Fig. 1h, BpFaeT132C−D143C activity increased with the extension of the post-induction period. The enzyme activity peaked between 30 and 36 h after induction, which was slightly longer compared to the post-induction period required for BpFae (Fu et al. 2020). This is also most likely because the enzyme’s mutation would have a different effect on the growth and expression of the cells. Additionally, it has been shown that after a too long post-induction period, a small amount of protease is released when some cells die, which can degrade the target protein, causing a decrease in the practical expression of exogenous proteins (Zhao et al. 2020). Therefore, it is crucial to determine the appropriate post-induction period for the expression of heterologous proteins.
Since surfactants are amphiphilic to cell membranes, they can alter permeability, which can alter the membrane’s capacity to transfer, transport, and absorb nutrients. Surfactants can also alter membrane protein activity, which can change the function of microbial cell metabolism (Jiang et al. 2005; Liu et al. 2004). Surfactants have been shown to affect enzyme production and accumulation of metabolites in natural microorganisms. However, fewer studies have been conducted on their effects on enzyme production in cloned expression strains (Ma et al. 2022; Cheng et al. 2023). The experimental results showed (Fig. 1i) that different surfactant types had different effects on the activity of BpFaeT132C−D143C, except that Tween-80 improved the activity of BpFaeT132C−D143C; all the other surfactants had an inhibitory effect on the enzyme activity, which may be related to the different effects of the different surfactants on the cell membrane (Fan et al. 2020a). The study on the concentration of Tween-80 addition revealed that the addition of different concentrations of Tween-80 enhanced the activity of BpFaeT132C−D143C compared with that of the unadded Tween-80 group. Still there was no significant difference between the different concentration groups, which may be attributed to the excellent compatibility of Tween-80 with E. coli cells (Fig. 1j). Based on cost consideration, the concentration of Tween-80 was selected as 0.5 g/l.
PB design
Based on the results of the single factor design, seven factors, including post-induction temperature, shaker rotational speed, inoculum size, IPTG concentration, initial pH, pre-induction period, and post-induction period, were selected for the PB experiments to assess the effects of these factors on the expression of BpFaeT132C−D143C in E. coli. As shown by the experimental results (Table 2), the activity of BpFaeT132C−D143C varied in the range of 0.01–1.54 U/ml, and there was a significant difference between the highest and the lowest activity, which indicated that different combinations of high and low levels among different factors had a large effect on BpFaeT132C−D143C activity. Analysis of significant differences (Table 3) showed that post-induction temperature (X1), pre-induction period (X6), and post-induction period (X7) had a substantial effect on BpFaeT132C−D143C activity. In contrast, shaker rotational speed (X2), inoculum size (X3), IPTG concentration (X4), and pH (X5) had no significant effect. For this reason, only the above three significant factors were further investigated in subsequent experiments. At the same time, shaker rotational speed (X2), inoculum size (X3), IPTG concentration (X4), and pH (X5) were set to 200 rpm, 3.2%, 0.05 mmol/l, and 8, respectively, according to the single factor design optimal results.
Steepest ascent path design
In order to determine the optimal region when optimizing the response surface of the three significance factors, the steepest ascent path design was preferred for optimization. According to the regression analysis of the PB design results, the levels of the three variables, including post-induction temperature, pre-induction period, and post-induction period, need to be continuously reduced in the steepest ascent path design. As indicated by the results of the steepest ascent path design (Table 4), BpFaeT132C−D143C activity was the highest in the fourth set of experiments, reaching 2.72 U/ml. Therefore, the levels corresponding to the factors in the fourth set of experiments were used as the center point of the subsequent response surface experiments.
RSM design
Based on the results of the PB design and the steepest ascent path design, the experimental Box-Behnken design was used to design a three-level response surface analysis experiment with three factors (post-induction temperature, pre-induction period, and post-induction period), with BpFaeT132C−D143C activity as the response value, and the central point of the design was repeated three times to estimate the error. The results of the experiment are shown in Table 5. It can be seen that there are significant differences in BpFaeT132C−D143C activities in different experimental groups. Among them, the highest BpFaeT132C−D143C activity was 3.52 U/ml in number 1, while the lowest BpFaeT132C−D143C activity was 1.54 U/ml in numbers 9 and 12.
By performing multiple regression analysis on the experimental data, the following regression equation was fitted:
Y = − 69.40 + 5.2803 × A + 3.9946 × B + 0.9667 × C − 0.0208 × AB + 0.0006 × AC + 0.0170 × BC − 0.1250 × A2 − 0.5553 × B2 − 0.0236 × C.2
where Y is the predicted response (BpFaeT132C−D143C activity).
From the results of ANOVA, the pattern P < 0.05 indicates that the model is significant (Table 6). The R2 of the quadratic regression equation is 0.8983, indicating that the equation is a good fit for the test with a small error (Table 6). Moreover, the non-significant lack-of-fit term suggests a strong correlation between the actual value and the predicted value, thereby indicating that the model can accurately reflect and predict BpFaeT132C−D143C activity. In the regression equation, the P values of the primary terms A (post-induction temperature) and B (pre-induction period) are less than 0.05, indicating that the effects of these two factors on the expression of BpFaeT132C−D143C in E. coli show a significant linear relationship; the interaction terms are all insignificant, indicating that the interaction of these three factors has a negligible effect on the expression of BpFaeT132C−D143C in E. coli; among the secondary terms, terms A2 and B2 were significant (P < 0.05), indicating a considerable surface effect between post-induction temperature and pre-induction period and BpFaeT132C−D143C activity (Table 6).
According to the above regression equation to plot the two-by-two interaction effect between the three factors in three-dimensional plots, it can be seen from Fig. 2 that BpFaeT132C−D143C activity with the increase of post-induction temperature, pre-induction period, and post-induction period all showed a trend of increasing and then decreasing. In the optimized region, BpFaeT132C−D143C activity has its highest value. Design-Expert 11 was used to predict the optimal value of BpFaeT132C−D143C activity under the conditions of A (post-induction temperature) = 21 ℃, B (pre-induction period) = 3.5 h, and C (post-induction period) = 22 h, under which the predicted value of enzyme activity was 3.46 U/ml. The predicted conditions were experimentally verified at the post-induction temperature of 21 ℃, pre-induction period of 3.5 h, post-induction period of 22 h, shaker speed of 200 rpm, inoculum size of 3.2% (v/v), IPTG concentration of 0.05 mmol/l, initial pH of 8, and Tween-80 concentration of 0.5 g/l for the induced expression, and the results showed that the BpFaeT132C−D143C activity was 3.58 U/ml, which was close to the predicted value, and proved that the model was more accurate and effective. This result was 7.6 times higher than that before optimization (0.47 U/ml) and higher than the activity of the original enzyme of BpFae expressed by IPTG-induced production in E. coli, which provides sufficient enzyme resources for the subsequent application of the enzyme (Fu et al. 2020).
Three-dimensional surface maps of the effects of pairwise interaction of various factors on BpFaeT132C−D143C activity. a Interaction of post-induction temperature and pre-induction period. b Interaction of post-induction temperature and post-induction period. c Interaction of pre-induction period and post-induction period
Batch fermentation result
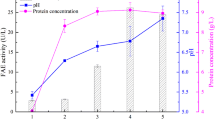
Based on the optimal conditions of shake flask culture, the recombinant bacterium was expanded in a 3-l fermenter and its enzyme production was studied. As depicted in Fig. 3, the pH of the fermentation broth was initially recorded at 7.56 at the onset of induction, subsequently rising to 8.85 after 18 h of induction. This increase in pH can be attributed to the consumption of nutrients, particularly the nitrogen source, by microorganisms, resulting in the production of alkaline substances. The stabilization of pH after that may be attributed to the microorganisms reaching a steady growth state in the medium (Liu et al. 2011). The amount of E. coli cell remained increasing during the pre-induction period and reached a maximum at 12–24 h of inducer addition, followed by a slight decrease. When IPTG was first added, BpFaeT132C−D143C activity was almost undetectable in the fermentation system, which indicated that the enzyme needed an inducer to induce its expression, which was in agreement with the results of most studies (Ran et al. 2016). Subsequently, BpFaeT132C−D143C was rapidly expressed and accumulated, and the highest level of 70% enzyme activity could be reached at 6 h of induction, which indicated that the induction of the expression of exogenous proteins in E. coli was very rapid after the growth state was in a good condition. Continuing to extend the induction time, the activity of BpFaeT132C−D143C increased gently, reaching a maximum value of 1.52 U/ml at 30 h of induction (Fig. 3). Then, due to the degradation of BpFaeT132C−D143C by the protease from the dead cells and the action of the shear force, its activity was reduced. The observed increase in extracellular BpFaeT132C−D143C activity with prolonged induction time can be attributed to the cells transitioning through growth stabilization and decay phases, as well as the shear effect of stirring paddles causing cell rupture and subsequent release of intracellular enzymes into the medium. This phenomenon is supported by the trend in protein concentration changes observed in electrophoretic graphs (Fig. 4). Compared with the BpFaeT132C−D143C activity in shake flask fermentation, the activity in the fermenter was low, which was similar to BpFae, which was related to the differences in cell growth and metabolism caused by the differences in dissolved oxygen, hydraulic pressure, and shear force (Fan et al. 2020b). In the two fermentation devices, the fermentation conditions optimized for shake flasks could not be simply scaled up to be used in the fermenter directly, but need to be optimized based on the optimization of the fermentation conditions in shake flasks (Fan et al. 2020b; Li et al. 2007).
Distribution of FAE activity
E. coli is currently the most efficient, convenient, and technologically mature in terms of operation engineering bacteria. However, it lacks an effective secretion system. The expressed proteins are often stored in the cytoplasm and need to be released after breaking the cell, resulting in the target protein mixed with a large number of heterogeneous proteins in the cytoplasm, which makes it difficult to purify the target protein. However, in recent years, it has been found that some exogenous proteins can be secreted to the extracellular space or transported to the periplasmic space between the inner and outer double membrane structures of E. coli for some unknown reasons, which can help to improve the expression level of target proteins and the purification efficiency of target proteins (Luo et al. 2014; Xu et al. 2019; Zhou et al. 2018; Li et al. 2022; Wang et al. 2022). Therefore, cellular localization of E. coli recombinant bacteria producing BpFaeT132C−D143C is necessary. As shown in Fig. 5, the supernatant was essentially devoid of the target protein and the corresponding BpFaeT132C−D143C activity, the periplasmic space contained a small amount of the target protein, more than 90% of the target protein was present in the intracellular space, and the distribution of the expression of BpFaeT132C−D143C was consistent with the distribution of the majority of the proteins in E. coli (Li et al. 2022; Li et al. 2017). In order to further improve the expression of BpFaeT132C−D143C and the efficiency of subsequent purification, we will study the replacement of signal peptides used in other research results that can guide the extracellular expression of exogenous proteins in the later stage.
SDS-PAGE analysis of different cell components. M, low molecular weight standard protein markers; lane 1, supernatant of culture medium (supernatant 1 mentioned in the “Materials and methods”); lanes 2 and 3, cytoplasm (supernatants 2 and 3 mentioned in the “Materials and methods”); lane 4, supernatant of lysate after cytoplasmic extraction; lane 5, pellet of lysate after cytoplasmic extraction; lane 6, supernatant of lysate without cytoplasmic extraction; lane 7, pellet of lysate without cytoplasmic extraction
Conclusion
In this study, we optimized the expression conditions of FAE BpFaeT132C−D143C, which has excellent heat resistance, in E. coli by using single factor design, PB design, steepest ascent path design, and RSM design. Under the optimized fermentation conditions, such as post-induction temperature of 21 ℃, pre-induction period of 3.5 h, post-induction period of 22 h, shaker rotational speed of 200 rpm, inoculum size of 3.2% (v/v), IPTG concentration of 0.05 mmol/l, pH 8, and Tween-80 concentration of 0.5 g/l, the activity of BpFaeT132C−D143C could reach up to 3.58 U/ml, which is 7.6 times of the pre-optimization (0.47 U/ml). This provided sufficient enzyme resources for the subsequent application of the enzyme, such as producing XOS and FA from agricultural wastes.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Asther M, Haon M, Roussos S, Record E, Delattre M, Lesage-Meessen L, Labat M, Asther M (2002) Feruloyl esterase from Aspergillus niger a comparison of the production in solid state and submerged fermentation. Process Biochem 38:685–691. https://doi.org/10.1016/S0032-9592(02)00196-6
Chen YL, Huang WQ, Zhou XB, Ling XP, Lu YH (2011) Medium optimization for β-1, 3–1, 4-glucanase productionby recombinant Escherichia coli. J Xiamen Univ Nat Sci. 50:896–902
Cheng RW, Wang FQ, Xu YR, Wei L, Ma JH, Gao P, Liu XY, Fan GS, Yang R (2023) Optimization of submerged fermentation conditions for glucanase production by Burkholderia pyrrocinia B1213 using Jiuzao. Emir J Food Agr 35:468–480. https://doi.org/10.9755/ejfa.2023.v35.i5.3091
Choi JH, Jeong KJ, Kim SC, Lee SY (2000) Efficient secretory production of alkaline phosphatase by high cell density culture of recombinant Escherichia coli using the Bacillus sp. endoxylanase signal sequence. Appl Microbiol Biot 53:640–645. https://doi.org/10.1007/s002530000334
Choi JH, Lee SY (2004) Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl Microbiol Biot 64:625–635. https://doi.org/10.1007/s00253-004-1559-9
Duan XG, Chen J, Wu J (2013) Optimization of pullulanase production in Escherichia coli by regulation of process conditions and supplement with natural osmolytes. Bioresource Technol 146:379–385. https://doi.org/10.1016/j.biortech.2013.07.074
Fan GS, Cheng LJ, Fu ZL, Sun BG, Teng C, Jiang XY, Li XT (2020a) Screening of yeasts isolated from Baijiu environments for 2-phenylethanol production and optimization of production conditions. 3 Biotech 10:275. https://doi.org/10.1007/s13205-020-02267-5
Fan GS, Zhu YT, Fu ZL, Sun BG, Teng C, Yang R, Li XT (2020b) Optimization of fermentation conditions for the production of recombinant feruloyl esterase from Burkholderia pyrrocinia B1213. 3 Biotech 10:216. https://doi.org/10.1007/s13205-020-02198-1
Fu ZL, Fan GS, Zhu YT, Teng C, Li HH, Liu Q, Yang R, Li XT (2020) Soluble expression of a novel feruloyl esterase from Burkholderia pyrrocinia B1213 in Escherichia coli and optimization of production conditions. Biotechnol Biotec Eq 34:732–746. https://doi.org/10.1080/13102818.2020.1803129
Fu ZL, Zhu YT, Teng C, Fan GS, Li XT (2022) Biochemical characterization of a novel feruloyl esterase from Burkholderia pyrrocinia B1213 and its application for hydrolyzing wheat bran. 3 Biotech 12:24. https://doi.org/10.1007/s13205-021-03066-2
Gadgil M, Kapur V, Hu WS (2005) Transcriptional response of Escherichia coli to temperature shift. Biotechnol Progr 21:689–699. https://doi.org/10.1021/bp049630l
Golotin VA, Balabanova LA, Noskova YA, Slepchenko LV, Bakunina IY, Vorobieva NS, Terenteva NA, Rasskazov VA (2016) Optimization of cold-adapted alpha-galactosidase expression in Escherichia coli. Protein Expres Purif 123:14–18. https://doi.org/10.1016/j.pep.2016.03.006
Huang YJ, Wang ZH, Zhao YW, Yang Y, Shi JG (2012) Optimization of culture medium and fermentation conditions of transglutaminase by recombinant Escherichia coli. China Brew 31:21–24. https://doi.org/10.3969/j.issn.0254-5071.2012.04.006
Jiang ZQ, Yang SQ, Yan QJ, Li LT, Tan SS (2005) Optimizing xylanase production by a newly isolated strain CAU44 of the thermophile Thermomyces lanuginosus. World J Microb Biot 21:863–867. https://doi.org/10.1007/s11274-004-5988-5
Ko JK, Jung MW, Kim KH, Choi IG (2009) Optimal production of a novel endo-acting β-1,4-xylanase cloned from Saccharophagus degradans 2–40 into Escherichia coli BL21(DE3). New Biotechnol 26:157–164. https://doi.org/10.1016/j.nbt.2009.07.009
Kosinski MJ, Rinas U, Bailey JE (1992) Isopropyl-β-d-thiogalactopyranoside influences the metabolism of Escherichia coli. Appl Microbiol Biot 36:782–784. https://doi.org/10.1007/BF00172194
Krause M, Neubauer A, Neubauer P (2016) The fed-batch principle for the molecular biology lab: controlled nutrient diets in ready-made media improve production of recombinant proteins in Escherichia coli. Microb Cell Fact 15:110. https://doi.org/10.1186/s12934-016-0513-8
Kroon PA, Garcia-Conesa MT, Fillingham IJ, Hazlewood GP, Williamson G (1999) Release of ferulic acid dehydrodimers from plant cell walls by feruloyl esterases. J Sci Food Agric 79:428–434. https://doi.org/10.1002/(SICI)1097-0010(19990301)79:3%3c428::AID-JSFA275%3e3.0.CO;2-J
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Li XN, Wang Y, Wang Z, Liu XL, Wu H, Zhou JZ, Xia XD (2022) Secretory expression of β-glucosidase based on signal peptide. Jiangsu J Agr Sci 38:223–231. https://doi.org/10.3969/j.issn.1000-4440.2022.01.027
Li YM, Chen AN, Yang YK, Bai ZH (2017) Study on secretory expression of recombination pullulanase from Bacillus acidopullulyticus in Escherichia coli. J Biol 34:23–29. https://doi.org/10.3969/j.issn.2095-1736.2017.01.023
Li Y, Cui FJ, Liu ZQ, Xu YY, Zhao H (2007) Improvement of xylanase production by Penicillium oxalicum ZH-30 using response surface methodology. Enzyme Microb Tech 40:1381–1388. https://doi.org/10.1016/j.enzmictec.2006.10.015
Li ZF, Su LQ, Wang L, Liu ZG, Gu ZB, Chen J, Wu J (2014) Novel insight into the secretory expression of recombinant enzymes in Escherichia coli. Process Biochem 49:599–603. https://doi.org/10.1016/j.procbio.2014.01.029
Liu J, Chen XD, Dai X, Tang B, Peng ZR (2004) Effect of surfactant on the production of thermophilic protease from Bacillus stearothermophilus. J Microbiol 24:58–59. https://doi.org/10.3969/j.issn.1005-7021.2004.06.017
Liu JF, Zhang ZJ, Li AT, Pan J, Xu JH (2011) Significantly enhanced production of recombinant nitrilase by optimization of culture conditions and glycerol feeding. Appl Microbiol Biot 89:665–672. https://doi.org/10.1007/s00253-010-2866-y
Liu WM, Yang ZJ, Luo JX, Zhuang XJ, Shen WH, Hu Y, Huang H (2015) Optimization of fermentation conditions of recombinant E. coli for coexpression of leucine dehydrogenase and formate dehydrogenase. Chin J Bioprocess Eng 13:23–28. https://doi.org/10.3969/j.issn.1672-3678.2015.04.005
Luo ZC, Zhang Y, Bao J (2014) Extracellular secretion of β-glucosidase in ethanologenic E. coli enhances ethanol fermentation of cellobiose. Appl Biochem Biotech 174:772–783. https://doi.org/10.1007/s12010-014-1108-7
Ma JH, Cheng LJ, Zhang YJ, Liu YC, Sun Q, Zhang J, Liu XY, Fan GS (2022) Screening of yeasts isolated from Baijiu environments for producing 3-methylthio-1-propanol and optimizing production conditions. Foods 11:3616. https://doi.org/10.3390/foods11223616
Oliveira DM, Mota TR, Oliva B, Segato F, Marchiosi R, Ferrarese O, Faulds CB, dos Santos WD (2019) Feruloyl esterases: biocatalysts to overcome biomass recalcitrance and for the production of bioactive compounds. Bioresource Technol 278:408–423. https://doi.org/10.1016/j.biortech.2019.01.064
Ran HY, Wu J, Wu D, Duan XG (2016) Enhanced production of recombinant Thermobifida fusca isoamylase in Escherichia coli MDS42. Appl Biochem Biotech 180:464–476. https://doi.org/10.1007/s12010-016-2110-z
Sadeghian-Rizi T, Behdani M, Naghavi-al-hosseini F, Dakhilpour SS, Khanahmad H, Jahanian-Najafabadi A (2020) Optimization of anti-CXCL10 nanobody expression using response surface methodology and evaluation of its anti-metastatic effect on breast cancer cells. Int J Pept Res Ther 26:1399–1407. https://doi.org/10.1007/s10989-019-09941-0
Sandoval-Basurto EA, Gosset G, Bolívar F, Ramírez OT (2005) Culture of Escherichia coli under dissolved oxygen gradients simulated in a two-compartment scale-down system: metabolic response and production of recombinant protein. Biotechnol Bioeng 89:453–463. https://doi.org/10.1002/bit.20383
Schulz K, Nieter A, Scheu AK, Copa-Patiño JL, Thiesing D, Popper L, Berger RG (2018) A type D ferulic acid esterase from Streptomyces werraensis affects the volume of wheat dough pastries. Appl Microbiol Biot 102:1269–1279. https://doi.org/10.1007/s00253-017-8637-2
Shu C, Luo SZ, Cai J, Jiang ST, Zheng Z (2016) Optimization of the fermentation conditions for transglutaminase by recombinant Escherichia coli. Sci Technol Food Ind 37:183–189. https://doi.org/10.13386/j.issn1002-0306.2016.12.027
Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL (2002) pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J Bacteriol 184:4246–4258. https://doi.org/10.1128/JB.184.15.4246-4258.2002
Su LQ, Huang Y, Wu J (2015) Enhanced production of recombinant Escherichia coli glutamate decarboxylase through optimization of induction strategy and addition of pyridoxine. Bioresource Technol 198:63–69. https://doi.org/10.1016/j.biortech.2015.08.153
Su LQ, Wu SX, Feng JY, Wu J (2019) High-efficiency expression of Sulfolobus acidocaldarius maltooligosyl trehalose trehalohydrolase in Escherichia coli through host strain and induction strategy optimization. Bioproc Biosyst Eng 42:345–354. https://doi.org/10.1007/s00449-018-2039-4
Ukkonen K, Veijola J, Vasala A, Neubauer P (2013) Effect of culture medium, host strain and oxygen transfer on recombinant Fab antibody fragment yield and leakage to medium in shaken E. Coli Cultures. Microb Cell Fact 12:73. https://doi.org/10.1186/1475-2859-12-73
Voulgaris I, Finka G, Uden M, Hoare M (2015) Enhancing the selective extracellular location of a recombinant E. coli domain antibody by management of fermentation conditions. Appl Microbiol Biot 99:8441–8453. https://doi.org/10.1007/s00253-015-6799-3
Vuillemin M, Malbert Y, Laguerre S, Remaud-Siméon M, Moulis C (2014) Optimizing the production of an α-(1→2) branching sucrase in Escherichia coli using statistical design. Appl Microbiol Biot 98:5173–5184. https://doi.org/10.1007/s00253-014-5627-5
Wang YZ, Sun T, Wu H, Ma JF (2022) Advances in expression of recombinant protein fusion signal peptide in Escherichia coli. Microbiol China 49:794–806. https://doi.org/10.13344/j.microbiol.china.210325
Xiao Y, Wang YH, Yang B (2023) Modeling and optimization of fermentation by lipase MAS1-producin recombinant Escherichia coli based on support vector machine. Mod Food Sci Technol 39:59–68. https://doi.org/10.13982/j.mfst.1673-9078.2023.1.0136
Xu ZS, Wang T, Zhang SS (2019) Extracellular secretion of feruloyl esterase derived from Lactobacillus crispatus in Escherichia coli and its application for ferulic acid production. Bioresource Technol 288:121526. https://doi.org/10.1016/j.biortech.2019.121526
Yang R, Ma JH, Wang ZC, Du YH, Tian SB, Fan GS, Liu XY, Teng C (2024) The identification of a strain for the biological purification of soy molasses to produce functional soy oligosaccharides and optimize purification conditions. Foods 13:296. https://doi.org/10.3390/foods13020296
Yang YN, Shan WX, Wang PW (2017) Upscale production of a recombinant cyclodextrin glycosyltransferase from Paenibacillus macerans in Escherichia coli. 3 Biotech 7:207. https://doi.org/10.1007/s13205-017-0838-y
Yin X, Li JF, Wang CJ, Hu D, Wu Q, Gu Y, Wu MC (2015) Improvement in the thermostability of a type A feruloyl esterase, AuFaeA, from Aspergillus usamii by iterative saturation mutagenesis. Appl Microbiol Biot 99:10047–10056. https://doi.org/10.1007/s00253-015-6889-2
You ZY, Zhang SP, Liu XX, Wang YJ (2018) Enhancement of prodigiosin synthetase (PigC) production from recombinant Escherichia coli through optimization of induction strategy and media. Prep Biochem Biotech 48:226–233. https://doi.org/10.1080/10826068.2017.1421965
Zamani M, Berenjian A, Hemmati S, Nezafat N, Ghoshoon MB, Dabbagh F, Mohkam M, Ghasemi Y (2015) Cloning, expression, and purification of a synthetic human growth hormone in Escherichia coli using responsesurface methodology. Mol Biotechnol. 57:241–250. https://doi.org/10.1007/s12033-014-9818-1
Zhao T, Huang LQ, Jin ZY, Yuan SQ, Liu J (2020) Fermentation optimization of acetolactate synthetase in recombinant Escherichia coli. Food Ferment Ind 46:156–162. https://doi.org/10.13995/j.cnki.11-1802/ts.023567
Zhou YL, Lu ZH, Wang X, Selvaraj JN, Zhang GM (2018) Genetic engineering modification and fermentation optimization for extracellular production of recombinant proteins using Escherichia coli. Appl Microbiol Biot 102:1545–1556. https://doi.org/10.1007/s00253-017-8700-z
Zhu DJ, Zhang JW, Yao Y, Shan YL, Yang ML, Shen W, Yang HQ, Xia YY, Chen L, Chen XZ (2022) Secretory expression of pullulanase gene from Bacillus subtilis and its application to vermicelli production. J Food Sci Biotechnol 41:82–93. https://doi.org/10.3969/j.issn.1673-1689.2022.02.011
Funding
This research was supported by the Beijing Natural Science Foundation (6222003) and the Open Research Fund Program of Henan Key Laboratory of Industrial Microbial Resources and Fermentation Technology (HIMFT20190205).
Author information
Authors and Affiliations
Contributions
Jinghao Ma carried out the experiments and wrote the manuscript. Xuan Zhao and Sihan Yuan carried out the experiments. Guangsen Fan conceived of the study, designed the experimental protocol, and wrote the manuscript. Jinghao Ma and Xiaoyan Liu analyzed the data. Rana Abdul Basit edited the language. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, J., Basit, R.A., Yuan, S. et al. Optimization of fermentation conditions for the production of recombinant feruloyl esterase BpFaeT132C−D143C. Folia Microbiol (2024). https://doi.org/10.1007/s12223-024-01197-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12223-024-01197-6