Abstract
Pichia pastoris, a methylotrophic yeast, is known to be an efficient host for heterologous proteins production. In this study, a recombinant P. pastoris Y11430 was found better for β-glucosidase activity in comparison with a wild type P. pastoris Y11430 strain, and thereby, subjected to methanol intermittent feed profiling for β-glucosidase production. The results showed that at 72 h of cultivation time, the cultures with 16.67% and 33.33% methanol feeding with constant rate could produce the total dry cell weight of 52.23 and 118.55 g/L, respectively, while the total mutant β-glucosidase activities were 1001.59 and 3259.82 units, respectively. The methanol feeding profile was kept at 33% with three methanol feeding strategies such as constant feed rate, linear feed rate, and exponential feed rate which were used in fed-batch fermentation. At 60 h of cultivation, the highest total mutant β-glucosidase activity was 2971.85 units for exponential feed rate culture. On the other hand, total mutant β-glucosidase activity of the constant feed rate culture and linear feed rate culture were 1682.25 and 1975.43 units, respectively. The kinetic parameters of exponential feed rate culture were specific growth rate on glycerol 0.228/h, specific growth of methanol 0.061/h, maximum total dry cell weight 196.73 g, yield coefficient biomass per methanol (\({Y_{x/m}}\)) 0.57 gcell/gMeOH, methanol consumption rate (\({Q_m}\)) 5.76 gMeOH/h, and enzyme productivity (\({Q_p}\)) 75.96 units/h. In conclusion, higher cell mass and β- glucosidase activity were produced under exponential feed rate than constant and linear feed rates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
β-glucosidases (EC 3.2.1.21) belong to a group of widely distributed hydrolytic enzymes that hydrolyze the β-O-glucosidic bonding between β-D-glucose and aglycone/another sugar (Kytidou et al. 2020). Dalcochinase, a β-glucosidase from Dalbergia cochinchinensis Pierre (Thai rosewood) (Ratananikom et al. 2013; Toonkool et al. 2006), carries an isoflavonoid glucosidase, dalcochinin-8′-O-β-D-glucosidase, as its natural substrate (Karnišová Potocká et al. 2021). Moreover, dalcochinase can also function in reverse hydrolysis (Paul et al. 2020), and transglucosylation (Kongsaeree et al. 2010) to synthesize oligosaccharides and glycosides. From this aspect, dalcochinase has a potential application for novel oligosaccharide biosynthesis. To obtain dalcochinase, the enzyme has to be extracted from the plant, which is not appropriate because of the time consuming process (to grow plant first) and not reliable (very little amount of enzyme from the plant). More reliable method has to be developed to produce the enzyme for industrial purpose.
Protein secretion is a multi-step mechanism, regulated by a number of proteins physically influenced by the presence of the disulfide bonds, hydrophobicity of protein molecules, and the molecular weight of it, whereas the post-translational modifications are particularly owing to glycosylation (Sultan et al. 2020). Pichia pastoris, methylotrophic yeast, is known to be a potent host for heterologous proteins production (Mastropietro et al. 2021). P. pastoris secretes extracellular protein, under the influence of alcohol oxidase (AOX1) promoter which is strongly induced by methanol (Türkanoğlu et al. 2019), glycosylation, and high cell density growth on inexpensive medium in bioreactor cultures (Wang et al. 2017). In bioreactor, cell growth plays a vital part in the secretion of protein, as concentration of the protein in the growth medium is almost proportional to its concentration in extracellular medium. The fed-batch fermentation provides a strong growth inhibition by methanol as a substrate (Liu et al. 2016).
P. pastoris involves three specific periods in fed-batch fermentation: (I) glycerol batch period: primary cell growth, (II) glycerol fed-batch period: AOX1 de-repression and increase cell density, and (III) induction period: recombinant proteins expression (Chang et al. 2018). Methanol, an inducer of heterologous gene expression, also works as a substrate for protein production at the expense of high oxygen utilization; nonetheless, unlimited methanol resource can cause abrupt depletion of oxygen. Although limitation of oxygen negatively affects the expression of foreign genes (Wang et al. 2017) and concentration of dissolved oxygen (DO) is an important variable for obtaining high cell density, oxygen-limited cultivation can reduce product modification or increase the specific product purity relative to the amount of total protein released to the medium (Gmeiner et al. 2015). One of the most common methods in protein fed-batch fermentation involves controlling the level of DO within the bioreactors through the controlled addition of substrate feeding (Krause et al. 2016; Poontawee et al. 2020).
The feeding strategy of methanol, dictating the specific growth rate during the induction period, is a significant approach to maximize the recombinant protein production (Azadi et al. 2017; Nieto-Taype et al. 2020). The methanol feeding strategy can be controlled by two methods. First, the methanol feed is controlled by a feedback regulation using DO signal to prevent oxygen limitation. When the DO signal increases than the given set point, the methanol is supplied to the bioreactor but halted as soon as the DO signal is lower/equal to the given set point (Jahic et al. 2002). Methanol-stat is another method for online concentration monitoring of methanol (Beiroti et al. 2019). The methanol feed is controlled by feedback regulation directed by a methanol signal to uphold its concentration at a specific set point. The methanol feeding rate is enhanced if its concentration is less than a given set point, whereas it decreases when the concentration of methanol is higher or equal to a specified set point. These two controlling methods provide high efficiency in protein production. However, they are required sophisticated instruments and software to operate feedback control. Feed forward control requires less sophisticated instruments and software though less efficiency. Nevertheless, this control is commonly used in recombinant Pichia pastoris fed-batch fermentation (Looser et al. 2015). The main aim of this research was to determine a specific methanol utilization rate through various feed forward strategies in order to generate a methanol feed profile based on.
Materials and methods
Strain collection
The methylotrophic yeast Pichia pastoris Y-11430 (Mut+ and His+) strain, which expresses the mutant Thai Rosewood β-glucosidase (A454N), was obtained from the lab of Asst. Prof. Prachumporn Kongsaeree, Faculty of Science, Department of Biochemistry, Kasetsart University, Bangkok, Thailand.
Inoculum preparation
For a primary inoculum, a single marginal colony of P. pastoris was inoculated into a yeast-peptone-dextrose medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L dextrose) broth (total volume: 20 mL), comprised of 100 μg/mL zeocin, and cultivated for 24 h at 30 ๐C with 250 rpm shaking in a shaker incubator. This primary inoculum was poured into an 80 mL BMGY medium (10 g/L yeast extract, 20 g/L peptone, 13.4 g/L yeast nitrogen base without amino acid, 2 mL/L biotin, 10 g/L glycerol, and 0.1 M potassium phosphate buffer to maintain pH 6) to get a secondary inoculum and further incubated for 24 h.
Fed-batch fermentation
A 100 mL propagated shake-flask secondary inoculum was used for inoculating a 2-L fermenter (BIOSTAT B; B. Braun Biotech International B), carrying 0.9 L basal salt medium (40 g/L glycerol, 18 g/L K2SO4, 27 mL/L H3PO4, 0.9 g/L CaSO4.2H2O, 14.9 g/L MgSO4.7H2O, 4.13 g/L KOH) and 4.35 mL PTM1 trace salts solution (6.0 g/L CuSO4.5H2O, 0.08 g/L KI, 3.0 g/L MnSO4.H2O, 0.2 g/L Na2MoO4. 2H2O, 0.02 g/L H3BO3, 0.9 g/L CoCl2.6H2O, 20 g/L ZnCl2, 13.7 g/L FeCl3.6 H2O, 0.2 g/L biotin, 5 mL H2SO4). The fermentation process was carried out using the given conditions: 30 °C, pH 5.5, and 50% dissolved oxygen (DO). For pH adjustment of media, NH4OH (25% w/v) was used in the medium; antifoam 1614 Dow Corning was used for regulation of foam, while DO concentration was controlled by manipulating the agitation rate cascading with the aeration rate (2–5 vvm).
The three phases of fed-batch fermentation were operated in the experiment. The initial stage was glycerol batch phase in which the entire glycerol (40 g/L) was used up, as exhibited by a sharp drop in agitation rate (around 18 h). In the starvation phase, the glycerol and their metabolite was completely consumed for preparation of AOX1 expression (approximately 3 h). In the last stage (methanol fed-batch phase or induction phase), the feed medium (absolute methanol with 12 mL/L PTM1 trace salts solution) was supplied at concentration of 4 g/L with intermittent feed pattern, and this stage was set for 42 h induction. The β-glucosidase activity (U/L), β-glucosidase productivity (QP; U/L/h), and yield coefficient of β-glucosidase from methanol (YP/m) were determined.
Different methanol feed profile for mutant β-glucosidase production from Pichia pastoris strain Y-11430
Constant feed rates with 16.67% and 33.33% methanol concentration
The similar experiment conditions were carried out mentioned above, except the induction phase. The two methanol concentrations (16.67% and 33.33% (w/v)) were fed with two constant rates. Based on the previous studies, the constant feed were calculated from the average of biomass concentration (\(\overline X\)) and the specific rate of methanol consumption (qm).
Feeding with methanol concentration of 16.67%
Assumption:
-
The specific rate of methanol consumption (qm) = 0.07 g/L/h.
-
The average of biomass concentration (\(\overline X\)) = 35.92 g/L.
-
The methanol consumption rate (QM) = 2.5 g/h or 3.18 mL/h.
The methanol feeding rate (F) was 19.48 mL/h for 51 h induction phase. The samples were collected after every 3 h throughout the three phases and were analyzed for β-glucosidase activity (U/L), methanol concentration (g/L), glycerol concentration (g/L), and dry cell mass concentration (g/L).
Feeding with methanol concentration of 33.33%
Assumption:
-
The specific rate of methanol consumption (qm) = 0.14 g/L/h.
-
The average of biomass concentration (\(\overline X\)) = 35.92 g/L.
-
The methanol consumption rate (QM) = 5.03 g/h or 6.37 mL/h.
The methanol feeding rate (F) was 18.75 mL/h for 51 h induction phase. The samples were collected after every 3 h throughout the three phases and were analyzed for β-glucosidase activity (U/L), methanol concentration (g/L), glycerol concentration (g/L), and dry cell mass concentration (g/L).
Linear feed rate
As described in the constant feed rate, the 33.33% methanol constant feed rate were used to calculate the linear methanol feed rate as:
whereas:
-
F = methanol feeding rate (mL/h).
-
qm = 0.14 g/L.
-
V = broth volume at sampling time (mL).
-
\(\overline X\)= the average of cell mass concentration (g/L) from the 33.33% methanol constant feed rate fed-batch fermentation.
-
S = the adding methanol concentration (g/L).
The samples were collected every 3 h throughout the three phases and were analyzed for β-glucosidase activity (U/L), methanol concentration (g/L), glycerol concentration (g/L), and dry cell mass concentration (g/L).
Exponential feed rate
Based on the 33.33% methanol constant feed rate, the methanol exponential feed rates were determined as:
whereas:
-
F= the methanol feed rate (mL/h).
-
V = broth volume at sampling time (mL).
-
\(\overline X\) = the average of cell mass concentration (g/L) from the methanol linear feed rate.
-
fed-batch fermentation.
-
YX/S = the yield coefficient cell mass per methanol (g/g methanol) from the methanol linear feed rate fed-batch fermentation.
-
S = the adding methanol concentration (g/L).
-
μ = the specific growth rate (h).
The samples were collected every 3 h throughout the three phases and were analyzed for β-glucosidase activity (U/L), methanol concentration (g/L), glycerol concentration (g/L), and dry cell mass concentration (g/L).
Cell mass concentration
Spectrophotometer was used for measuring cell growth at 600 nm (OD600 nm). For dry cell weight, a 5 mL culture medium was centrifuged at 5000 rpm for 10 min to obtain pallet which was washed twice with deionized water and dried at 105 °C until constant weight.
β-glucosidases activity
Modified hydrolytic activity of recombinant mutant β-glucosidases toward 3.3 mM p-nitrophenyl-β-D-Glucopyranoside (pNP-Glc) was carried out in 0.1 M sodium acetate (pH 5.0) for 45 min, at 30 °C in a 1.0 mL total volume (Torres et al. 2019). The reaction was stopped by using 2 mL of 2 M sodium carbonate, and the release of p-nitrophenol was analyzed at 400 nm. One unit of enzyme was defined as the amount of enzyme releasing 1 μmol pNP/min (Li et al. 2018; Tissopi et al. 2022).
Glycerol concentration
Culture samples from the fermenter were centrifuged for 5 min at 10,000 rpm and filtered through 0.2 μm filter nylon. Glycerol was analyzed by an HPLC method. Separation of 10 μL of sample was accomplished on a Lichrocart-C18 column (250 mm × 4.0 mm, Merck) equilibrated at 25 °C and eluted with 0.1% phosphoric acid as the mobile phase at a flow rate of 0.9 mL/min. Glycerol was detected using a refractive index Model Refracto Monitor IV (LDC, USA). The concentration of glycerol was estimated through comparing the peak area of sample to the glycerol reference standard.
Methanol concentration
Culture samples from the fermenter were centrifuged for 10 min at 5000 rpm and filtered through 0.2 μm nylon filter. Methanol concentration was assessed with a Hewlett Packard 6890 gas chromatography with the help of Factor Four Capillary column (Model VF-1 ms, Varian). Nitrogen, oxygen, helium, and hydrogen were used as carrier gas. The injector and the flame ionization detector and were set at 160 °C and 250 °C, respectively. The temperature of column oven was kept constant at 45 °C.
Results and discussion
Methanol intermittent feed profile for mutant β-glucosidase production from Pichia pastoris strain Y11430
The mutant β-glucosidase production from recombinant Pichia pastoris strain Y11430 is shown in Fig. 1. The results found were 252.45 U/L for β-glucosidase activity, 5.94 U/L/h for β-glucosidase productivity (QP), and 2.34 U/L/g for yield coefficient of β-glucosidase (YP/m). In glycerol batch phase, 38 g/L of glycerol was used as a carbon and energy source for the initial cell growth. The phase was continued untill a sharp decrease in agitation was observed, which indicated the complete utilization of glycerol (18 h). The dry weight of cells was significantly increased from 0.82 to 18.22 g/L after the end of the first phase. In the starvation phase, the glycerol and its metabolites were completely consumed for preparation of AOX1 expression (approximately 3 h.). At the end of the starvation phase, the cell concentration was 19.12 g/L. Moreover, in the induction phase, methanol was fed at the concentration of 4 g/L with intermittent feed pattern since the methanol concentration higher than 5 g/L may inhibite the cell growth.
Production of mutant β-glucosidase (A454N) from recombinant Pichia pastoris strain Y11430 with methanol intermittent feeding pattern (black circle, dry cell concentration (g/L); white square, glycerol concentration (g/L); black square, methanol accumulation (g/L); black triangle, β-glucosidase activity (U/L)
Methanol constant feed rates by the methanol concentration at 16.67% and 33.33% for cell mass concentration and β-glucosidase production by recombinant Pichia pastoris strain Y11430
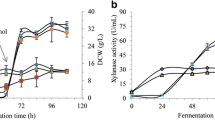
For feeding, the methanol concentration at 16.67 and 33.33% with constant rate, the maximum cell weights during glycerol batch phase at 0–8 h were 15.98 and 25.20 g, as glycerol fed-batch phase increases cell density (Fig. 2) and the specific growth rates (µgly) were 0.134 and 0.205/h, respectively (Table 1). Although both feed rates were run under the same condition, the differences between the dry cell weights and the specific growth rates of recombinant Pichia pastoris strain Y11430 might obtain from the initial cell concentration and another factors error during the experiments. In the induction phase, methanol could be used to stimulate β-glucosidase production and for cell growth. The dry cell concentration profiles of methanol constant feed rate for methanol concentrations of 16.67 and 33.33% at 72 h were 52.23 and 118.55 g/L, respectively (Fig. 2). The specific growth rates from methanol (µMeOH) were 0.023 and 0.036/h at 16.67 and 33.33% methanol concentrations and the yield coefficient biomass per methanol (Yx/m) were 0.273 and 0.383 g/g, respectively (Table 1). As the methanol concentration was increased, the yeast could use the methanol for cell mass production.
The rate of methanol consumption (Qm) at 16.67 (Fig. 3A) and 33.33% (Fig. 3B) displayed that the recombinant yeast was adjusted to the medium because of the low rate of methanol consumption during 21–23 h. After that, the rates of methanol consumption were almost the same. The specific rates of methanol consumption (qm) (Fig. 3C, D) were gradually decreased because of high cell concentration. The high cell concentration consumed methanol rapidly for the cell growth; meanwhile methanol was insufficient for cell consumption during the end of fermentation.
With the constant feed rate of methanol concentration at 16.67 and 33.33%, β-glucosidase activity profiles increased continuously (Fig. 2C, D). At 72 h, the maximum enzyme activity was observed as 3289.52 units for methanol 16.67%, while the maximum enzyme activity was 1001.59 units for 33.33% methanol. It was noticed that the enzyme activity and production, cell density, and methanol metabolism would highly rely on the strategy of methanol induction (Gunes et al. 2016; Nieto-Taypeet al. 2020). The results from the enzyme activity were related to the enzyme productivity rate (Qp) and the specific enzyme productivity rate (qp). The trends of the enzyme productivity rate (Fig. 4A) and the specific enzyme productivity rate (Fig. 4C) of constant feeding methanol at 33.33% were higher than those of constant feeding methanol 16.67% (Fig. 4B, D). The yield coefficient product per methanol (Yp/m) of constant feeding methanol at 33.33% was 13.035 (U/g), which was greater than the yield coefficient product per methanol of constant feeding methanol 16.67% (7.601 U/g) presented in Table 1. These results have indicated that the recombinant yeast could use the higher methanol concentration (33.33%) for cell growth and enzyme production.
Different feed patterns for cell growth and β-glucosidase production by recombinant Pichia pastoris strain Y11430
At 33.33% methanol concentration, methanol was differently fed into the fermenter with constant feed rate, linear feed rate, and exponential feed rate. During glycerol batch phase at 18 h, the dry cell weights for three different feedings were 25.2, 24.14, and 21.69 g (Fig. 5), and the specific growth rates of glycerol (µgly) were 0.205, 0.112, and 0.228/h, respectively (Table 2). Although the three feeding patterns were run under the same condition, the differences between the dry cell weights and the specific growth rates of recombinant Pichia pastoris strain Y11430 might be obtained from the initial cell concentration or may be due to some other factors during the experiments. Increase in glycerol concentration inhibits the expression of genes driven by the AOX1 (Kastilan et al. 2017). Additionally, derepression can be achieved by limiting the glycerol fed in manner to increase the cell density (Cos et al. 2006).
During 21–60 h with constant feed rate, linear feed rate, and exponential feed rate in the induction phase, the dry cell weights were 86.80, 135.91, and 196.73 g (Fig. 5), while the specific growth rates of methanol (µMeOH) were 0.04, 0.04, and 0.061 h−1, respectively (Table 2). The same results were obtained from the yield coefficient biomass per methanol: 0.34, 0.39, and 0.57 g.g−1, respectively (Table 2).
The rate of methanol consumption (Qm) and the specific rate of methanol consumption (qm) with three methanol feeding patterns are depicted in Fig. 6. The slopes of the methanol consumption rates of linear feed rate (Fig. 6B) and of exponential feed rate (Fig. 6C) were higher than the methanol consumption rate of constant feed rate (Fig. 6A). The specific rate of methanol consumption (qm) of constant feed rate (Fig. 6A) was declined; whereas those of linear feed rate and exponential feed rate were inclined (Fig. 6B, C). The recombinant yeast consumed methanol more and efficiency in linear feed rate and exponential feed rate than constant feed rate and the insufficient of methanol over time did not occur in linear feed rate and exponential feed rate like in constant feed rate. It was indicated that the calculation of feed rate from linear and exponential profiles was more correlated to growth of yeast than constant profiles. Parameters such as the average cell quantity (Xt) and the specific rate of methanol consumption (qm) in constant feed rate were used to calculate the linear methanol feed rate, similar to the addition of methanol with exponential feed rate, which was calculated from the specific rate of methanol consumption (qm) and yield coefficient biomass per methanol (Yx/m) from the linear feed rate. Thus, the rate of methanol consumption (Qm) and the specific rate of methanol consumption (qm) of linear and exponential feed rate were increased according to growth cell yeast equilibrated with the feeding methanol quantity.
Since the maximum volume of the fermenter is 2 L, the volumes of fermentation broth of linear feed rate and exponential feed rate were reached the maximum volume at 60 h. Therefore, β-glucosidase activities from constant feed rate, linear feed rate, and exponential feed rate at 60 h were 1629.86, 2088.50, and 2971.85 units, respectively (Fig. 7) and the yield coefficients product per methanol (Yp/m) from three feeding patterns were 8.43, 7.74, and 9.68 U/g, respectively (Table 2). The profiles of enzyme productivity rate (Qp) and specific enzyme productivity rate (qp) with different feeding methods are exhibited in Fig. 8. The slopes of the both rates from exponential feed rate were found slightly higher than the slope of those from constant feed rate and linear feed rate due to the highest added-volume of methanol (Table 2) during an adequate induction phase.
Discussion
In this study, glycerol was used as a carbon and energy source. As glycerol enhances biomass, thereby, the dry weight of cells was significantly increased from 0.82 to 18.22 g/L. Glycerol also helps in transition to methanol metabolism phase (Liu et al. 2016) which is assumed to inhibit the cell growth at its higher concentration of more than 5 g/L. Glycerol accumulation leads to cell growth inhibition and generation of by-products such as ethanol, to repress AOX1 promoter and expression of recombinant protein during methanol induction phase (Jia et al. 2021).
Moreover, in the induction phase, methanol was fed at the concentration of 4 g/L with intermittent feed pattern since the methanol concentration higher than 5 g/L may inhibit the cell growth. Likewise, Nguyen et al. (2020) also found that methanol at 5% or more causes inhibition in cell proliferation.
During glycerol batch phase with methanol constant feeding rate, the methanol concentration of 33.33% in contrast with 16.67% gave maximum cell weights and the specific growth rates (µgly) of 15.98 and 25.20 g, and 0.134 and 0.205/h, respectively. However, at constant methanol feeding, a high ratio of glycerol may lead to a higher percentage of methanol directly dissimilating to carbon dioxide, which may create energy imbalance and hence, decompose the recombinant protein (Liu et al. 2019).
In addition, the β-glucosidase activity profiles observed were 3289.52 units for methanol 16.67%, whereas, 1001.59 units for 33.33% methanol which may be attributable to the fact that P. pastoris is sensitive to methanol concentration which must be controlled between 2 and 3.5 g/L. However, induction of AOX1 promoter by methanol may lead to Pichia cell growth inhibition when methanol concentration is greater than 4 g/L (Liu et al. 2020).
At 33.33% methanol concentration (optimized level), methanol was fed with constant feed rate, linear feed rate, and exponential feed rate. While comparing all three conditions for the dry cell weights, the specific growth rates of glycerol (µgly), the yield coefficient biomass per methanol, rate of methanol consumption (Qm), β-glucosidase activities, enzyme productivity rate (Qp), and specific enzyme productivity rate (qp) were all at its maximum level during exponential feed rate.
Glycerol goes for transition successfully from glycerol to methanol metabolism phase, while during this transition phase, glycerol is utilized completely. This step involves either the sole supply of glycerol or mixed feed of methanol and glycerol. The last stage (production phase) is marked with the addition of methanol to induce protein expression. AOXI promoter is used commonly since it is repressed by glucose; hence, glycerol is used as a carbon source to generate biomass while repressing gene expression (Liu et al. 2019).
It is recommended that a long pause between initial growth on glucose and methanol induction is needed to prevent inhibition of the AOX promoter by glucose, glycerol, or the formed overflow metabolite, ethanol (Wollborn et al. 2022).
Conclusion
Production of protein with recombinant methylotrophic yeast Pichia pastoris is trending in academic research these days. The prime purpose of this work was to determine a specific methanol utilization rate through various feed forward strategies including constant feed rate, linear feed rate, and exponential feed rate, in order to generate a methanol based feeding profile through fed-batch fermentation. The methanol feeding strategy can be controlled by two methods, (i) feedback regulation using dissolved oxygen signal to prevent oxygen limitation. When the DO signal surpasses the given set point, the methanol is supplied to the bioreactor but halted as soon as the DO signal is lower/equal to the given set point; (ii) methanol-stat, for monitoring online concentration of methanol. The kinetic parameters, i.e., specific growth rate of glycerol, specific growth of methanol, maximum total dry cell weight, yield coefficient biomass per methanol, methanol consumption rate, specific methanol consumption rate, enzyme productivity, specific rate of enzyme formation, and yield coefficient enzyme per methanol, favored the exponential feed rate owing to the production of highest cell mass and mutant β-glucosidase activity than that by constant and linear feed rates.
Availability of data and materials
All the data has been declared.
References
Azadi S, Mahboubi A, Naghdi N, Solaimanian R, Mortazavi SA (2017) Evaluation of sorbitol-methanol co-feeding strategy on production of recombinant human growth hormone in Pichia pastoris. Iran J Pharm Res 16:1555–1564. PMID: 29552064
Beiroti A, Hosseini SN, Aghasadeghi MR, Norouzian D (2019) Comparative study of μ-stat methanol feeding control in fed-batch fermentation of Pichia pastoris producing HBsAg: an open-loop control versus recurrent artificial neural network-based feedback control. J Chem Technol Biotechnol 94:3924–3931. https://doi.org/10.1002/jctb.6192
Chang CH, Hsiung HA, Hong KL, Huang CT (2018) Enhancing the efficiency of the Pichia pastoris AOX1 promoter via the synthetic positive feedback circuit of transcription factor Mxr1. BMC Biotechnol 18:81. https://doi.org/10.1186/s12896-018-0492-4
Cos O, Ramon R, Montesinos JL, Valero F (2006) Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: a review. Microb Cell Fact 5:17. https://doi.org/10.1186/1475-2859-5-17
Gmeiner C, Saadati A, Maresch D, Krasteva S, Frank M, Altmann F, Herwig C, Spadiut O (2015) Development of a fed-batch process for a recombinant Pichia pastoris Δoch1 strain expressing a plant peroxidase. Microb Cell Fact 14:1. https://doi.org/10.1186/s12934-014-0183-3
Gunes H, Boy E, Ata O, Zerze GH, Calik P, ve Ozdamar TH (2016) Methanol feeding strategy design enhances recombinant human growth hormone production by Pichia pastoris. J Chem Technol Biotechnol 91:664–671. https://doi.org/10.1002/jctb.4619
Jahic M, Rotticci-Mulder J, Martinelle M, Hult K, Enfors SO (2002) Modeling of growth and energy metabolism of Pichia pastoris producing a fusion protein. Bioprocess Biosyst Eng 24:385–393. https://doi.org/10.1007/s00449-001-0274-5
Jia L, Li T, Wu Y, Wu C, Li H, Huang A (2021) Enhanced human lysozyme production by Pichia pastoris via periodic glycerol and dissolved oxygen concentrations control. Appl Microbiol Biotechnol 105:1041–1050. https://doi.org/10.1007/s00253-021-11100-9
Karnišová Potocká E, Mastihubová M, Mastihuba V (2021) Apiose-Relevant Glycosidases Catalysts 11:1251. https://doi.org/10.3390/catal11101251
Kastilan R, Boes A, Spiegel H, Voepel N, Chudobová I, Hellwig S, Fischer R (2017) Improvement of a fermentation process for the production of two PfAMA1-DiCo-based malaria vaccine candidates in Pichia pastoris. Sci Rep 7:11991. https://doi.org/10.1038/s41598-017-11819-4
Kongsaeree PT, Ratananikom K, Choengpanya K, Tongtubtima N, Sujiwattanarat P, Porncharoennop C, Onpium A, Svasti J (2010) Substrate specificity in hydrolysis and transglucosylation by family 1 β-glucosidases from cassava and Thai rosewood. J Mol Catal B Enzym 67:257–265. https://doi.org/10.1016/j.molcatb.2010.09.003
Krause M, Neubauer A, Neubauer P (2016) The fed-batch principle for the molecular biology lab: controlled nutrient diets in ready-made media improve production of recombinant proteins in Escherichia coli. Microb Cell Fact 15:110. https://doi.org/10.1186/s12934-016-0513-8
Kytidou K, Artola KM, Overkleeft HS, Aerts JMFG (2020) Plant glycosides and glycosidases: a treasure-trove for therapeutics. Front Plant Sci 11:357. https://doi.org/10.3389/fpls.2020.00357
Li T, Zhang W, Hao J, Sun M, Lin SX (2018) Cold-active extracellular lipase: expression in Sf9 insect cells, purification, and catalysis. Biotechnol Rep (amst) 21:e00295. https://doi.org/10.1016/j.btre.2018.e00295
Liu W, Gong T, Wang QH, Liang X, Chen JJ, Zhu P (2016) Scaling-up fermentation of Pichia pastoris to demonstration-scale using new methanol-feeding strategy and increased air pressure instead of pure oxygen supplement. Sci Rep 6:18439. https://doi.org/10.1038/srep18439
Liu W, Xiang H, Zhang T, Pang X, Su J, Liu H, Ma B, Yu L (2020) Development of a new high-cell density fermentation strategy for enhanced production of a fungus β-glucosidase in Pichia pastoris. Front Microbiol 11:1988. https://doi.org/10.3389/fmicb.2020.0198
Liu W, Sarah I, Ting G, Ashish S, Li-Yan Y, Ping Z (2019) Fed-batch high-cell-density fermentation strategies for Pichia pastoris growth and production. Crit Rev Biotechnol 39:258–271. https://doi.org/10.1080/07388551.2018.1554620
Looser V, Bruhlmann B, Bumbak F, Stenger C, Costa M, Camattari A, Fotiadis D, Kovar K (2015) Cultivation strategies to enhance productivity of Pichia pastoris. A Review Biotechnol Adv 33:1177–1193. https://doi.org/10.1016/j.biotechadv.2015.05.008
Mastropietro G, Aw R, Polizzi KM (2021) Expression of proteins in Pichia pastoris. Methods Enzymol 660:53–80. https://doi.org/10.1016/bs.mie.2021.07.004
Nguyen S, Nguyen H, Truong K (2020) Comparative cytotoxic effects of methanol, ethanol and DMSO on human cancer cell lines. Biomed Res Ther 7:3855–3859. https://doi.org/10.15419/bmrat.v7i7.614
Nieto-Taype MA, Garcia-Ortega X, Albiol J, Montesinos-Seguí JL, Valero F (2020) Continuous cultivation as a tool toward the rational bioprocess development with Pichia pastoris cell factory. Front Bioeng Biotechnol 8:632. https://doi.org/10.3389/fbioe.2020.00632
Paul L, Mudogo C, Mtei K, Machunda R, Ntie-Kang F (2020) A computer-based approach for developing linamarase inhibitory agents. Phys Sci Rev 5:20190098. https://doi.org/10.1515/psr-2019-0098
Poontawee R, Limtong S (2020) Feeding strategies of two-stage fed-batch cultivation processes for microbial lipid production from sugarcane top hydrolysate and crude glycerol by the oleaginous red yeast Rhodosporidiobolus fluvialis. Microorganisms 8:151. https://doi.org/10.3390/microorganisms8020151
Ratananikom K, Choengpanya K, Tongtubtim N, Charoenrat T, Withers SG, Kongsaeree PT (2013) Mutational analysis in the glycone binding pocket of Dalbergia cochinchinensis β-glucosidase to increase catalytic efficiency toward mannosides. Carbohydr Res 373:35–41. https://doi.org/10.1016/j.carres.2012.10.018
Sultan IN, Keawsompong S, Kongsaeree P, Parakulsuksatid P (2020) Formulation of an efficient combinatorial cellulase cocktail by comparative analysis of gibson assembly and NEBuilder HiFi DNA assembly modus operandi. Int J Emerging Technol 11:490–495
Tissopi T, Kumar S, Sadhu A, Mutturi S (2022) Surface display of novel transglycosylating α-glucosidase from Aspergillus neoniger on Pichia pastoris for synthesis of isomaltooligosaccharides. Biochem Eng J 181:108400. https://doi.org/10.1016/j.bej.2022.108400
Toonkool P, Metheenukul P, Sugiwattanrat P, Paiboon P, Tongtubtim N, Ketudat-Cairns M, Ketudat-Cairsns J, Svasti J (2006) Expression and purification of dalcochinase, a β-glucosidase from Dalbergia cochinchinesis Pierre, in yeast and bacteria hosts. Protein Expr Purif 48:195–204. https://doi.org/10.1016/j.pep.2006.05.011
Torres P, Saa PA, Albiol J, Ferrer P, Agosin E (2019) Contextualized genome-scale model unveils high-order metabolic effects of the specific growth rate and oxygenation level in recombinant Pichia pastoris. Metab Eng Commun 9:e00103. https://doi.org/10.1016/j.mec.2019.e00103
Türkanoğlu Özçelik A, Yılmaz S, Inan M (2019) Pichia pastoris promoters. Methods Mol Biol 1923:97–112. https://doi.org/10.1007/978-1-4939-9024-5_3
Wang J, Wang X, Shi L, Qi F, Zhang P, Zhang Y, Zhou X, Song Z, Cai M (2017) Methanol-independent protein expression by AOX1 promoter with trans-acting elements engineering and glucose-glycerol-shift induction in Pichia pastoris. Sci Rep 7:41850. https://doi.org/10.1038/srep41850
Wollborn D, Müller RL, Munkler LP, Horstmann R, Germer A, Blank LM, Büchs J (2022) Auto-induction screening protocol for ranking clonal libraries of Pichia pastoris MutS strains. Biotechnol Bioproc E 27:572–585. https://doi.org/10.1007/s12257-022-0006-z
Acknowledgements
This research was supported by the Department of Biotechnology, Faculty of Agro-Industry, Kasetsart University, Thailand. The authors also acknowledge the support of Associate Professor Prachumporn Kongsaree, Department of Biochemistry, Kasetsart University, Bangkok, Thailand.
Author information
Authors and Affiliations
Contributions
Sirirat Changming: investigation and writing-original draft. Prachumporn Kongsaree: resource person and supervision. Imrana Niaz Sultan: validation and review and editing the draft. Afrasiab Khan Tareen: formal analysis, conceptualization, and review and editing draft. Wirat Vanichsriratana: project administration and resources. Sarote Sirisansaneeyakul: supervision and formal analysis. Pramuk Parakulsuksatid: conceptualization, supervision, project administration, and resources.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Changming, S., Kongsaree, P., Sultan, I.N. et al. β-glucosidase production by recombinant Pichia pastoris strain Y1433 under optimal feed profiles of fed-batch cultivation. Folia Microbiol 68, 245–256 (2023). https://doi.org/10.1007/s12223-022-01008-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-022-01008-w