Abstract
Mixed infections and heteroresistance of Helicobacter pylori contribute to decreased efficacy of treatments. This study aimed to investigate frequency of clarithromycin heteroresistance and its link with mixed infections, medication history, and disease severity. A total of 40 pairs of H. pylori strains were isolated from the antrum and corpus of 97 patients. Susceptibility of the strains to clarithromycin was measured by agar dilution method. Site-specific mutations of 23S rRNA at A2143G, A2142G, and A2142C positions were analyzed by PCR and genomic relatedness of pairs of the strains was determined by random amplified polymorphic DNA (RAPD)-PCR. The results showed a prevalence of 35% (14/40) clarithromycin resistance. Diversity of the antrum and corpus isolates in resistance to clarithromycin was detected among 17.5% (7/40) of the patients. Similarly, diversity in MIC value was also detected in two patients infected with the sensitive strains. Significant difference in frequency of resistance was detected among patients with peptic ulcer disease (PUD) (MIC90 32 μg/mL) and severe gastritis (MIC90 16 μg/mL), compared with those who suffered from non-ulcer dyspepsia (NUD) (MIC90 8 μg/mL) and chronic gastritis (MIC90 0.25 μg/mL). MIC values showed 8–32 folds increased levels in the corpus. A2142G, A2143G, and A2142C mutations were detected in three, two, and two patients, respectively, but not observed in 46% of the resistant strains. RAPD-PCR fingerprints showed identical molecular patterns for the isolates of the corpus and antrum in each patient. In conclusion, microevolution of H. pylori strains during chronic infection, rather than mixed infection, and inappropriate medication appear to be main reasons of treatment failure in adults.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Helicobacter pylori (H. pylori) is a motile, curved, and Gram-negative bacterium that colonizes the human stomach. H. pylori is the major cause of gastric disorders ranging from chronic gastritis to gastric adenocarcinoma and its frequency accounts for nearly 50% in the world and as high as 80–90% in developing countries (Peek and Blaser 2002; Sgouras et al. 2015). Chronic infection with this bacterium promotes the multistep carcinogenic process of gastric cancer, which evolves through chronic gastritis, acute gastritis, gastric atrophy, intestinal metaplasia, dysplasia, and finally gastric adenocarcinoma (Kao et al. 2016; Shimizu et al. 2015; Wroblewski et al. 2014). Although eradication of H. pylori infection could not reverse histological changes of intestinal metaplasia and dysplasia, its significant effect on improvement of gastritis and gastric atrophy was shown by several studies (Kang et al. 2012; Kong et al. 2014). This improvement is not site specific and could occur in both the antrum and corpus (Norazah et al. 2009; Selgrad et al. 2014).
Failure of eradication therapy regimens is a therapeutic challenge in areas with high prevalence of antibiotic resistance rates. This failure could be explained by H. pylori genomic evolution in the stomach during chronic infection or colonization of this tissue with resistant strains (Alebouyeh et al. 2015; Shokrzadeh et al. 2015). Emergence of heteroresistance in H. pylori population was previously described (Farzi et al. 2015). Main causes of this emergence included consecutive evolution in H. pylori genome, high mutation rate and inefficient DNA mismatch repair systems (Huang et al. 2011; Teh et al. 2014), and high rate of recombination events among different strains within a host (Dorer et al. 2011). The patterns of H. pylori colonization in different gastric sites and the occurrence of associated disorders depend mainly on the level of acid output. In patients who have normal gastric acid production, bacterial colonization is predominantly confined to the antrum; however, involvement of gastric body and occurrence of corpus-predominant pangastritis could be observed in patients with decreased gastric acid output (Huang et al. 2017; Waldum et al. 2016). The colonized strains in the corpus generally evolved from the same lineage of strains that initially colonized the antrum. At these conditions, failures of eradication therapies can cause co-infection with genetically related or unrelated H. pylori strains displaying different antimicrobial susceptibilities in different parts of the stomach (Ayala et al. 2011).
Although efficacy of medication somewhat depends on host factors, selection of appropriate antimicrobial regimen should be based on knowledge about resistance rates of H. pylori strains and the presence of heteroresistance, especially in regions with a high infection rate. In developing countries, such as Iran, the prevalence of clarithromycin resistance rate altered during last years (Khademi et al. 2015; Shokrzadeh et al. 2015). Similarly, global clarithromycin resistance rates have increased from 9% in 1998 to 17.6% in 2008 in Europe and from 7% in 2000 to 27.7% in 2006 in Japan (Hu et al. 2017; Nishizawa and Suzuki 2014). This resistance rate was increased as high as > 40% in some countries, such as Turkey and China (Thung et al. 2016).
Failure of conventional regimens against H. pylori depends mainly on dominance of clarithromycin-resistant strains in each population. However, new cases of patients who have failed eradication regimens are emerging in areas with appropriate treatment history. Microevolution seems to be the main cause for this emergence. Resistance to clarithromycin is generally caused by point mutations in 23s rRNA gene, mainly through mutations at A2143G, followed by A2142G and A2142C positions (Boyanova 2017). These mutations can arise in the stomach of each patient during the chronic infection. In current study, to understand the microevolution of clarithromycin-resistant strains of H. pylori in a single host, we investigated development of these variants among H. pylori strains isolated from the antrum and corpus of the gastric tissue in each patient. Furthermore, differences of molecular fingerprints, minimal inhibitory concentrations (MIC) of the isolates, and their association with noted mutations were analyzed.
Patients and methods
Patients and data collection
The received biopsy samples were from patients that underwent endoscopy at Taleghani Hospital in Tehran, Iran, during the period from May 2014 to February 2015. All the patients with recent history of medication were excluded from the study. Demographic data and endoscopic findings of the patients were recorded in a standard questionnaire. This study was approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences.
Microbiological and histopathological analyses
Three separate gastric biopsy specimens were taken from the antrum and from the corpus of the stomach. One of the biopsy samples from each site was sent to the pathology laboratory for histological analysis, and the other one was placed into a transport medium (thioglycolate agar plus yeast extract, 3%, Merck Co., Darmstadt, Germany, and Oxoid, Hampshire, UK, respectively) and sent to the microbiology laboratory for culture in a specific culture medium after homogenization. Growth of H. pylori colonies was screened on Brucella agar medium supplemented with 10% (v/v) fetal calf serum, 7% horse blood, and selective antimicrobials (vancomycin 10 mg/L; polymyxin B 0.25 mg/L; trimethoprim 5 mg/L; and amphotericin B 3 mg/L (Merck, Germany). The homogenized biopsies in BHI broth were inoculated and the plates were incubated at 37 °C for 3 to 5 days at microaerobic conditions (5% O2, 10% CO2, and 85% N2). The suspected grown colonies were evaluated by rapid urease test, Gram staining, and biochemical tests, including oxidase and catalase reactions. Pure subcultures of H. pylori strains were prepared and stocked in − 70 °C for molecular investigation and antimicrobial susceptibility testing. H. pylori strain RIGLD 742 was used as a positive control strain in all the reactions.
Molecular characterization and mutation analysis
Genomic DNAs of the H. pylori strains were extracted by QIAamp DNA extraction kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). Polymerase chain reaction (PCR) was done for molecular characterization of the Helicobacter strains at species level (glmM and 16S rRNA) as described before (Farzi et al. 2015). To detect site-specific mutations of 23S rRNA at A2143G, A2142G, and A2142C positions, a volume of 25-μL reaction mixture containing 1× PCR buffer, 0.3 μM of each primer, 1 μL of genomic DNA, 200 μM of dNTPs mix, 0.63 mmol of MgCl2, and 0.2 U/μL of Taq DNA polymerase was used. PCRs were done separately in an automated thermal cycler (AG 22331; Eppendorf, Hamburg, Germany) under the following conditions: 1 cycle of denaturation for 5 min at 94 °C, annealing for 5 min at 36 °C, extension for 5 min at 72 °C, followed by 30 cycles of denaturation for 1 min at 94 °C, annealing for 1 min at 36 °C, and extension for 2 min at 72 °C. Oligonucleotide sequences of the primers used in this study and size of the PCR products are shown in Table 1. DNA extracts of three reference strains encoding A2142G, A2143G, and A2142C (GenBank accession numbers JQ765438, JQ765441, and JQ765440) were used as control strains in this experiment.
Random amplified polymorphic DNA-PCR analysis
Random amplified polymorphic DNA (RAPD)-PCR typing was done for detection of related and distinct strains of H. pylori that dominantly colonized the antrum and corpus of the stomach in each patient. Accordingly, the more discriminative primer 1283 was used (Table 1). A volume of 25 μL containing 1× PCR buffer, 1 μmol/L of primer, 1 μL of genomic DNA (approximately 150 ng), 200 μmol/L of dNTPs mix, 2 mmol of MgCl2, and 0.05 U/μL of Taq DNA polymerase was used for each reaction. RAPD-PCR amplification was performed in an automated thermal cycler (AG 22331; Eppendorf, Hamburg, Germany) as described before (Farzi et al. 2015). PCR products were electrophoresed in 1.8% agarose gel. Similarity of all RAPD banding profiles was analyzed by GelCompar II Software (TX, USA). Lack of polymorphisms or existence of ≤ 2 and > 2 different RAPD-PCR bands was considered as a definitive criterion for detection of identical or related and different strains, respectively.
Determination of MICs for clarithromycin
MIC of clarithromycin was examined by the agar dilution method based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Different amounts of clarithromycin, at final concentration of 0.016 to 32 μg/mL (Sigma, St. Louis, MO), were added to Mueller-Hinton agar medium (Merck, Germany) containing 10% defibrinated horse blood. Freshly prepared H. pylori suspensions were prepared with turbidity equivalent to three McFarland standards, and then 10 μL of each suspension was inoculated on the prepared plates. The MIC values were determined after 72 h of incubation at 37 °C under microaerobic conditions. Epidemiological cut-off value > 0.25 μg/mL was used for detection of the resistant strains. Quality control was performed by using reference clarithromycin-resistant and clarithromycin-susceptible strains (RIGLD OC359 and RIGLD OC248, respectively) in each experiment.
Statistical analysis
SPSS for Windows (version 16.0; SPSS, Chicago, IL, USA) was used for the statistical analysis; baseline demographic data of the infected patients were analyzed and association of MIC values and site-specific mutations were compared by chi-square test. All the findings were considered statistically significant when the p values were less than 0.05.
Results
Prevalence of H. pylori infection
Out of 97 patients subjected to endoscopy, infection with H. pylori was detected among 42 (43.3%) patients. A total of 40 isolates from the antrum and 40 isolates from the corpus of the same patients were included in the study, and two patients were excluded, because of the lack of H. pylori colonization in one of the two stomach parts (the antrum or corpus). H. pylori-positive patients consisted of 20 males and 20 females, with their ages ranging between 10 and 70 years (mean age, 40–50 years). Identity of the isolates was confirmed as H. pylori by positive PCR results for glmM and 16S rRNA genes. The infected patients suffered from severe gastritis (6/40, 15%), chronic gastritis (7/40, 17.5%), peptic ulcer disease (5/40, 12.5%), erythema (20/40, 50%), and erosion (1/40, 2.5%) (Table 2).
Diversity of clarithromycin resistance phenotype in the antrum and corpus
Resistance to clarithromycin was detected among 35% (14/40) of the patients. Diversity in resistance to clarithromycin between paired of the isolates from the antrum and corpus of a single patient was characterized among 17.5% of the patients (7/40). Similarly, diversity in MIC values was also detected in two patients infected with clarithromycin-sensitive strains (Table 3). A correlation was found between higher age groups and the resistance rate. A significant difference in frequency of resistance was detected among patients with PUD and severe gastritis compared with patients who suffered from NUD and chronic gastritis, respectively (Table 2). The resistant strains were mostly isolated from infected patients with recent history of clarithromycin-based therapeutic regimen.
Diversity of the minimum inhibitory concentration
Different MIC values were determined, as highest ones were characterized among patients with PUD (MIC90 32 μg/mL) and severe active gastritis (MIC90 16 μg/mL) compared with those who suffered from NUD (MIC90 8 μg/mL) and chronic gastritis (MIC90 0.25 μg/mL), respectively (Table 2). Diversity of MIC values for the isolates from the antrum and corpus was also confirmed in nine patients (Table 3). While analysis of MIC values showed 8–32 folds increased levels in the corpus, a higher MIC value was detected for one isolate in the antrum.
Clarithromycin mutation rates
Screening of three common mutations in 23S rRNA (Dp4, Dp5, and Dp6) was done using site-specific primers. Results of the screening showed the existence of A2142G, A2143G, and A2142C mutations in three, two, and two patients respectively. Co-existence of A2142G and A2142C mutations was also detected in one patient. The A2142C mutation was detected in the corpus of one patient compared with wild-type allele in the H. pylori strain from the antrum (MIC 8 vs 0.25 μg/mL). All of these variants were obtained from patients with recent history of medication by clarithromycin-based regimens (Table 3).
RAPD typing
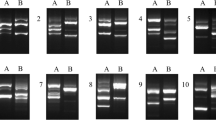
RAPD-PCR fingerprinting was performed on DNA extracts of the strains obtained from individual patients. Accordingly, the results showed identical molecular patterns for the isolates of the corpus and antrum in each patient. However, different RAPD patterns were detected among the strains from different patients (Fig. 1).
RAPD-PCR fingerprinting patterns of Helicobacter pylori strains obtained from different parts of the stomach. Each number represents two strains from the antrum (A) and corpus (C) of a single patient. The primer 1283 was used in this analysis. Lanes 1–5 are related to pairs of strains HC748, HC754, HC739, HC749, and HC753, respectively. ZipRuler™ Express Mix DNA Ladder (Thermo Fisher Scientific, USA) was shown in the first and last lanes
Discussion
In this study, our results showed that 17.5% of the patients harbored clarithromycin heteroresistance, including those with mutations at 23S rRNA gene (10%, 4/40). The observed resistance rate of 35% in the studied population proposed 65% effectiveness for clarithromycin-based medication in these patients. This estimate was supported by recent studies, where clarithromycin-based regimens achieve eradication rates of approximately 79–88% in Iran (Fakheri et al. 2016; Mokhtare et al. 2015). While this is lower in developed countries, its worldwide rate is similar (75–82%) (25).
Colonization of heteroresistant H. pylori strains in the stomach plays a critical role in the failure of eradication. Long-term infection with H. pylori in this tissue, exposure with sub-lethal or ineffective doses of antibiotics, and the capacity for genomic evolution are possibly involved in the emergence of resistant variants in a single host. Mixed infection, i.e., co-infection with different strains in a single patient, also affects the efficacy of current regimens. In a study by Jae J. Kim et al., a frequency of 2.7% clarithromycin heteroresistance was reported among the two biopsy sites from each patient. This rate was lower compared with those obtained in our study. However, in patients infected with clarithromycin-resistant strains, the same rate of heteroresistance was described (46%) (Kim et al. 2003). Frequency of clarithromycin heteroresistance in our study was also higher compared with that of a study conducted by Cheng-Yen Kao et al. They showed difference in antibiotic resistance of H. pylori strains between the two samples of a single host in 26.3% of patients, which clarithromycin heteroresistance constituted 0.5% of them (Kao et al. 2014).
Infection with clarithromycin resistance strains is a predictive marker for prescription of alternative therapies, including concomitant and sequential therapy. Clarithromycin resistance is generally due to point mutations in the 23S rRNA, mainly nucleotides A2142G and A2143G (Kao et al. 2014). In our study, while similar to other studies that mutation at A2143G nucleotide position was more common, no association was found between the mutation sites and the occurrence of heteroresistance phenotype (Vianna et al. 2016). Furthermore, increase in MIC values was not linked to the studied nucleotide changes. While in our study point mutations A2143G and A2142G were found in 53% of all isolates with CLA MIC > 2 mg/L, this frequency was reported in all CLA-resistant isolates by Zerbetto De Palma et al. (2017). Existence of other mutations in 23S rRNA gene and their association with resistance phenotype of clarithromycin were reported in some countries (Rimbara et al. 2007). In addition, this difference could be explained by mutations in transcription initiation factor IF-2 (infB), 50S ribosomal protein L22 (rpl22), expression of efflux pumps of RND family (resistance modulation cell division), or alteration of outer membrane proteins (OMPs), which similarly confer resistance to clarithromycin in this bacterium (Kang et al. 2012; Hu et al. 2017; Hu et al. 2016). Further investigations are needed to find these relationships in the studied strains.
Our results showed resistance to clarithromycin, most commonly among those patients who belonged to the older age groups (> 50 years old). The observed association between resistance to clarithromycin and age was described previously by Ji Z. et al. (Agudo et al. 2010; Ji et al. 2016). They showed the highest antibiotic resistance rate in patients’ ages 71 to 80 years old. Extensive use of clarithromycin during childhood illnesses was proposed as the main reason. The higher rate of resistance in patients who were treated ineffectively compared with those not receiving medication supports this idea somewhat (Agudo et al. 2010). The observed link between antibiotic resistance and disease severity (e.g., PUD and severe active gastritis) is not well known. Exacerbation of gastric disease after ineffective treatment occurs in most of the patients, which accompanied with dominance of resistant strains in the stomach. In this view, the resistant strains may act as an innocent bystander, and their role as a trigger for severe disorders remains somewhat controversial.
Analyses of RAPD-PCR results did not show alteration in the fingerprints of pairs of the strains in each patient. This finding proposed that heteroresistant strains were mostly derived from pre-existing strains in each patient rather than from mixed-type infection. This finding was similarly supported by Jae J. Kim et al. and Cheng-Yen Kao et al. (Kao et al. 2014; Kim et al. 2003).
The presence of heteroresistant H. pylori infection suggested failure of current treatments for most of the studied patients. Administration of distinct non-standard regimens possibly plays an auxiliary role in this failure, since no internationally defined regimens were prescribed for the studied patients. Previous studies in Iran showed that triple therapy (PPI plus two antibiotics for 1 week) is not optimal, while furazolidone-based or clarithromycin-based quadruple therapy (furazolidone or clarithromycin plus PPI, bismuth subcitrate, amoxicillin for 2 weeks) was recommended as the first-line treatment regimen (Malekzadeh et al. 2004; Saberi-Firoozi and Nejabat 2006). Due to failure rates of 20–40% in Iran, second-line therapy that included tetracycline-based quadruple therapy (containing a PPI, bismuth, tetracycline, and two antibiotics, furazolidone or metronidazole, for 2 weeks) was recommended in the failed treatments. Prescription of this regimen was observed in only two of our patients. Inappropriateness of the medication was also observed by the administration of clarithromycin-based regimens for all the patients with 23S rRNA mutations. To eliminate the occurrence of untreatable infection through the emergence of highly resistant H. pylori strains, there is a need for harmonization of treatment regimens by the publication of a national guideline.
Conclusion
Our results showed the presence of clarithromycin heteroresistance in our patients. They were mostly derived from pre-existing strains in each patient. The results showed the same rate of H. pylori colonization at the antrum and the corpus of the stomach of most of the patients. While higher MIC levels were observed in the corpus, resistance to clarithromycin was observed in both sites of the stomach. The higher MIC levels were not correlated with mutations detected at nucleotide positions 2142 and 2143 of 23S rRNA gene. Inappropriate medication can support recurrence of the infection in patients with resistant variants of clarithromycin. Upon failure of treatments, providing multiple biopsy specimens from different parts of the stomach seems to be necessary for doing the antimicrobial susceptibility testing.
References
Agudo S, Pérez-Pérez G, Alarcón T, López-Brea M (2010) High prevalence of clarithromycin-resistant Helicobacter pylori strains and risk factors associated with resistance in Madrid, Spain. J Clin Microbiol 48(10):3703–3707
Alebouyeh M, Yadegar A, Farzi N, Miri M, Zojaji H, Gharibi S, Fazeli Z, Ebrahimi Daryani N, Asadzadeh Aghdaei H, Zali MR (2015) Impacts of H. pylori mixed-infection and heteroresistance on clinical outcomes. Gastroenterology and Hepatology from bed to bench 8(Suppl.1):S1–S5
Ayala G, Galván-Portillo M, Chihu L, Fierros G, Sánchez A, Carrillo B, Román A, López-Carrillo L, Silva-Sánchez J, Study Group J (2011) Resistance to antibiotics and characterization of Helicobacter pylori strains isolated from antrum and body from adults in Mexico. Microb Drug Resist 17(2):149–155
Bohr UR, Primus A, Zagoura A, Glasbrenner B, Wex T, Malfertheiner P (2002) A group specific PCR assay for the detection of Helicobacteraceae in human gut. Helicobacter 7:378–383
Boyanova L (2017) Amoxicillin/clarithromycin. Reactions 1646:34–38
Dorer MS, Sessler TH, Salama NR (2011) Recombination and DNA repair in Helicobacter pylori. Annu Rev Microbiol 65:329–348
Fakheri H, Bakhshi Z, Bari Z, Alhooei S (2016) Effects of clarithromycin-containing quadruple therapy on Helicobacter Pylori eradication after nitroimidazole-containing quadruple therapy failure. Middle East J Dig Dis 8(1):51–56
Farzi N, Malekian T, Alebouyeh M, Vaziri F, Zali MR (2015) Genotype diversity and quasispecies development of Helicobacter pylori in a single host. Jpn J Infect Dis 68(3):176–180
Finger SA, Velapatiño B, Kosek M, Santivañez L, Dailidiene D, Quino W, Balqui J, Herrera P, Berg DE, Gilman RH (2006) Effectiveness of enterobacterial repetitive intergenic consensus PCR and random amplified polymorphic DNA fingerprinting for Helicobacter pylori strain differentiation. Appl and Environ Microbiol 72(7):4713–4716
Hu Y, Zhang M, Lu B, Dai J (2016) Helicobacter pylori and antibiotic resistance, a continuing and intractable problem. Helicobacter 21:349–363
Hu Y, Zhu Y, Lu NH (2017) Novel and effective therapeutic regimens for Helicobacter pylori in an era of increasing antibiotic resistance. Front Cell Infect Microbiol 7:168
Huang JY, Sweeney EG, Guillemin K, Amieva MR (2017) Multiple acid sensors control Helicobacter pylori colonization of the stomach. PLoS Pathog 13(1):e1006118
Huang XW, Luo RH, Zhao Q, Shen ZZ, Huang LL, An XY, Zhao LJ, Wang J, Huang YZ (2011) Helicobacter pylori induces mitochondrial DNA mutation and reactive oxygen species level in AGS cells. Int J Med Sci 8(1):56–67
Ji Z, Han F, Meng F, Tu M, Yang N, Zhang J (2016) The association of age and antibiotic resistance of Helicobacter Pylori: a study in Jiaxing City, Zhejiang Province, China. Medicine 95(8):e2831
Kang JM, Kim N, Shin CM, Lee HS, Lee DH, Jung HC, Song IS (2012) Predictive factors for improvement of atrophic gastritis and intestinal metaplasia after Helicobacter Pylori eradication: a three-year follow-up study in Korea. Helicobacter 17(2):86–95
Kao CY, Lee AY, Huang AH, Song PY, Yang YJ, Sheu SM, Chang WL6, Sheu BS, Wu JJ (2014) Heteroresistance of Helicobacter pylori from the same patient prior to antibiotic treatment. Infect Genet Evol 23:196–202
Kao CY, Sheu BS, Wu JJ (2016) Helicobacter pylori infection: an overview of bacterial virulence factors and pathogenesis. Biom J 39(1):14–23
Kauser F, Hussain MA, Ahmed I, Srinivas S, Devi, SM, Majeed AA, Rao KR, Khan AA, Sechi LA, Ahmed N (2005) Comparative genomics of Helicobacter pylori isolates recovered from ulcer disease patients in England. BMC Microbiol 5:32
Khademi F, Poursina F, Hosseini E, Akbari M, Safaei HG (2015) Helicobacter pylori in Iran: a systematic review on the antibiotic resistance. Iran J Basic Med Sci 18(1):2–7
Kim JJ, Kim JG, Kwon DH (2003) Mixed-infection of antibiotic susceptible and resistant Helicobacter pylori isolates in a single patient and underestimation of antimicrobial susceptibility testing. Helicobacter 8(3):202–206
Kong YJ, Yi HG, Dai JC, Wei MX (2014) Histological changes of gastric mucosa after Helicobacter pylori eradication: a systematic review and meta-analysis. World J Gastroenterol 20(19):5903–5911
Malekzadeh R, Mohamadnejad M, Siavoshi F, Massarrat S (2004) Treatment of Helicobacter pylori in Iran: low efficacy of recommended western regimens. Arch Iranian Med 7(1):1–8
Mokhtare M, Agah S, Fakheri H, Hosseini V, Hemami MR, Ghafoori SMS (2015) Efficacy of clarithromycin containing bismuth–based regimen as a second-line therapy in Helicobacter pylori eradication. Middle East J Dig Dis 7(2):75–81
Nishizawa T, Suzuki H (2014) Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing. Front Mol Biosci 1
Norazah A, Rasinah WZ, Zaili Z, Aminuddin A, Ramelah M (2009) Analysis of PCR-RAPD DNA and antibiotic susceptibility profiles of antrum and corpus isolates of Helicobacter pylori from Malaysian patients. Malays J Pathol 31(1):29–34
Pan ZJ, Su WW, Tytgat GNJ, Dankert J, van der Ende A (2002) Assessment of clarithromycin-resistant Helicobacter pylori among patients in Shanghai and Guangzhou, China, by primer-mismatch PCR. J Clin Microbiol 40(1):259–261
Peek RM, Blaser MJ (2002) Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer 2(1):28–37
Rimbara E, Noguchi N, Kijima H, Yamaguchi T, Kawai T, Sasatsu M (2007) Mutations in the 23S rRNA gene of clarithromycin-resistant Helicobacter pylori from Japan. Int J Antimicrob Agents 30(3):250–254
Saberi-Firoozi M, Nejabat M (2006) Experiences with Helicobacter pylori treatment in Iran. Iran J Med Sci 31(4):181–185
Selgrad M, Tammer I, Langner C, Bornschein J, Meißle J, Kandulski A, Varbanova M, Wex T, Schlüter D, Malfertheiner P (2014) Different antibiotic susceptibility between antrum and corpus of the stomach, a possible reason for treatment failure of Helicobacter pylori infection. World J Gastroenterol 20(43):16245–16251
Sgouras DN, Trang TTH, Yamaoka Y (2015) Pathogenesis of Helicobacter pylori infection. Helicobacter 20(S1):8–16
Shimizu T, Marusawa H, Watanabe N, Chiba T (2015) Molecular pathogenesis of Helicobacter pylori-related gastric cancer. Gastroenterol Clin N Am 44(3):625–638
Shokrzadeh L, Alebouyeh M, Mirzaei T, Farzi N, Zali MR (2015) Prevalence of multiple drug-resistant Helicobacter pylori strains among patients with different gastric disorders in Iran. Microb Drug Resist 21(1):105–110
Teh X, Khosravi Y, Lee WC, Leow AHR, Loke MF, Vadivelu J, Goh KL (2014) Functional and molecular surveillance of Helicobacter pylori antibiotic resistance in Kuala Lumpur. PLoS One 9(7):e101481
Thung I, Aramin H, Vavinskaya V, Gupta S, Park J, Crowe S, Valasek M (2016) The global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther 43(4):514–533
Vianna JS, Ramis IB, Ramos DF, Von Groll A, Silva PEAD (2016) Drug resistance in Helicobacter pylori. Arq Gastroenterol 53(4):215–223
Waldum HL, Kleveland PM, Sørdal ØF (2016) Helicobacter pylori and gastric acid: an intimate and reciprocal relationship. Ther Adv Gastroenterol 9(6):836–844
Wroblewski LE, Piazuelo MB, Chaturvedi R, Schumacher M, Aihara E, Feng R, Noto JM, Delgado A, Israel DA, Zavros Y, Montrose MH, Shroyer N, Correa P, Wilson KT, Peek RMJR (2014) Helicobacter pylori targets cancer-associated apical-junctional constituents in gastroids and gastric epithelial cells. Gut 64(5):720–730
Zerbetto De Palma G, Mendiondo N, Wonaga A, Viola L, Ibarra D, Campitelli E, Salim N, Corti R, Goldman C, Catalano M (2017) Occurrence of mutations in the antimicrobial target genes related to levofloxacin, clarithromycin, and amoxicillin resistance in Helicobacter pylori isolates from Buenos Aires city. Microb Drug Resist 23(3):351–358
Acknowledgments
The authors of this article like to thank all staff of endoscopy unit of Ayatollah Taleghani Hospital and Foodborne and Waterborne Diseases Research Center for their sincere help and assistance.
Funding
This study was part of a fellowship dissertation and financially supported by a grant from Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This study was approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Farzi, N., Behzad, C., Hasani, Z. et al. Characterization of clarithromycin heteroresistance among Helicobacter pylori strains isolated from the antrum and corpus of the stomach. Folia Microbiol 64, 143–151 (2019). https://doi.org/10.1007/s12223-018-0637-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-018-0637-9