Abstract
In 2011–2012, a survey was performed in three regional hospitals in the Czech Republic to determine the incidence of Clostridium difficile infections (CDIs) and to characterize bacterial isolates. C. difficile isolates were characterized by PCR ribotyping, toxin genes detection, multiple-locus variable-number tandem-repeat analysis (MLVA), and antimicrobial susceptibility testing to fidaxomicin, vancomycin, metronidazole, clindamycin, LFF571, and moxifloxacin using agar dilution method. The incidence of CDI in three studied hospitals was 145, 146, and 24 cases per 100,000 inhabitants in 2011 and 177, 258, and 67 cases per 100,000 inhabitants in 2012. A total of 64 isolates of C. difficile was available for molecular typing and antimicrobial susceptibility testing. 60.9% of the isolates were classified as ribotype 176. All 41 isolates of ribotypes 176 and 078 were positive for the presence of binary toxin genes. Ribotype 176 also carried 18-bp deletion in the regulatory gene tcdC. Tested isolates of C. difficile were fully susceptible to vancomycin and metronidazole, whereas 65.1% of the isolates were resistant to moxifloxacin. MLVA results indicated that isolates from three different hospitals were genetically related, suggesting transmission between healthcare facilities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Clostridium difficile is the most common nosocomial pathogen of diarrhea in humans. C. difficile infection (CDI) is often triggered by antimicrobial therapy. The prevalence and severity of the disease increased worldwide in the past 10–15 years (Rupnik et al. 2009). Stubbs et al. (1999) described more than 100 different PCR ribotypes. The so-called hypervirulent ribotype 027 of C. difficile is associated with a more severe disease course, a higher production of toxins, presence of a binary toxin, and a higher resistance to fluoroquinolones than other ribotypes (Razavi et al. 2007). In Central European countries, such as Poland and the Czech Republic, C. difficile ribotype 176 is frequently found (Krutova et al. 2014; Obuch-Woszczatynski et al. 2014). C. difficile ribotype 176 is highly related to ribotype 027 and differs only in one band by PCR ribotyping (Nyc et al. 2011; Valiente et al. 2012). In contrast to the worldwide spread of ribotype 027, ribotype 176 was reported in Poland, the Czech Republic, and now also in Croatia (Rupnik et al. 2016).
Since 2011, we noticed an increase in the CDI incidence in three hospitals in Eastern Bohemia (Czech Republic). A retrospective analysis to the incidence of CDI was performed with characterization of the cultured C. difficile isolates. C. difficile isolates were characterized by PCR ribotyping, toxin genes detection, multiple-locus variable-number tandem-repeat analysis (MLVA), and antimicrobial susceptibility testing to fidaxomicin, vancomycin, metronidazole, clindamycin, LFF571, and moxifloxacin using the CLSI (Clinical and Laboratory Standards Institute) agar dilution method.
Material and methods
The study design
In 2011, the Czech Anaerobic Bacteria Reference Laboratory in Ostrava requested diagnostic laboratories in the Czech Republic to send C. difficile isolates for typing and characterization. In the period 2011–2012, three hospitals in Eastern Bohemia participated to this survey; a regional hospital in Nachod (609 beds), a regional hospital in Litomysl (632 beds), and a regional hospital in Pardubice (932 beds). All three hospitals are located within a radius of about 100 km. Inclusion criteria for testing patients for CDI were diarrhea and previous antimicrobial treatment. Diagnosis of patients with CDI was performed at the microbiological departments with toxin A/B and glutamate dehydrogenase (GDH) detection using rapid tests (C. DIFF QUIK CHEK COMPLETE®, Techlab) as a routine part of CDI diagnostic algorithm. Toxin A/B and/or GDH positive stool samples were cultured anaerobically at 37 °C for 48 h. The stool samples were tested only in patients on the physician request. The microbiological laboratories were asked to send strain and patient information to the Department of Biomedical Sciences, Faculty of Medicine at the University of Ostrava. Patients diagnosed with CDI were used to determine CDI incidence rate per 100,000 inhabitants in Eastern Bohemia region.
Culture of isolates and DNA isolation
C. difficile isolates were transferred from the hospital laboratories to our laboratory at the University of Ostrava and then from the Czech Republic to Leiden University Medical Center in Leiden, Netherlands, using Amies transport swabs (COPAN, Italy). Swabs were cultured on selective C. difficile agar (M836, Himedia, India) at 37 °C in anaerobic workstation for 48 h. DNA was extracted using the QIAamp DNA Blood Mini Kit (Qiagen).
PCR ribotyping
PCR ribotyping using both the agarose gel electrophoresis and capillary electrophoresis detection system (automated sequencer and fragment analysis system ABI-PRISM™ 3100, POP-4™ Polymer, Applied Biosystems) was performed as described by Bidet et al. (1999) and Fawley et al. (2015). Fragment analysis was performed using GeneMapper® Software Version 5.0 (Applied Biosystems) and identification of ribotypes was carried out using the PCR ribotyping library (the database of the National Reference Laboratory for Clostridium difficile of the Netherlands in Leiden) with BioNumerics® Software Version 7.1 (Applied Maths).
Detection of toxin genes
Multiplex PCR was employed for the detection of gluD (C. difficile identification) and toxin genes tcdA, tcdB, cdtA, and cdtB as described by Paltansing et al. (2007) and Persson et al. (2008).
Detection of deletions in the tcdC gene and the presence of ermB
A monoplex PCR was used for the detection of tcdC gene deletions according to Spigaglia and Mastrantonio (2002) modified by the National Reference Laboratory in Leiden as follows: primers CD-tcdC-1-S (5′-CATATCCTTCTTCTCCTCTTC-3′) and CD-tcdC-2-AS (5′-AATTGTCTGATGCTGAACC-3′) were used. External control strains of C. difficile were used with no deletion (159 bp, strain 630), ∆18 bp (141 bp, strain UK 027), and ∆39 bp (120 bp, strain ∆39) deletion.
A monoplex PCR was used for the detection of ermB genes according to Farrow et al. (2000).
MLVA typing
Multiple-locus variable-number tandem-repeat analysis (MLVA) was performed according to van den Berg et al. (2007). The genetic relationship among the genotypes was determined by clustering them according to MLVA-type using the number of differing loci and the summed absolute distance as coefficients for calculating the minimum-spanning tree, as described by Marsh et al. (2006) using the BioNumerics software program (version 7.1, Applied Maths, Belgium). Briefly, the summed absolute distance between two MLVA-typed isolates is the summed tandem-repeat difference (STRD) at all seven variable number of tandem-repeat (CDR) loci. Isolates with a STRD ≤10 were defined as genetically related, irrespective of the number of differing loci. Clonal complexes were defined by an STRD ≤2, provided that isolates were single locus variants (SLV’s) or double locus variants (DLV’s) of each other (Marsh et al. 2006).
Antimicrobial susceptibility testing
Susceptibility of C. difficile strains to six antimicrobial agents was tested: fidaxomicin, vancomycin, metronidazole, clindamycin, moxifloxacin, and LFF571 (Aplichem or Fluka, Germany). The strains were tested using the agar dilution method according to the CLSI guidelines (2007). The antimicrobials were diluted into Brucella blood agar (pancreatic digest of casein, peptic digest of animal tissue, yeast extract, glucose, sodium chloride, sodium bisulfite, and agar) supplemented with 5% sheep blood, hemin, CaCl2, and vitamin K1. Bacterial isolates were cultured on blood agar plates at 37 °C and after 48 h, suspended to a concentration of 0.5 McFarland in phosphate-buffered saline (PBS). The strains were inoculated onto solid media using multipoint inoculator to a final concentration of 104 CFU per spot. Plates were incubated in an anaerobic chamber (Don Whitley Scientific; 10% H2, 5% CO2, 85% N2) at 37 °C and read after 24 and 48 h for growth. The MIC50 and MIC90 were determined. The following antimicrobial concentration series were used: 8, 4, 2, 1, 0.5, 0.25, 0.125, and 0.06 μg/mL (except of clindamycin: 64, 32, 16, 8, 4, 2, 1, 0.5 μg/mL). The breakpoints for C. difficile and Gram-positive anaerobes according to EUCAST (2016) are (mg/L) vancomycin (2), metronidazole (2), moxifloxacin (4), clindamycin (4), fidaxomicin (−), and LFF571 (−).
Statistical analysis
Frequency distribution of selected results was statistically compared in groups of strains in order to find significant differences. Statistical analysis was processed with NCSS software (Hintze 2012). Dependence of binary toxin gene and ∆tcdC gene occurrence and moxifloxacin resistance on ribotype 176 occurrence was assessed by Fisher’s exact two-side test in four-fold tables. Variable ribotyping, ∆tcdC, and moxifloxacin were dichotomized according to the rules: ribotyping = 176 vs. others, ∆tcdC = 18 bp vs. others, and moxifloxacin resistance by cutpoint 4.
Results and discussion
Patient characteristics
The incidence of CDI in three studied hospitals was 145, 146, and 24 cases per 100,000 inhabitants in 2011 and 177, 258, and 67 cases per 100,000 inhabitants in 2012. In 2012, the average incidence of CDI in these three hospitals was 136 cases per 100,000 persons per year. This is higher than that of reported recently in Germany, where an incidence rate was found of 5–20 cases per 100,000 persons per year (Luebbert et al. 2014). In the USA, the incidence of CDI was 35 cases per 100,000 persons in 2009 (Burke and Lamont 2014). Recent results of an extensive surveillance program in the USA estimated the CDI prevalence at 0.5% in acute care hospitals. In our study, we probably found out a local increase in the number of cases compared with that of normal occurrence, but data from other hospitals in the Czech Republic are not comparable because of different assessment methodologies.
In total, 817 patients with CDI were diagnosed of which 64 (7.8%) were included in this study. Only limited clinical and demographic data were available. CDI was most commonly diagnosed in the departments of internal medicine (61.5%) and long-term care departments (26.2%). CDI was mainly diagnosed in elderly (mean age 76 years) and at a higher rate in women (72.3%). According to the primary diagnosis (before the onset of diarrhea), 44.6% of patients had infectious etiology, of which 62.1% had gastroenteritis and diarrhea of various infectious etiologies.
C. difficile ribotypes and toxin detection
Of 817 patients diagnosed with CDI in this study, 64 (7.8%) C. difficile isolates were available for further analysis. These isolates derived from hospitals Nachod (n = 28), Litomysl (n = 32), and Pardubice (n = 4). Of 64 isolates, 39 belonged to ribotype 176, six to ribotype 014, and 4 to ribotype 002. Ribotype 078 was found in two cases (Fig. 1). The frequency of ribotype 014 (9.4%) and 002 (6.3%) was similar as reported for instance in the USA (Tickler et al. 2014) or England (Wilcox et al. 2012). C. difficile ribotype 176 is according to some researchers recently considered as a “hypervirulent ribotype” since it resembles genetically ribotype 027 very much and clinical findings suggest increased virulence (Drabek et al. 2015; Polivkova et al. 2016).
The occurrence of C. difficile ribotype 176 in the Czech Republic was first described by Nyc et al. (2011). A recent report indicated that ribotype 176 had increased in the Czech Republic, since 40% of 624 typed isolates in a study performed in 2013 belonged to ribotype 176 (Krutova et al. 2014). This ribotype was found also in Poland (Obuch-Woszczatynski et al. 2014) and in the USA (Valiente et al. 2012). Another ribotype, which is considered as a “hypervirulent” (Goorhuis et al. 2008) and was found in our study, is ribotype 078 (3.1%). Researchers in the USA in a comprehensive study reported 2.0% prevalence of this ribotype (Tickler et al. 2014) and in Europe, even 8% (Bauer et al. 2011). Goorhuis et al. (2008) report even 13% incidence in the Netherlands and note that the occurrence of ribotype 078 is more closely associated with community-acquired disease and younger patients.
Of all 64 isolates, 62 contained the gene for toxin A (tcdA) and 63 the gene for toxin B (tcdB). A single isolate of ribotype 017 contained only tcdB and not tcdA and also had no genes for binary toxin cdtA and cdtB or a deletion in the gene tcdC. All 39 isolates of ribotype 176 and two isolates of ribotype 078 were positive for the presence of binary toxin genes (cdtA and cdtB). All other samples were negative for the presence of the binary toxin genes except one undetermined ribotype. This difference was statistically significant (p < 0.001). Isolates belonging to ribotype 176 also carried a possible 18-bp deletion in the regulatory gene tcdC in most cases (with the exception of two isolates), whereas ribotype 078 had a possible 39-bp deletion and other ribotypes had no deletion in this gene except for ribotype 015 (18-bp deletion). This difference was also statistically significant (p < 0.001).
Both C. difficile ribotypes 176 and 078 have genes for binary toxin, which would correspond with their increased virulence (Cowardin et al. 2016). Stewart et al. (2013) suggested that binary toxin could be responsible for recurrent colitis since statistically significant association was found between the presence of this virulence factor and recurrent disease. Similarly, both ribotypes have deletions in the gene tcdC. tcdC gene is responsible for negative regulation of expression of genes for toxins and a deletion therein causes an increased production of toxins (Matamouros et al. 2007). According to Bakker et al. (2012), the role of tcdC in regulation of toxin expression is unclear.
MLVA analysis
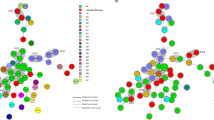
Of 39 isolates belonging to ribotype 176, 30 were selected for MLVA analysis; 13 from Litomysl, 16 from Nachod and 1 from Pardubice. MLVA results are depicted in Fig. 2 and revealed that isolates originating from all three hospitals were genetically related (STRD ≤10), whereas a second complex of genetically related strains only had isolates from one hospital (Litomysl). Five clonal complexes (STRD ≤2) were found of which one contained isolates from two hospitals. Ribotype 176 was the most commonly found ribotype (60.9%) and MLVA results indicated that isolates from three different hospitals were genetically related, suggesting transmission between healthcare facilities.
MLVA typing of 30 isolates (ribotype 176) from three hospitals. A total of seven loci were used. A clonal complex is defined as isolates differing in two or less summed tandem-repeats (dark gray area). Strains are genetically related if the isolates differ in 10 or less summed tandem-repeats (light gray area)
Antimicrobials susceptibility testing
All 64 tested isolates of C. difficile were susceptible to vancomycin and metronidazole. In contrast, 65.1% of the isolates were resistant to moxifloxacin. Ribotype 176 had a high level of resistance to moxifloxacin (100%) compared with other ribotypes (12%; p < 0.001). Only two isolates of ribotype other than 176 were resistant to clindamycin. MIC50 and MIC90 values are depicted in Table 1.
Clinical breakpoints presented in the database EUCAST (2016) are listed for Gram-positive anaerobes and C. difficile separately. For newly introduced antimicrobials, such as fidaxomicin and LFF571 (new semisynthetic thiopeptide), breakpoints have not yet been set. However, the MIC values determined for these antimicrobials are very low. MIC ranges for fidaxomicin and LFF571 are 0.06–0.5 mg/L, for both antimicrobials (Debast et al. 2013). Corbett et al. (2015) provide reference MIC for fidaxomicin 0.25 mg/L. For clindamycin, there is no breakpoint regarding C. difficile, but it is shown in Table 1 for Gram-positive anaerobes.
Susceptibility testing of C. difficile revealed no or low resistance to vancomycin and metronidazole (Freeman et al. 2015). In addition, LFF571 and fidaxomicin that were tested in some studies display MICs even lower than those for vancomycin and metronidazole (Debast et al. 2013). Interestingly, a significant difference in resistance to moxifloxacin was found between ribotype 176 and other ribotypes (p < 0.001). High resistance of ribotype 176 to moxifloxacin was also observed by Lachowicz et al. (2015) and by Krutova et al. (2015). This finding is in agreement with ribotype 027 which is also frequently high-level resistant to new fluoroquinolones (Razavi et al. 2007). Only 3.2% of all isolates in our study were resistant to clindamycin. One previous study in the Czech Republic (Beran et al. 2014) states resistance of C. difficile isolates to clindamycin (4.8%). Published reports from the Central European countries showed resistance to clindamycin generally higher even up to 57% (Fenner et al. 2008; Indra et al. 2008; Terhes et al. 2009). The ermB gene, the marker for resistance to macrolides, lincosamides, and streptogramin B, was detected in two isolates only (MIC to clindamycin was in these isolates 64 and 4 μg/mL). Another isolate, which had MIC to clindamycin 64 μg/mL, did not contain this gene. Interestingly, the resistance to clindamycin was observed in non-RT027 isolates and not in presumptive RT027 isolates in the study of Beran et al. (2014). Conversely, in this work, a higher resistance to moxifloxacin was confirmed in ribotype 176.
Ribotype 176 was the most frequently found ribotype in three hospitals participating in a CDI survey. The close relatedness among ribotype 176 isolates determined by MLVA suggests the transmission among health-care facilities. Our isolates corresponding to the ribotype 176 showed resistance to moxifloxacin.
References
Bakker D, Smits WK, Kuijper EJ, Corver J (2012) TcdC does not significantly repress toxin expression in Clostridium difficile 630ΔErm. PLoS One. doi:10.1371/journal.pone.0043247
Bauer MP, Notermans DW, van Benthem BH, ECDIS Study Group et al (2011) Clostridium difficile infection in Europe: a hospital-based survey. Lancet. doi:10.1016/S0140-6736(10)61266-4
Beran V, Chmelar D, Vobejdova J et al (2014) Sensitivity to antibiotics of Clostridium difficile toxigenic nosocomial strains. Folia Microbiol (Praha). doi:10.1007/s12223-013-0283-1
van den Berg RJ, Schaap I, Templeton KE et al (2007) Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J Clin Microbiol 45:1024–1028
Bidet P, Barbut F, Lalande V et al (1999) Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett 175:261–266
Burke KE, Lamont JT (2014) Clostridium difficile infection: a worldwide disease. Gut Liver. doi:10.5009/gnl.2014.8.1.1
Clinical and Laboratory Standards Institute (2007) Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard–seventh edition. CLSI document M11-A7 [ISBN 1–56238–626-3]. Clinical and Laboratory Standards Institute, Wayne
Corbett D, Wise A, Birchall S et al (2015) In vitro susceptibility of Clostridium difficile to SMT19969 and comparators, as well as the killing kinetics and post-antibiotic effects of SMT19969 and comparators against C. difficile. J Antimicrob Chemother. doi:10.1093/jac/dkv006
Cowardin CA, Buonomo EL, Saleh MM et al (2016) The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat Microbiol. doi:10.1038/nmicrobiol.2016.108
Debast SB, Bauer MP, Sanders IM, ECDIS Study Group et al (2013) Antimicrobial activity of LFF571 and three treatment agents against Clostridium difficile isolates collected for a pan-European survey in 2008: clinical and therapeutic implications. J Antimicrob Chemother. doi:10.1093/jac/dkt013
Drabek J, Nyc O, Krutova M et al (2015) Clinical features and characteristics of Clostridium difficile PCR-ribotype 176 infection: results from a 1-year university hospital internal ward study. Ann Clin Microbiol Antimicrob. doi:10.1186/s12941-015-0114-0
Farrow KA, Lyras D, Rood JI (2000) The macrolide-lincosamide-streptogramin B resistance determinant from Clostridium difficile 630 contains two erm (B) genes. Antimicrob Agents Chemother 44:411–413
Fawley WN, Knetsch CW, MacCannell DR et al (2015) Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile. PLoS One. doi:10.1371/journal.pone.0118150
Fenner L, Frei R, Gregory M et al (2008) Epidemiology of Clostridium difficile-associated disease at university hospital Basel including molecular characterisation of the isolates 2006-2007. Eur J Clin Microbiol Infect Dis. doi:10.1007/s10096-008-0564-9
Freeman J, Vernon J, Morris K, Pan-European Longitudinal Surveillance of Antibiotic Resistance among Prevalent Clostridium difficile Ribotypes’ Study Group et al (2015) Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect. doi:10.1016/j.cmi.2014.09.017
Goorhuis A, Bakker D, Corver J et al (2008) Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis. doi:10.1086/592257
Hintze J (2012) NCSS 8. NCSS, LLC, Kaysville www.ncss.com
Indra A, Schmid D, Huhulescu S et al (2008) Characterization of clinical Clostridium difficile isolates by PCR ribotyping and detection of toxin genes in Austria, 2006-2007. J Med Microbiol. doi:10.1099/jmm.0.47476-0
Krutova M, Matejkova J, Nyc O (2014) C. difficile Ribotype 027 or 176? Folia Microbiol (Praha). doi:10.1007/s12223-014-0323-5
Krutova M, Matejkova J, Tkadlec J, Nyc O (2015) Antibiotic profiling of Clostridium difficile ribotype 176--a multidrug resistant relative to C. difficile ribotype 027. Anaerobe. doi:10.1016/j.anaerobe.2015.07.009
Lachowicz D, Pituch H, Obuch-Woszczatynski P (2015) Antimicrobial susceptibility patterns of Clostridium difficile strains belonging to different polymerase chain reaction ribotypes isolated in Poland in 2012. Anaerobe. doi:10.1016/j.anaerobe.2014.09.004
Luebbert C, John E, von Mueller L (2014) Clostridium difficile infection: guideline-based diagnosis and treatment. Dtsch Arztebl Int. doi:10.3238/arztebl.2014.0723
Marsh JW, O’Leary MM, Shutt KA et al (2006) Multilocus variable-number tandem-repeat analysis for investigation of Clostridium difficile transmission in hospitals. J Clin Microbiol 44:2558–2566
Matamouros S, England P, Dupuy B (2007) Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol Microbiol 64:1274–1288
Nyc O, Pituch H, Matejkova J et al (2011) Clostridium difficile PCR ribotype 176 in the Czech Republic and Poland. Lancet. doi:10.1016/S0140-6736(11)60575-8
Obuch-Woszczatynski P, Lachowicz D, Schneider A et al (2014) Occurrence of Clostridium difficile PCR-ribotype 027 and it’s closely related PCR-ribotype 176 in hospitals in Poland in 2008-2010. Anaerobe. doi:10.1016/j.anaerobe.2014.04.007
Paltansing S, van den Berg RJ, Guseinova RA et al (2007) Characteristics and incidence of Clostridium difficile-associated disease in the Netherlands, 2005. Clin Microbiol Infect 13:1058–1064
Persson S, Torpdahl M, Olsen KE (2008) New multiplex PCR method for the detection of Clostridium difficile toxin a (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect. doi:10.1111/j.1469-0691.2008.02092.x
Polivkova S, Krutova M, Petrlova K et al (2016) Clostridium difficile ribotype 176 - a predictor for high mortality and risk of nosocomial spread? Anaerobe. doi:10.1016/j.anaerobe.2016.05.002
Razavi B, Apisarnthanarak A, Mundy LM (2007) Clostridium difficile: emergence of hypervirulence and fluoroquinolone resistance. Infection 35:300–307
Rupnik M, Wilcox MH, Gerding DN (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. doi:10.1038/nrmicro2164
Rupnik M, Tambic Andrasevic A, Trajkovska Dokic E et al (2016) Distribution of Clostridium difficile PCR ribotypes and high proportion of 027 and 176 in some hospitals in four south eastern European countries. Anaerobe. doi:10.1016/j.anaerobe.2016.10.005
Spigaglia P, Mastrantonio P (2002) Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J Clin Microbiol 40:3470–3475
Stewart DB, Berg A, Hegarty J (2013) Predicting recurrence of C. difficile colitis using bacterial virulence factors: binary toxin is the key. J Gastrointest Surg. doi:10.1007/s11605-012-2056-6
Stubbs SL, Brazier JS, O'Neill GL, Duerden BI (1999) PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol 37:461–463
Terhes G, Urban E, Soki J et al (2009) Assessment of changes in the epidemiology of Clostridium difficile isolated from diarrheal patients in Hungary. Anaerobe. doi:10.1016/j.anaerobe.2009.01.010
The European Committee on Antimicrobial Susceptibility Testing (2016) Breakpoint tables for interpretation of MICs and zone diameters. Version 6.0. http://www.eucast.org
Tickler IA, Goering RV, Whitmore JD, Healthcare Associated Infection Consortium et al (2014) Strain types and antimicrobial resistance patterns of Clostridium difficile isolates from the United States, 2011 to 2013. Antimicrob Agents Chemother. doi:10.1128/AAC.02775-13
Valiente E, Dawson LF, Cairns MD et al (2012) Emergence of new PCR ribotypes from the hypervirulent Clostridium difficile 027 lineage. J Med Microbiol. doi:10.1099/jmm.0.036194-0
Wilcox MH, Shetty N, Fawley WN et al (2012) Changing epidemiology of Clostridium difficile infection following the introduction of a national ribotyping-based surveillance scheme in England. Clin Infect Dis 55:1056–1063
Acknowledgments
This work was supported by the project Postdoc 2, University of Ostrava [Strengthening research institutions at the University of Ostrava, grant number CZ.1.07/2.3.00/30.0047]. The project was co-financed by the European Social Fund in the Czech Republic; the European Union and the Ministry of Education, Youth and Sports of the Czech Republic under the Operational Programme Education for Competitiveness.
Authors’ contributions
VB: the author of the article and scenario of experiments, and article writing.
EJK: organization of experimental work in Leiden laboratories, suggestions, and checking the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Beran, V., Kuijper, E.J., Harmanus, C. et al. Molecular typing and antimicrobial susceptibility testing to six antimicrobials of Clostridium difficile isolates from three Czech hospitals in Eastern Bohemia in 2011–2012. Folia Microbiol 62, 445–451 (2017). https://doi.org/10.1007/s12223-017-0515-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-017-0515-x